Reviving Natural Rubber Synthesis via Native/Large Nanodiscs
Abstract
1. Introduction
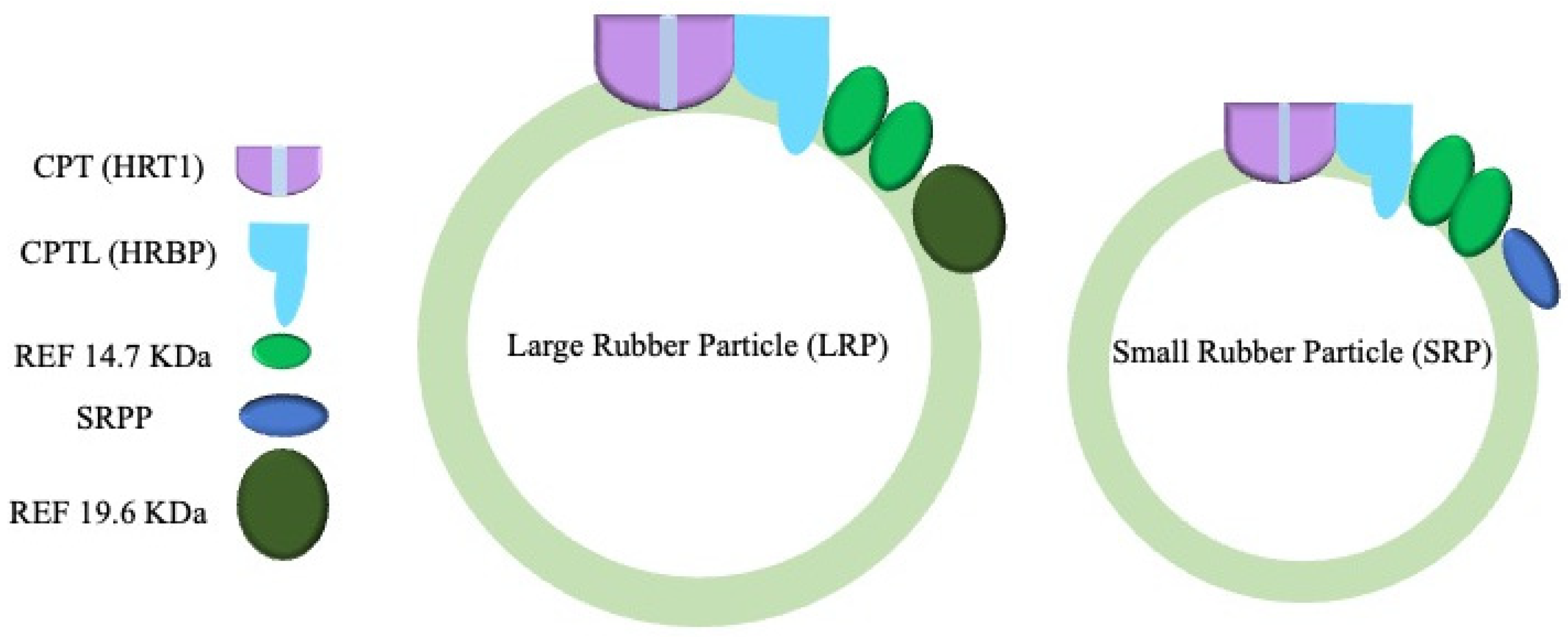
2. CRISPR/Cas9 Mutagenesis: CPT Alone Does Not Solely Determine NR Length
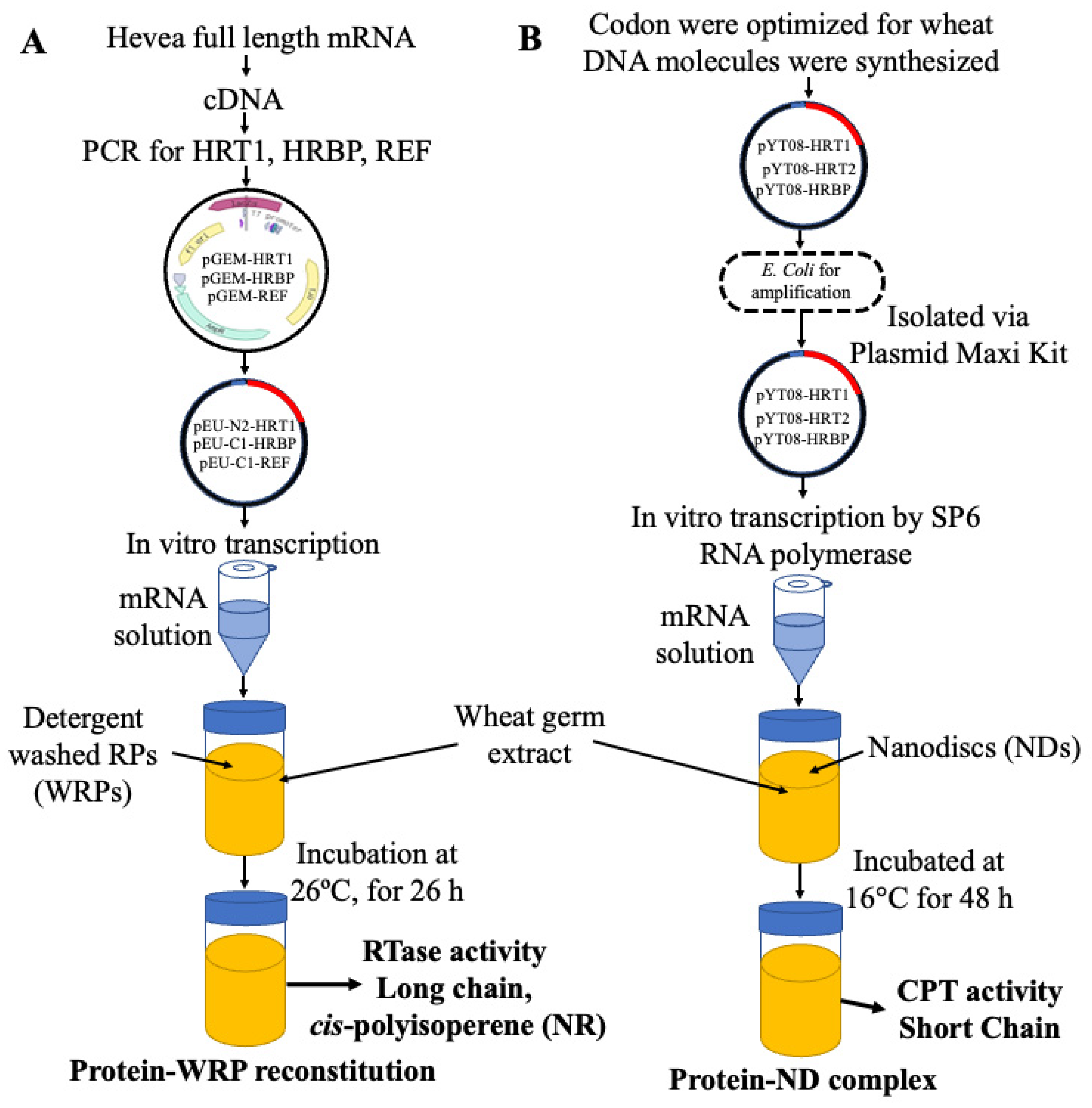
3. Cell-Free System for Membrane Proteins
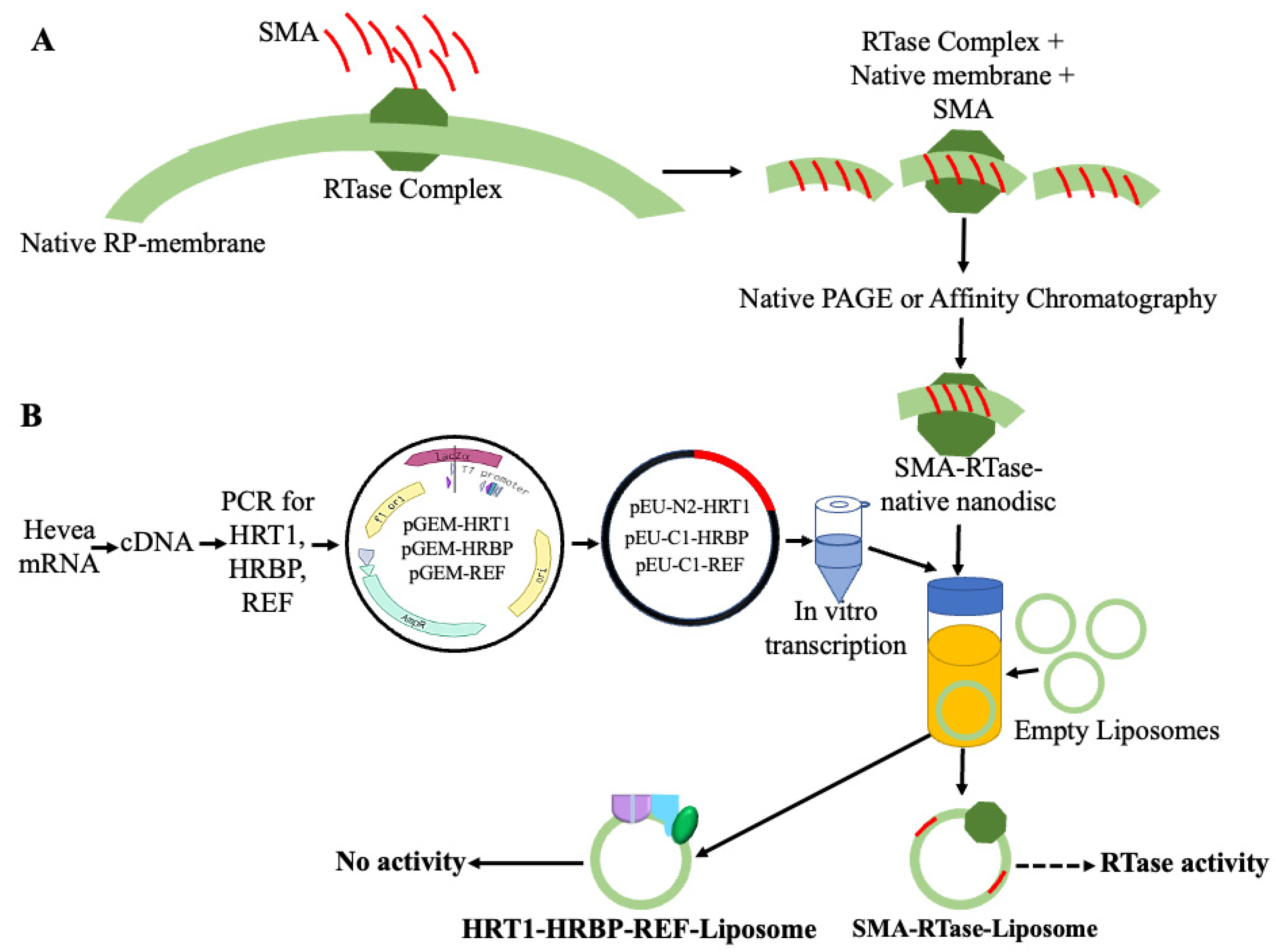
4. Detergent-Free Native-like Membrane Proteins’ Reconstitution in Liposome
5. Large-Sized DNA-Corralled Nanodiscs for Investigating Membrane Proteins
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castillón, J.; Cornish, K. Regulation of initiation and polymer molecular weightof cis-1,4-polyisoprene synthesized in vitro by particlesisolated from Parthenium argentatum (Gray). Phytochemistry 1999, 51, 43–51. [Google Scholar] [CrossRef]
- Umar, A.W.; Park, J.C.; Ling, T.; Ryu, S.B. Plant molecular engine out of the chassis: Natural rubber synthesis in cell-free systems. Ind. Crops Prod. 2023, 195, 116166. [Google Scholar] [CrossRef]
- Tanaka, Y.; Aik-Hwee, E.; Ohya, N.; Nishiyama, N.; Tangpakdee, J.; Kawahara, S.; Wititsuwannakul, R. Initiation of rubber biosynthesis in Hevea brasiliensis: Characterization of initiating species by structural analysis. Phytochemistry 1996, 41, 1501–1505. [Google Scholar] [CrossRef]
- Yamashita, S.; Takahashi, S. Molecular Mechanisms of Natural Rubber Biosynthesis. Annu. Rev. Biochem. 2020, 89, 821–851. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.F.; Cornish, K. Microstructure of purified rubber particles. Int. J. Plant Sci. 2000, 161, 435–445. [Google Scholar] [CrossRef]
- Cornish, K.; Wood, D.F.; Windle, J.J. Rubber particles from four different species, examined by transmission electron microscopy and electron-paramagnetic-resonance spin labeling, are found to consist of a homogeneous rubber core enclosed by a contiguous, monolayer biomembrane. Planta 1999, 210, 85–96. [Google Scholar] [CrossRef]
- Wei, Y.-C.; Zhu, D.; Xie, W.-Y.; Xia, J.-H.; He, M.-F.; Liao, S. In-situ observation of spatial organization of natural rubber latex particles and exploring the relationship between particle size and mechanical properties of natural rubber. Ind. Crops Prod. 2022, 180, 114737. [Google Scholar] [CrossRef]
- Yamashita, S.; Mizuno, M.; Hayashi, H.; Yamaguchi, H.; Miyagi-Inoue, Y.; Fushihara, K.; Koyama, T.; Nakayama, T.; Takahashi, S. Purification and characterization of small and large rubber particles from Hevea brasiliensis. Biosci. Biotechnol. Biochem. 2018, 82, 1011–1020. [Google Scholar] [CrossRef]
- Light, D.R.; A Lazarus, R.; Dennis, M.S. Rubber Elongation by Farnesyl Pyrophosphate Synthases Involves a Novel Switch in Enzyme Stereospecificity. J. Biol. Chem. 1989, 264, 18598–18607. [Google Scholar] [CrossRef]
