Electrospun Scaffolds Enriched with Nanoparticle-Associated DNA: General Properties, DNA Release and Cell Transfection
Abstract
:1. Introduction
2. Materials and Methods
2.1. pDNA Production
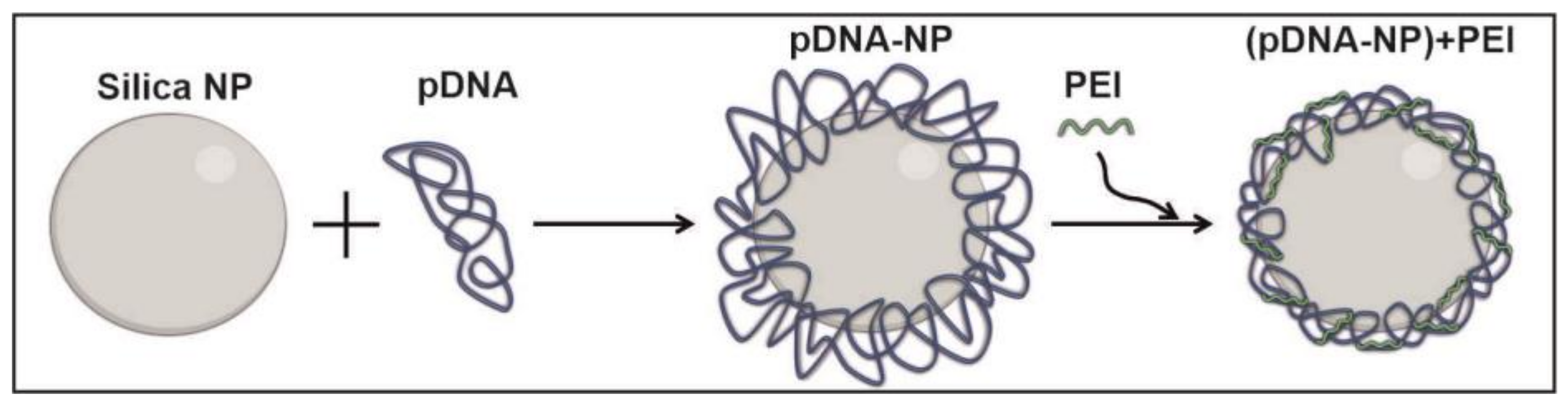
2.2. Preparation of Nanoparticles-Containing pDNA
2.3. Characterization of DNA-Nanoparticles
2.4. Fabrication of Electrospun Scaffolds
2.5. Characterization of Electrospun Scaffolds
2.6. In Vitro pDNA Release from Electrospun Scaffolds
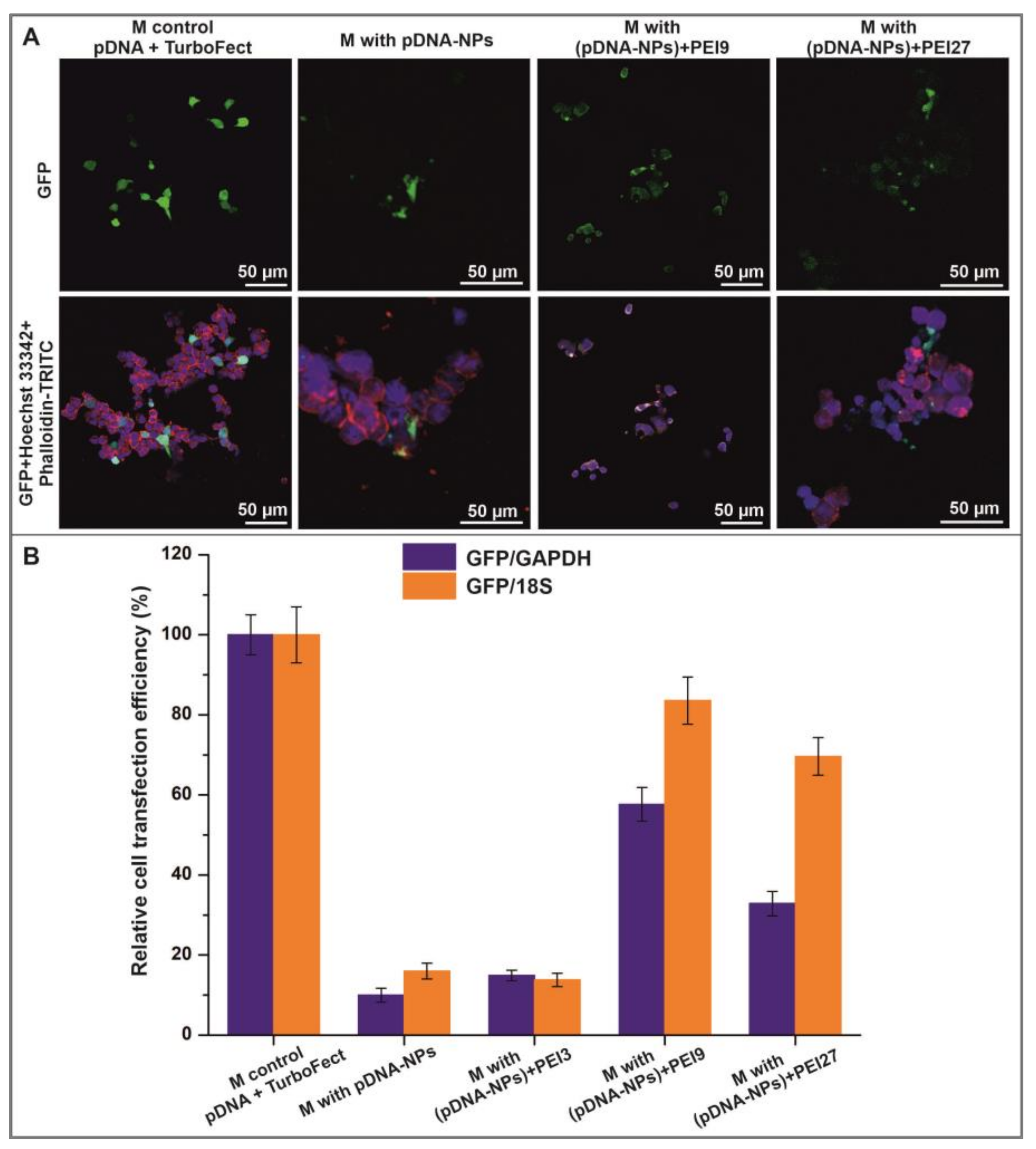
2.7. Cultivation, Viability and Transfection Efficiency of Cells on Fibrous Scaffolds
3. Results and Discussion
3.1. Synthesis and Characterization of silica NPs Containing pDNA
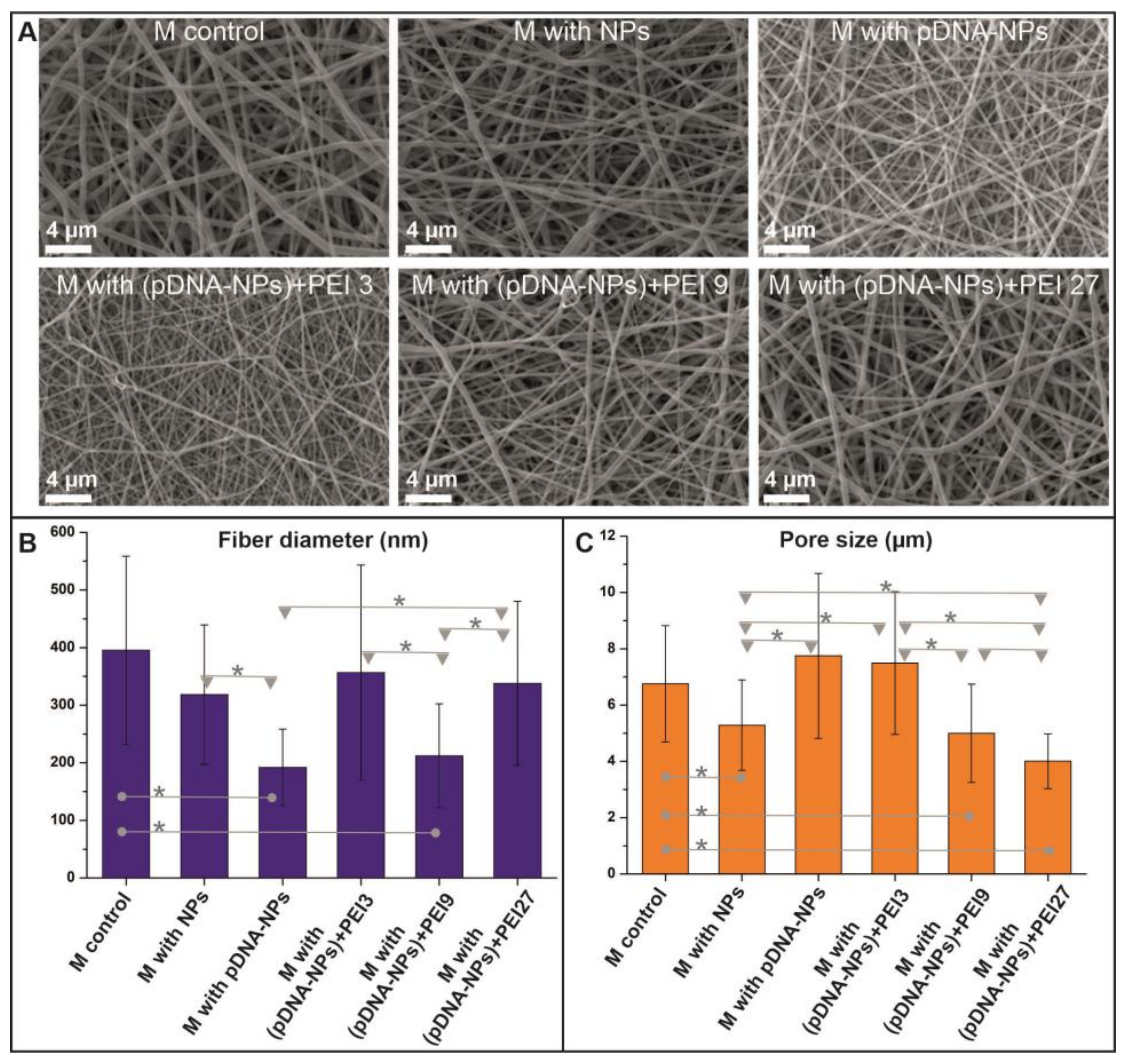
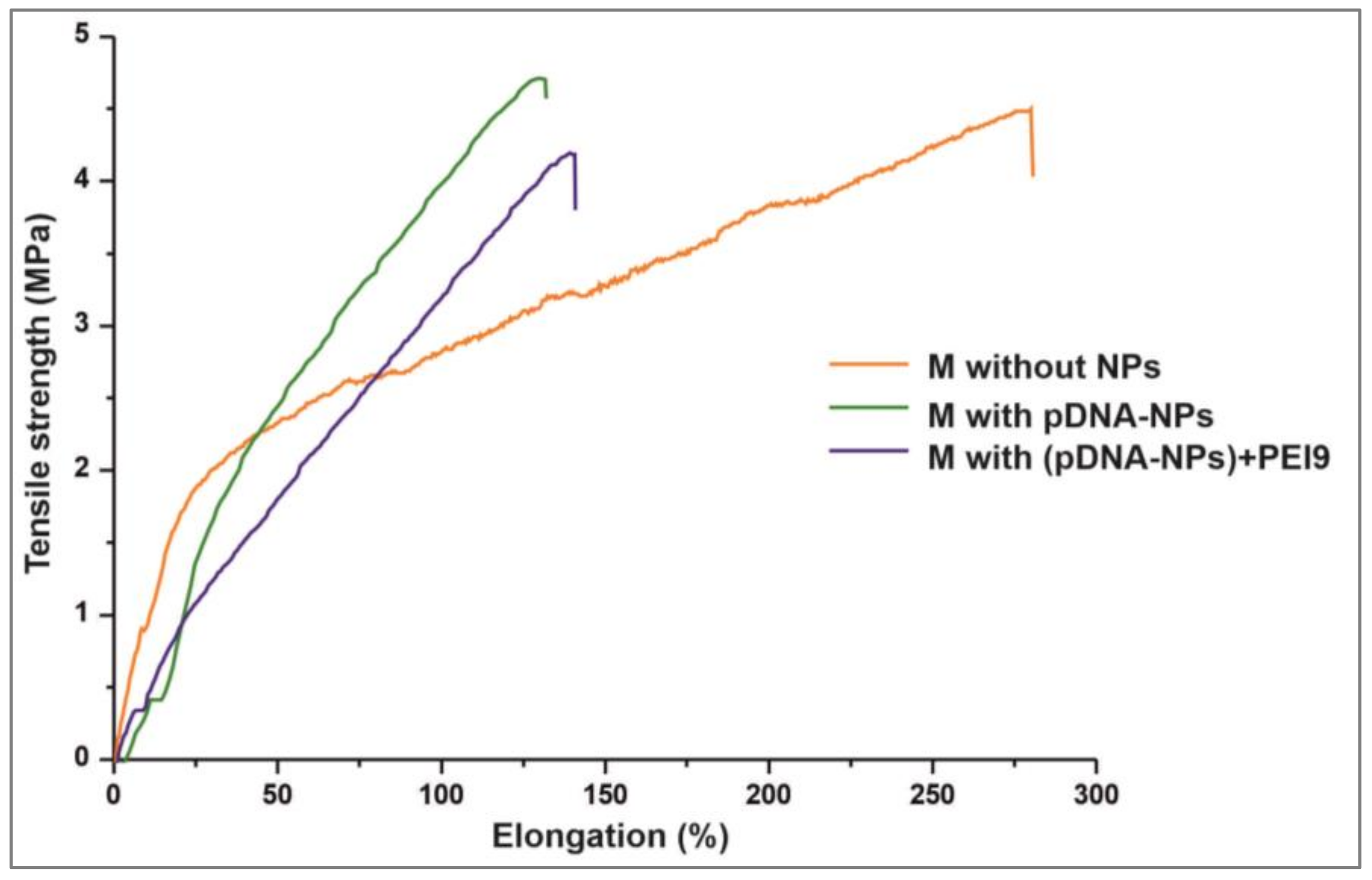
3.2. Properties of DNA-Enriched Electrospun Scaffolds
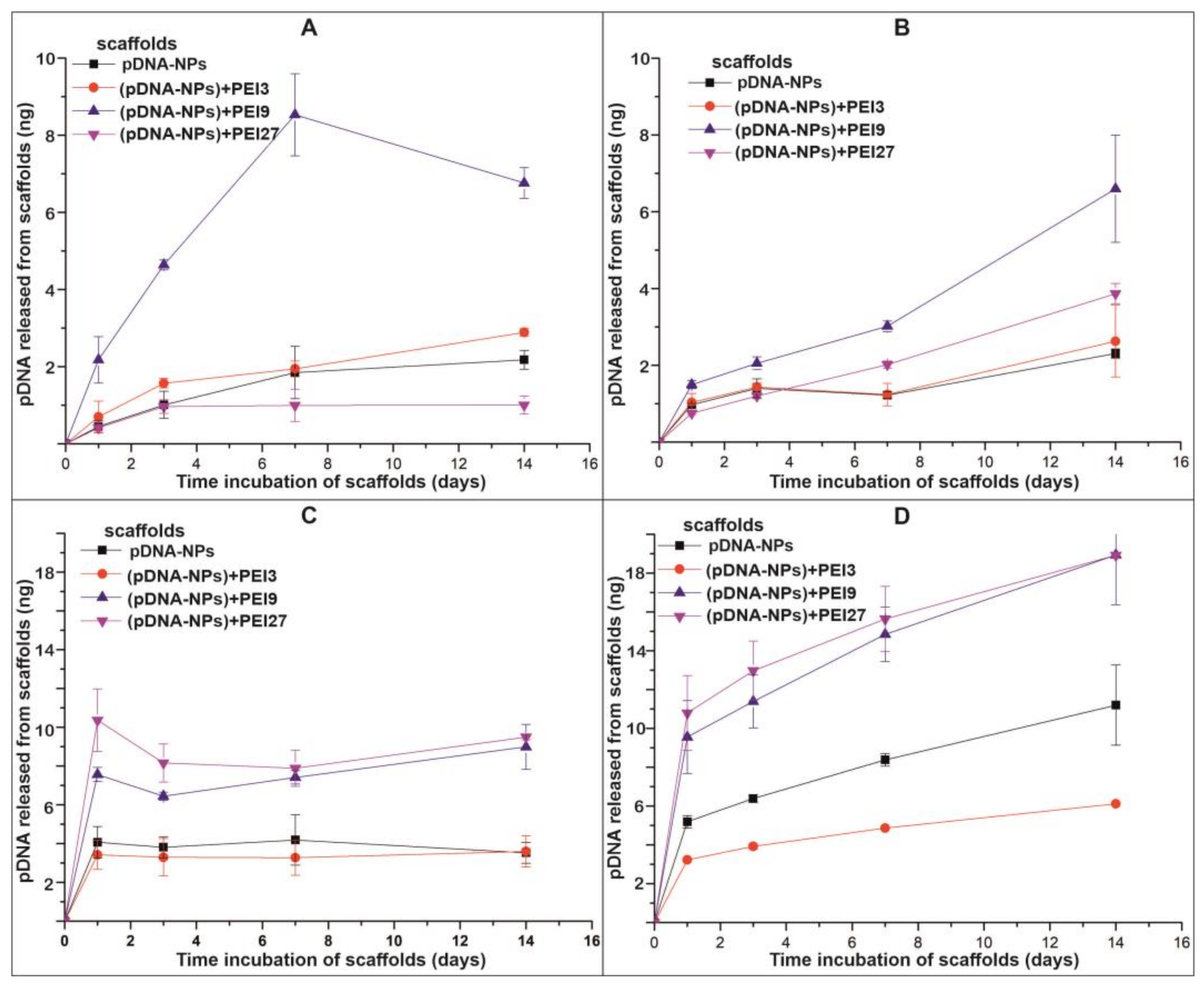
3.3. pDNA Release from Electrospun Scaffolds
3.4. Interaction of HEK293T Cells with Electrospun Scaffolds
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernhard, J.C.; Vunjak-Novakovic, G. Should we use cells, biomaterials, or tissue engineering for cartilage regeneration? Stem Cell Res. Ther. 2016, 7, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroni, L.; Elisseeff, J.H. Biomaterials engineered for integration. Mater. Today 2008, 11, 44–51. [Google Scholar] [CrossRef]
- Johnston, A.; Callanan, A. Recent Methods for Modifying Mechanical Properties of Tissue-Engineered Scaffolds for Clinical Applications. Biomimetics 2023, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, M.P.; Chavali, M.S. Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 2019, 4, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.P.; Sadia, M.; Jaganathan, S.K.; Khudzari, A.Z.; Supriyanto, E.; Saidin, S.; Ramakrishna, S.; Ismail, A.F.; Faudzi, A.A.M. A review on 3D printing in tissue engineering applications. J. Polym. Eng. 2022, 42, 243–265. [Google Scholar] [CrossRef]