- Oh, S.K.; Kang, H.; Shin, D.H.; Yang, J.; Chow, K.-S.; Yeang, H.Y.; Wagner, B.; Breiteneder, H.; Han, K.-H. Isolation, Characterization, and Functional Analysis of a Novel cDNA Clone Encoding a Small Rubber Particle Protein from Hevea brasiliensis. J. Biol. Chem. 1999, 274, 17132–17138. [Google Scholar] [CrossRef]
- Asawatreratanakul, K.; Zhang, Y.W.; Wititsuwannakul, D.; Wititsuwannakul, R.; Takahashi, S.; Rattanapittayaporn, A.; Koyama, T. Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis: A key factor participating in natural rubber biosynthesis. Eur. J. Biochem. 2003, 270, 4671–4680. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; Feeney, M.; Ahmadi, M.; Lonoce, C.; Sajari, R.; Di Cola, A.; Frigerio, L. Subcellular localization and interactions among rubber particle proteins from Hevea brasiliensis. J. Exp. Bot. 2017, 68, 5045–5055. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Yamaguchi, H.; Waki, T.; Aoki, Y.; Mizuno, M.; Yanbe, F.; Ishii, T.; Funaki, A.; Tozawa, Y.; Miyagi-Inoue, Y. Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. elife 2016, 5, e19022. [Google Scholar] [CrossRef]
- Wu, S.; Guyot, R.; Bocs, S.; Droc, G.; Oktavia, F.; Hu, S.; Tang, C.; Montoro, P.; Leclercq, J. Structural and Functional Annotation of Transposable Elements Revealed a Potential Regulation of Genes Involved in Rubber Biosynthesis by TE-Derived siRNA Interference in Hevea brasiliensis. Int. J. Mol. Sci. 2020, 21, 4220. [Google Scholar] [CrossRef]
- Xiang, Q.; Xia, K.; Dai, L.; Kang, G.; Li, Y.; Nie, Z.; Duan, C.; Zeng, R. Proteome analysis of the large and the small rubber particles of Hevea brasiliensis using 2D-DIGE. Plant Physiol. Biochem. 2012, 60, 207–213. [Google Scholar] [CrossRef]
- Hillebrand, A.; Post, J.J.; Wurbs, D.; Wahler, D.; Lenders, M.; Krzyzanek, V.; Prüfer, D.; Gronover, C.S. Down-Regulation of Small Rubber Particle Protein Expression Affects Integrity of Rubber Particles and Rubber Content in Taraxacum brevicorniculatum. PLoS ONE 2012, 7, e41874. [Google Scholar] [CrossRef]
- Matsui, M. Rubber Genome & Transcriptome Database; RIKEN: Tokyo, Japan, 2022; Available online: http://ricefox.psc.riken.jp/ (accessed on 7 January 2023).
- Lau, N.-S.; Makita, Y.; Kawashima, M.; Taylor, T.D.; Kondo, S.; Othman, A.S.; Shu-Chien, A.C.; Matsui, M. The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci. Rep. 2016, 6, 28594. [Google Scholar] [CrossRef]
- Connerly, P. How do proteins move through the golgi apparatus. Nat. Educ. 2010, 3, 60. [Google Scholar]
- Harbers, M. Wheat germ systems for cell-free protein expression. FEBS Lett. 2014, 588, 2762–2773. [Google Scholar] [CrossRef]
- Kuroiwa, F.; Nishino, A.; Mandal, Y.; Honzawa, M.; Suenaga-Hiromori, M.; Suzuki, K.; Takani, Y.; Miyagi-Inoue, Y.; Yamaguchi, H.; Yamashita, S.; et al. Reconstitution of prenyltransferase activity on nanodiscs by components of the rubber synthesis machinery of the Para rubber tree and guayule. Sci. Rep. 2022, 12, 3734. [Google Scholar] [CrossRef]
- Cornish, K.; Scott, D.J.; Xie, W.; Mau, C.J.; Zheng, Y.F.; Liu, X.-H.; Prestwich, G.D. Unusual subunits are directly involved in binding substrates for natural rubber biosynthesis in multiple plant species. Phytochemistry 2018, 156, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Liguori, L.; Marques, B.; Villegas-Méndez, A.; Rothe, R.; Lenormand, J.-L. Production of membrane proteins using cell–free expression systems. Expert Rev. Proteom. 2007, 4, 79–90. [Google Scholar] [CrossRef]
- Schwarz, D.; Dötsch, V.; Bernhard, F. Production of membrane proteins using cell-free expression systems. Proteomics 2008, 8, 3933–3946. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, F.; Tozawa, Y. Cell-free expression—Making a mark. Curr. Opin. Struct. Biol. 2013, 23, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Nanamiya, H.; Miyata, T.; Linka, N.; Endo, Y.; Weber, A.P.M.; Tozawa, Y. A Cell-Free Translation and Proteoliposome Reconstitution System for Functional Analysis of Plant Solute Transporters. Plant Cell Physiol. 2007, 48, 1815–1820. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, V.; Zunabovic, M.; Nehm, L.; Bergmair, J.; Kneifel, W. Influence of argon modified atmosphere packaging on the growth potential of strains of Listeria monocytogenes and Escherichia coli. Food Control. 2016, 59, 513–523. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef]
- Denisov, I.G.; McLean, M.A.; Shaw, A.W.; Grinkova, Y.V.; Sligar, S.G. Thermotropic Phase Transition in Soluble Nanoscale Lipid Bilayers. J. Phys. Chem. B 2005, 109, 15580–15588. [Google Scholar] [CrossRef]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef]
- Kuroiwa, F.; Suda, H.; Yabuki, M.; Atsuzawa, K.; Yamaguchi, H.; Toyota, M.; Kaneko, Y.; Yamashita, S.; Takahashi, S.; Tozawa, Y. Cell-free translation system with artificial lipid-monolayer particles as a unique tool for characterizing lipid-monolayer binding proteins. Biosci. Biotechnol. Biochem. 2024, 88, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Hodgins, C.L.; Salama, E.M.; Dias, K.R.; Parikh, A.; Mackey, A.V.; Catenza, K.F.; Vederas, J.C.; Ro, D.-K. New insights into natural rubber biosynthesis from rubber-deficient lettuce mutants expressing goldenrod or guayule cis-prenyltransferase. New Phytol. 2023, 239, 1098–1111. [Google Scholar] [CrossRef] [PubMed]
- Bushman, B.S.; Scholte, A.A.; Cornish, K.; Scott, D.J.; Brichta, J.L.; Vederas, J.C.; Ochoa, O.; Michelmore, R.W.; Shintani, D.K.; Knapp, S.J. Identification and comparison of natural rubber from two Lactuca species. Phytochemistry 2006, 67, 2590–2596. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Burke, I.C.; Neff, M.M. Genetic and biochemical evaluation of natural rubber from Eastern Washington prickly lettuce (Lactuca serriola L.). J. Agric. Food Chem. 2015, 63, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.-J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Cherian, S.; Ryu, S.B.; Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019, 17, 2041–2061. [Google Scholar] [CrossRef]