- Kantaros, A. 3D Printing in Regenerative Medicine: Technologies and Resources Utilized. Int. J. Mol. Sci. 2022, 23, 14621. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.; Mikos, A.G. Advances in In Vitro and In Vivo Bioreactor-Based Bone Generation for Craniofacial Tissue Engineering. BME Front. 2023, 4, 4. [Google Scholar] [CrossRef]
- Stephenson, M.; Grayson, W. Recent advances in bioreactors for cell-based therapies. F1000Research 2018, 7, 517. [Google Scholar] [CrossRef]
- Crupi, A.; Costa, A.; Tarnok, A.; Melzer, S.; Teodori, L. Inflammation in tissue engineering: The Janus between engraftment and rejection. Eur. J. Immunol. 2015, 45, 3222–3236. [Google Scholar] [CrossRef]
- Tu, Z.; Zhong, Y.; Hu, H.; Shao, D.; Haag, R.; Schirner, M.; Lee, J.; Sullenger, B.; Leong, K.W. Design of therapeutic biomaterials to control inflammation. Nat. Rev. Mater. 2022, 7, 557–574. [Google Scholar] [CrossRef] [PubMed]
- Xi, K.; Gu, Y.; Tang, J.; Chen, H.; Xu, Y.; Wu, L.; Cai, F.; Deng, L.; Yang, H.; Shi, Q.; et al. Microenvironment-responsive immunoregulatory electrospun fibers for promoting nerve function recovery. Nat. Commun. 2020, 11, 4504. [Google Scholar] [CrossRef]
- Yanez, M.; Blanchette, J.; Jabbarzadeh, E. Modulation of Inflammatory Response to Implanted Biomaterials Using Natural Compounds. Curr. Pharm. Des. 2017, 23, 6347–6357. [Google Scholar] [CrossRef] [Green Version]
- Giri, J.; Li, W.-J.; Tuan, R.S.; Cicerone, M.T. Stabilization of proteins by nanoencapsulation in sugar-glass for tissue engineering and drug delivery applications. Adv. Mater. 2011, 23, 4861–4867. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Biedrzycki, A.H.; Khalil, A.S.; Hess, D.; Umhoefer, J.M.; Markel, M.D.; Murphy, W.L. Nanostructured Mineral Coatings Stabilize Proteins for Therapeutic Delivery. Adv. Mater. 2017, 29, 1701255. [Google Scholar] [CrossRef] [PubMed]
- Uludag, H.; Ubeda, A.; Ansari, A. At the Intersection of Biomaterials and Gene Therapy: Progress in Non-viral Delivery of Nucleic Acids. Front. Bioeng. Biotechnol. 2019, 7, 131. [Google Scholar] [CrossRef]