- Epping, J.; van Deenen, N.; Niephaus, E.; Stolze, A.; Fricke, J.; Huber, C.; Eisenreich, W.; Twyman, R.M.; Prüfer, D.; Gronover, C.S. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 2015, 1, 15048. [Google Scholar] [CrossRef]
- Shimizu, N.; Koyama, T.; Ogura, K. Molecular Cloning, Expression, and Purification of Undecaprenyl Diphosphate Synthase: No sequence similarity between e-andz-prenyl diphosphate synthases. J. Biol. Chem. 1998, 273, 19476–19481. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, T.A.; Surowiecki, P.; Siekierska, H.; Kania, M.; Van Gelder, K.; Rea, K.A.; Virta, L.K.; Vatta, M.; Gawarecka, K.; Wojcik, J.; et al. Polyprenols Are Synthesized by a Plastidial cis-Prenyltransferase and Influence Photosynthetic Performance. Plant Cell 2017, 29, 1709–1725. [Google Scholar] [CrossRef]
- Baddiley, J. Bacterial cell wall biosynthesis. In Ciba Foundation Symposium 7—Polymerization in Biological Systems; Wolstenholme, G.E.W., O’Connor, M., Eds.; J. & A. Churchill: London, UK, 1972; pp. 87–107. [Google Scholar]
- Manat, G.; Roure, S.; Auger, R.; Bouhss, A.; Barreteau, H.; Mengin-Lecreulx, D.; Touzé, T. Deciphering the Metabolism of Undecaprenyl-Phosphate: The Bacterial Cell-Wall Unit Carrier at the Membrane Frontier. Microb. Drug Resist. 2014, 20, 199–214. [Google Scholar] [CrossRef]
- Virta, L. Polyisoprenoid Alcohols Influence Plastidial Membrane Dynamics and Photosynthetic Performance in Solanum lycopersicum and Arabidopsis thaliana; University of Guelph: Guelph, ON, Canada, 2018. [Google Scholar]
- Brasher, M.I.; Surmacz, L.; Leong, B.; Pitcher, J.; Swiezewska, E.; Pichersky, E.; Akhtar, T.A. A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 2015, 82, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Grabińska, K.A.; Guan, Z.; Stránecký, V.; Hartmannová, H.; Hodaňová, K.; Barešová, V.; Sovová, J.; Jozsef, L.; Ondrušková, N.; et al. Mutation of Nogo-B Receptor, a Subunit of cis-Prenyltransferase, Causes a Congenital Disorder of Glycosylation. Cell Metab. 2014, 20, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Kwon, E.-J.; Ro, D. cis-Prenyltransferase and polymer analysis from a natural rubber perspective. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 576, pp. 121–145. [Google Scholar]
- Zhang, H.; Ohyama, K.; Boudet, J.; Chen, Z.; Yang, J.; Zhang, M.; Muranaka, T.; Maurel, C.; Zhu, J.-K.; Gong, Z. Dolichol Biosynthesis and Its Effects on the Unfolded Protein Response and Abiotic Stress Resistance in Arabidopsis. Plant Cell 2008, 20, 1879–1898. [Google Scholar] [CrossRef] [PubMed]
- Surowiecki, P.; Onysk, A.; Manko, K.; Swiezewska, E.; Surmacz, L. Long-Chain Polyisoprenoids Are Synthesized by AtCPT1 in Arabidopsis thaliana. Molecules 2019, 24, 2789. [Google Scholar] [CrossRef] [PubMed]
- Lakusta, A.M.; Kwon, M.; Kwon, E.-J.G.; Stonebloom, S.; Scheller, H.V.; Ro, D.-K. Molecular studies of the protein complexes involving cis-prenyltransferase in guayule (Parthenium argentatum), an alternative rubber-producing plant. Front. Plant Sci. 2019, 10, 428199. [Google Scholar] [CrossRef] [PubMed]
- Post, J.; van Deenen, N.; Fricke, J.; Kowalski, N.; Wurbs, D.; Schaller, H.; Eisenreich, W.; Huber, C.; Twyman, R.M.; Prüfer, D.; et al. Laticifer-Specific cis-Prenyltransferase Silencing Affects the Rubber, Triterpene, and Inulin Content of Taraxacum brevicorniculatum. Plant Physiol. 2012, 158, 1406–1417. [Google Scholar] [CrossRef]
- Qu, Y.; Chakrabarty, R.; Tran, H.T.; Kwon, E.-J.G.; Kwon, M.; Nguyen, T.-D.; Ro, D.-K. A Lettuce (Lactuca sativa) Homolog of Human Nogo-B Receptor Interacts with cis-Prenyltransferase and Is Necessary for Natural Rubber Biosynthesis. J. Biol. Chem. 2015, 290, 1898–1914. [Google Scholar] [CrossRef]
- Barnes, E.K.; Kwon, M.; Hodgins, C.L.; Qu, Y.; Kim, S.-W.; Yeung, E.C.; Ro, D.-K. The promoter sequences of lettuce cis-prenyltransferase and its binding protein specify gene expression in laticifers. Planta 2021, 253, 1–12. [Google Scholar] [CrossRef]
- Hua, Y.; Wang, C.; Huang, J.; Wang, K. A simple and efficient method for CRISPR/Cas9-induced mutant screening. J. Genet. Genom. 2017, 44, 207–213. [Google Scholar] [CrossRef]
- Collins-Silva, J.; Nural, A.T.; Skaggs, A.; Scott, D.; Hathwaik, U.; Woolsey, R.; Schegg, K.; McMahan, C.; Whalen, M.; Cornish, K.; et al. Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry 2012, 79, 46–56. [Google Scholar] [CrossRef]
- Cornish, K.; Castillón, J.; Scott, D.J. Rubber Molecular Weight Regulation, in Vitro, in Plant Species that Produce High and Low Molecular Weights in Vivo. Biomacromolecules 2000, 1, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, Y.; Liu, X.; Ahmad, N.; Wang, N.; Jin, L.; Yao, N.; Liu, X. A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 5393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ahmad, N.; Li, Z.; He, J.; Wang, N.; Naeem, M.; Jin, L.; Yao, N.; Liu, X. CtCYP71A1 promotes drought stress tolerance and lignin accumulation in safflower and Arabidopsis. Environ. Exp. Bot. 2023, 213, 105430. [Google Scholar] [CrossRef]