- De Laporte, L.; Shea, L.D. Matrices and scaffolds for DNA delivery in tissue engineering. Adv. Drug Deliv. Rev. 2007, 59, 292–307. [Google Scholar] [CrossRef] [Green Version]
- Simpson, D.J.; Olova, N.N.; Chandra, T. Cellular reprogramming and epigenetic rejuvenation. Clin. Epigenetics 2021, 13, 170. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Babos, K.; Ichida, J.K. Small Molecules Take a Big Step by Converting Fibroblasts into Neurons. Cell Stem Cell 2015, 17, 127–129. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.A.; Shafieian, R.; Alipour, F. Role of growth factors and cytokines in therapeutic angiogenesis. In Biomaterials for Vasculogenesis and Angiogenesis; Elsevier: Amsterdam, The Netherlands, 2022; pp. 85–111. ISBN 9780128218679. [Google Scholar]
- Metzemaekers, M.; Gouwy, M.; Proost, P. Neutrophil chemoattractant receptors in health and disease: Double-edged swords. Cell Mol. Immunol. 2020, 17, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, L. An overview on electrospinning and its advancement toward hard and soft tissue engineering applications. Colloid Polym. Sci. 2022, 300, 875–901. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Chen, Y.; Wang, X.; Xu, X.; Shen, Y.; Khan, A.u.R.; Aldalbahi, A.; Fetz, A.E.; Bowlin, G.L.; El-Newehy, M.; et al. Electrospinning nanofiber scaffolds for soft and hard tissue regeneration. J. Mater. Sci. Technol. 2020, 59, 243–261. [Google Scholar] [CrossRef]
- Eivazi Zadeh, Z.; Solouk, A.; Shafieian, M.; Haghbin Nazarpak, M. Electrospun polyurethane/carbon nanotube composites with different amounts of carbon nanotubes and almost the same fiber diameter for biomedical applications. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 118, 111403. [Google Scholar] [CrossRef]
- Caldiroli, A.; Pederzani, E.; Pezzotta, M.; Azzollini, N.; Fiori, S.; Tironi, M.; Rizzo, P.; Sangalli, F.; Figliuzzi, M.; Fiore, G.B.; et al. Hybrid fibroin/polyurethane small-diameter vascular grafts: From fabrication toin vivopreliminary assessment. Biomed. Mater. 2022, 17, 55015. [Google Scholar] [CrossRef]