- Ahmad, N.; Jianyu, L.; Xu, T.; Noman, M.; Jameel, A.; Na, Y.; Yuanyuan, D.; Nan, W.; Xiaowei, L.; Fawei, W.; et al. Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Stonebloom, S.H.; Scheller, H.V. Transcriptome analysis of rubber biosynthesis in guayule (Parthenium argentatum gray). BMC Plant Biol. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Karumamkandathil, R.; Jayasree, P.K.; Radha, J.; Uthup, T.K.; Mathew, S.A.; Sathik, M.B.M. Genetics and Genomics of Abiotic Stress in Rubber Tree (Hevea brasiliensis). In Genomic Designing for Abiotic Stress Resistant Technical Crops; Springer: Cham, Switzerland, 2022; pp. 245–298. [Google Scholar]
- Muthusamy, M.; Lee, S.I. Abiotic stress-induced secondary metabolite production in Brassica: Opportunities and challenges. Front. Plant Sci. 2024, 14, 1323085. [Google Scholar] [CrossRef]
- Hein, C.; Henrich, E.; Orbán, E.; Dötsch, V.; Bernhard, F. Hydrophobic supplements in cell-free systems: Designing artificial environments for membrane proteins. Eng. Life Sci. 2014, 14, 365–379. [Google Scholar] [CrossRef]
- Genji, T.; Nozawa, A.; Tozawa, Y. Efficient production and purification of functional bacteriorhodopsin with a wheat-germ cell-free system and a combination of Fos-choline and CHAPS detergents. Biochem. Biophys. Res. Commun. 2010, 400, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Fujimoto, R.; Matsuoka, H.; Tsuboi, T.; Tozawa, Y. Cell-free synthesis, reconstitution, and characterization of a mitochondrial dicarboxylate–tricarboxylate carrier of Plasmodium falciparum. Biochem. Biophys. Res. Commun. 2011, 414, 612–617. [Google Scholar] [CrossRef]
- Tadaki, D.; Yamaura, D.; Araki, S.; Yoshida, M.; Arata, K.; Ohori, T.; Ishibashi, K.-I.; Kato, M.; Ma, T.; Miyata, R.; et al. Mechanically stable solvent-free lipid bilayers in nano- and micro-tapered apertures for reconstitution of cell-free synthesized hERG channels. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Horn, P.J.; James, C.N.; Gidda, S.K.; Kilaru, A.; Dyer, J.M.; Mullen, R.T.; Ohlrogge, J.B.; Chapman, K.D. Identification of a New Class of Lipid Droplet-Associated Proteins in Plants. Plant Physiol. 2013, 162, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.; Ryu, S.B.; Kwak, Y.S.; Kang, H. A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J. Exp. Bot. 2004, 55, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid Cleavage Dioxygenase (CmCCD4a) Contributes to White Color Formation in Chrysanthemum Petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Chamoun, Z.; Vacca, F.; Parton, R.G.; Gruenberg, J. PNPLA3/adiponutrin functions in lipid droplet formation. Biol. Cell 2013, 105, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Kirchman, D.; K’Nees, E.; Hodson, R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 1985, 49, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Kory, N.; Farese, R.V., Jr.; Walther, T.C. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 2016, 26, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Bulankina, A.V.; Deggerich, A.; Wenzel, D.; Mutenda, K.; Wittmann, J.G.; Rudolph, M.G.; Burger, K.N.; Höning, S. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 2009, 185, 641–655. [Google Scholar] [CrossRef]
- Baker, M.R.; Fan, G.; Serysheva, I.I. Single-Particle Cryo-EM of the Ryanodine Receptor Channel in an Aqueous Environment. Eur. J. Transl. Myol. 2015, 25, 4803. [Google Scholar] [CrossRef]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta (BBA)-Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef]
- Zoonens, M.; Popot, J.-L. Amphipols for each season. J. Membr. Biol. 2014, 247, 759–796. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, T.; Grinkova, Y.; Bayburt, T.; Denisov, I.; Zolnerciks, J.; Atkins, W.; Sligar, S. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009, 464, 211–231. [Google Scholar]
- Rigaud, J.-L.; Lévy, D. Reconstitution of membrane proteins into liposomes. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2003; Volume 372, pp. 65–86. [Google Scholar]
- Smirnova, I.A.; Ädelroth, P.; Brzezinski, P. Extraction and liposome reconstitution of membrane proteins with their native lipids without the use of detergents. Sci. Rep. 2018, 8, 14950. [Google Scholar] [CrossRef] [PubMed]
- Scheidelaar, S.; Koorengevel, M.C.; Pardo, J.D.; Meeldijk, J.D.; Breukink, E.; Killian, J.A. Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophys. J. 2015, 108, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Jamshad, M.; Grimard, V.; Idini, I.; Knowles, T.J.; Dowle, M.R.; Schofield, N.; Sridhar, P.; Lin, Y.; Finka, R.; Wheatley, M.; et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2014, 8, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.M.; Schäfer, M.; Koorengevel, M.C.; Killian, J.A. Detergent-Free Isolation, Characterization and Functional Reconstitution of a K+ Channel: The Power of Native Nanodiscs. Biophys. J. 2016, 110, 421a. [Google Scholar] [CrossRef]
- Orwick-Rydmark, M.; Lovett, J.E.; Graziadei, A.; Lindholm, L.; Hicks, M.R.; Watts, A. Detergent-Free Incorporation of a Seven-Transmembrane Receptor Protein into Nanosized Bilayer Lipodisq Particles for Functional and Biophysical Studies. Nano Lett. 2012, 12, 4687–4692. [Google Scholar] [CrossRef]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; Van Der Cruijsen, E.A.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef]
- Smirnova, I.A.; Sjostrand, D.; Li, F.; Bjorck, M.; Schafer, J.; Ostbye, H.; Hogbom, M.; von Ballmoos, C.; Lander, G.C.; Adelroth, P.; et al. Isolation of yeast complex IV in native lipid nanodiscs. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 2984–2992. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, M.; Hogle, J.M.; Shih, W.M.; Wagner, G.; Nasr, M.L. DNA-Corralled Nanodiscs for the Structural and Functional Characterization of Membrane Proteins and Viral Entry. J. Am. Chem. Soc. 2018, 140, 10639–10643. [Google Scholar] [CrossRef]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.