- Mohammadalizadeh, Z.; Bahremandi-Toloue, E.; Karbasi, S. Synthetic-based blended electrospun scaffolds in tissue engineering applications. J. Mater. Sci. 2022, 57, 4020–4079. [Google Scholar] [CrossRef]
- Owida, H.A.; Al-Nabulsi, J.I.; Alnaimat, F.; Al-Ayyad, M.; Turab, N.M.; Al Sharah, A.; Shakur, M. Recent Applications of Electrospun Nanofibrous Scaffold in Tissue Engineering. Appl. Bionics Biomech. 2022, 2022, 1953861. [Google Scholar] [CrossRef] [PubMed]
- Klabukov, I.; Balyasin, M.; Krasilnikova, O.; Tenchurin, T.; Titov, A.; Krasheninnikov, M.; Mudryak, D.; Sulina, Y.; Shepelev, A.; Chvalun, S.; et al. Angiogenic Modification of Microfibrous Polycaprolactone by pCMV-VEGF165 Plasmid Promotes Local Vascular Growth after Implantation in Rats. Int. J. Mol. Sci. 2023, 24, 1399. [Google Scholar] [CrossRef]
- Graceffa, V. Physical and mechanical cues affecting biomaterial-mediated plasmid DNA delivery: Insights into non-viral delivery systems. J. Genet. Eng. Biotechnol. 2021, 19, 90. [Google Scholar] [CrossRef]
- Achille, C.; Sundaresh, S.; Chu, B.; Hadjiargyrou, M. Cdk2 silencing via a DNA/PCL electrospun scaffold suppresses proliferation and increases death of breast cancer cells. PLoS ONE 2012, 7, e52356. [Google Scholar] [CrossRef] [Green Version]
- Kobsa, S.; Kristofik, N.J.; Sawyer, A.J.; Bothwell, A.L.M.; Kyriakides, T.R.; Saltzman, W.M. An electrospun scaffold integrating nucleic acid delivery for treatment of full-thickness wounds. Biomaterials 2013, 34, 3891–3901. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Jia, L.-N.; Xu, H.-Y.; Hu, X.-G.; Wang, W.; Jia, J. Fabrication of Core-Shell PEI/pBMP2-PLGA Electrospun Scaffold for Gene Delivery to Periodontal Ligament Stem Cells. Stem Cells Int. 2016, 2016, 5385137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, P.; Padmashali, R.M.; Andreadis, S.T. Cell-controlled and spatially arrayed gene delivery from fibrin hydrogels. Biomaterials 2009, 30, 3790–3799. [Google Scholar] [CrossRef] [Green Version]
- Luu, Y.K.; Kim, K.; Hsiao, B.S.; Chu, B.; Hadjiargyrou, M. Development of a nanostructured DNA delivery scaffold via electrospinning of PLGA and PLA-PEG block copolymers. J. Control. Release 2003, 89, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Wang, C.-H. Fabrication and characterization of PLGA/HAp composite scaffolds for delivery of BMP-2 plasmid DNA. J. Control. Release 2007, 120, 111–121. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Cheng, L.; He, S.; Zou, J.; Chen, F.; Zhang, Z. Core-sheath structured fibers with pDNA polyplex loadings for the optimal release profile and transfection efficiency as potential tissue engineering scaffolds. Acta Biomater. 2011, 7, 2533–2543. [Google Scholar] [CrossRef] [PubMed]
- Ravi, N.; Gupta, G.; Milbrandt, T.A.; Puleo, D.A. Porous PLGA scaffolds for controlled release of naked and polyethyleneimine-complexed DNA. Biomed. Mater. 2012, 7, 55007. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Liu, Y.; Luo, X.; Chen, F.; Guo, X.; Li, X. Electrospun fibrous scaffolds with continuous gradations in mineral contents and biological cues for manipulating cellular behaviors. Acta Biomater. 2012, 8, 1576–1585. [Google Scholar] [CrossRef]
- Tamkovich, S.N.; Laktionov, P.P.; Rykova, E.Y.; Vlassov, V.V. Simple and rapid procedure suitable for quantitative isolation of low and high molecular weight extracellular nucleic acids. Nucleosides Nucleotides Nucleic Acids 2004, 23, 873–877. [Google Scholar] [CrossRef]
- Vandeventer, P.E.; Mejia, J.; Nadim, A.; Johal, M.S.; Niemz, A. DNA adsorption to and elution from silica surfaces: Influence of amino acid buffers. J. Phys. Chem. B 2013, 117, 10742–10749. [Google Scholar] [CrossRef] [Green Version]
- Esser, K.-H.; Marx, W.H.; Lisowsky, T. maxXbond: First regeneration system for DNA binding silica matrices. Nat. Methods 2006, 3, i–ii. [Google Scholar] [CrossRef]