-Y.J.; Grigoriu, S.; Huser, S.; Plückthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Das, D.; Courtney, N.A.; Jiang, Y.; Briguglio, J.S.; Lou, X.; Roston, D.; Cui, Q.; Chanda, B.; Chapman, E.R. Dynamics and number of trans-SNARE complexes determine nascent fusion pore properties. Nature 2018, 554, 260–263. [Google Scholar] [CrossRef] [PubMed]
- McGoldrick, L.L.; Singh, A.K.; Saotome, K.; Yelshanskaya, M.V.; Twomey, E.C.; Grassucci, R.A.; Sobolevsky, A.I. Opening of the human epithelial calcium channel TRPV6. Nature 2018, 553, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Nasr, M.L.; Wagner, G. Covalently circularized nanodiscs; challenges and applications. Curr. Opin. Struct. Biol. 2018, 51, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Bush, J.; Singh, S.; Vargas, M.; Oktay, E.; Hu, C.-H.; Veneziano, R. Synthesis of DNA Origami Scaffolds: Current and Emerging Strategies. Molecules 2020, 25, 3386. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yang, Y.R.; Johnson-Buck, A.; Liu, M.; Liu, Y.; Walter, N.G.; Woodbury, N.W.; Yan, H. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 2014, 9, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Derr, N.D.; Goodman, B.S.; Jungmann, R.; Leschziner, A.E.; Shih, W.M.; Reck-Peterson, S.L. Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold. Science 2012, 338, 662–665. [Google Scholar] [CrossRef]
- Sun, W.; Boulais, E.; Hakobyan, Y.; Wang, W.L.; Guan, A.; Bathe, M.; Yin, P. Casting inorganic structures with DNA molds. Science 2014, 346, 1258361. [Google Scholar] [CrossRef]
- Miehling, J.; Goricanec, D.; Hagn, F. A Split-Intein-Based Method for the Efficient Production of Circularized Nanodiscs for Structural Studies of Membrane Proteins. ChemBioChem 2018, 19, 1927–1933. [Google Scholar] [CrossRef]
- Das, K.M.P.; Shih, W.M.; Wagner, G.; Nasr, M.L. Large Nanodiscs: A Potential Game Changer in Structural Biology of Membrane Protein Complexes and Virus Entry. Front. Bioeng. Biotechnol. 2020, 8, 539. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umar, A.W.; Ahmad, N.; Xu, M. Reviving Natural Rubber Synthesis via Native/Large Nanodiscs. Polymers 2024, 16, 1468. https://doi.org/10.3390/polym16111468
Umar AW, Ahmad N, Xu M. Reviving Natural Rubber Synthesis via Native/Large Nanodiscs. Polymers. 2024; 16(11):1468. https://doi.org/10.3390/polym16111468
Chicago/Turabian StyleUmar, Abdul Wakeel, Naveed Ahmad, and Ming Xu. 2024. "Reviving Natural Rubber Synthesis via Native/Large Nanodiscs" Polymers 16, no. 11: 1468. https://doi.org/10.3390/polym16111468
APA StyleUmar, A. W., Ahmad, N., & Xu, M. (2024). Reviving Natural Rubber Synthesis via Native/Large Nanodiscs. Polymers, 16(11), 1468. https://doi.org/10.3390/polym16111468