- Popova, V.; Poletaeva, Y.; Chubarov, A.; Pyshnyi, D.; Dmitrienko, E. Doxorubicin-Loaded Silica Nanocomposites for Cancer Treatment. Coatings 2023, 13, 324. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Kiseleva, E.; Pyshnaya, I.A.; Laktionov, P.P. Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation-Precipitation Approach for Downstream miRNAs Detection. Diagnostics 2021, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- ISO 7198:2016; Cardiovascular Implants and Extracorporeal Systems—Vascular Prostheses—Tubular Vascular Grafts and Vascular Patches. International Organization for Standardization: Geneva, Switzerland, 2016. Available online: https://www.iso.org/ru/standard/50661.html (accessed on 10 October 2022).
- Chernonosova, V.S.; Kvon, R.I.; Stepanova, A.O.; Larichev, Y.V.; Karpenko, A.A.; Chelobanov, B.P.; Kiseleva, E.V.; Laktionov, P.P. Human serum albumin in electrospun PCL fibers: Structure, release, and exposure on fiber surface. Polym. Adv. Technol. 2017, 28, 819–827. [Google Scholar] [CrossRef]
- Chernonosova, V.S.; Gostev, A.A.; Chesalov, Y.A.; Karpenko, A.A.; Karaskov, A.M.; Laktionov, P.P. Study of hemocompatibility and endothelial cell interaction of tecoflex-based electrospun vascular grafts. Int. J. Polym. Mater. Polym. Biomater. 2019, 68, 34–43. [Google Scholar] [CrossRef]
- Chernonosova, V.S.; Kuzmin, I.E.; Shundrina, I.K.; Korobeynikov, M.V.; Golyshev, V.M.; Chelobanov, B.P.; Laktionov, P.P. Effect of Sterilization Methods on Electrospun Scaffolds Produced from Blend of Polyurethane with Gelatin. J. Funct. Biomater. 2023, 14, 70. [Google Scholar] [CrossRef]
- Chernonosova, V.; Gostev, A.; Murashov, I.; Chelobanov, B.; Karpenko, A.; Laktionov, P. Assessment of Electrospun Pellethane-Based Scaffolds for Vascular Tissue Engineering. Materials 2021, 14, 3678. [Google Scholar] [CrossRef]
- Chernonosova, V.S.; Laktionov, P.P.; Murashov, I.S.; Karpenko, A.A.; Laktionov, P.P. Comparative gene expression profiling of human primary endotheliocytes cultivated on polyurethane-based electrospun 3D matrices and natural decellularized vein. Biomed. Mater. 2020, 15, 45012. [Google Scholar] [CrossRef]
- Kelly, D.C.; Raftery, R.M.; Curtin, C.M.; O’Driscoll, C.M.; O’Brien, F.J. Scaffold-Based Delivery of Nucleic Acid Therapeutics for Enhanced Bone and Cartilage Repair. J. Orthop. Res. 2019, 37, 1671–1680. [Google Scholar] [CrossRef]
- Pinese, C.; Lin, J.; Milbreta, U.; Li, M.; Wang, Y.; Leong, K.W.; Chew, S.Y. Sustained delivery of siRNA/mesoporous silica nanoparticle complexes from nanofiber scaffolds for long-term gene silencing. Acta Biomater. 2018, 76, 164–177. [Google Scholar] [CrossRef]
- Elangovan, S.; D’Mello, S.R.; Hong, L.; Ross, R.D.; Allamargot, C.; Dawson, D.V.; Stanford, C.M.; Johnson, G.K.; Sumner, D.R.; Salem, A.K. The enhancement of bone regeneration by gene activated matrix encoding for platelet derived growth factor. Biomaterials 2014, 35, 737–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reckhenrich, A.K.; Koch, C.; Egaña, J.T.; Plank, C. The use of non-viral gene vectors for bioactive poly-(D,L-lactide) implant surfaces in bone tissue engineering. Eur. Cell Mater. 2012, 23, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, E.; Nikishin, I.; Bogdanova, A.; Klinov, D.; Bagrov, D. The miscibility and spatial distribution of the components in electrospun polymer-protein mats. RSC Adv. 2020, 10, 4672–4680. [Google Scholar] [CrossRef] [PubMed]
- Topuz, F.; Uyar, T. Influence of Hydrogen-Bonding Additives on Electrospinning of Cyclodextrin Nanofibers. ACS Omega 2018, 3, 18311–18322. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.O.; Laktionov, P.P.; Cherepanova, A.V.; Chernonosova, V.S.; Shevelev, G.Y.; Zaporozhchenko, I.A.; Karaskov, A.M.; Laktionov, P.P. General Study and Gene Expression Profiling of Endotheliocytes Cultivated on Electrospun Materials. Materials 2019, 12, 4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, K.A.; Murashov, I.S.; Chernonosova, V.S.; Chelobanov, B.P.; Stepanova, A.O.; Sergeevichev, D.S.; Karpenko, A.A.; Laktionov, P.P. Vascular Stents Coated with Electrospun Drug-Eluting Material: Functioning in Rabbit Iliac Artery. Polymers 2020, 12, 1741. [Google Scholar] [CrossRef]
- Nazarkina, Z.K.; Chelobanov, B.P.; Chernonosova, V.S.; Romanova, I.V.; Karpenko, A.A.; Laktionov, P.P. Sirolimus-Eluting Electrospun-Produced Matrices as Coatings for Vascular Stents: Dependence of Drug Release on Matrix Structure and Composition of the External Environment. Materials 2020, 13, 2692. [Google Scholar] [CrossRef]
- Campora, S.; Ghersi, G. Recent developments and applications of smart nanoparticles in biomedicine. Nanotechnol. Rev. 2022, 11, 2595–2631. [Google Scholar] [CrossRef]
- Vallet-Regí, M. Our contributions to applications of mesoporous silica nanoparticles. Acta Biomater. 2022, 137, 44–52. [Google Scholar] [CrossRef]
- Manzano, M.; Vallet-Regí, M. Mesoporous Silica Nanoparticles for Drug Delivery. Adv. Funct. Mater. 2020, 30, 1902634. [Google Scholar] [CrossRef]
- Liu, H.-J.; Xu, P. Smart Mesoporous Silica Nanoparticles for Protein Delivery. Nanomaterials 2019, 9, 511. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, Z.; Subramaniam, S.; Khan, G.M.; Abeer, M.M.; Qu, Z.; Janjua, T.; Kumeria, T.; Batra, J.; Popat, A. Encapsulation and Controlled Release of Resveratrol Within Functionalized Mesoporous Silica Nanoparticles for Prostate Cancer Therapy. Front. Bioeng. Biotechnol. 2019, 7, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valente, J.F.A.; Pereira, P.; Sousa, A.; Queiroz, J.A.; Sousa, F. Effect of Plasmid DNA Size on Chitosan or Polyethyleneimine Polyplexes Formulation. Polymers 2021, 13, 793. [Google Scholar] [CrossRef] [PubMed]
- Mady, M.M.; Mohammed, W.A.; El-Guendy, N.M.; Elsayed, A.A. Effect of polymer molecular weight on the DNA/PEI polyplexes properties. Rom. J. Biophys. 2011, 21, 151–165. [Google Scholar]
- Bettinger, T.; Remy, J.S.; Erbacher, P. Size reduction of galactosylated PEI/DNA complexes improves lectin-mediated gene transfer into hepatocytes. Bioconjug. Chem. 1999, 10, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Clima, L.; Craciun, B.F.; Gavril, G.; Pinteala, M. Tunable Composition of Dynamic Non-Viral Vectors over the DNA Polyplex Formation and Nucleic Acid Transfection. Polymers 2019, 11, 1313. [Google Scholar] [CrossRef] [Green Version]
- Wagner, E.; Cotten, M.; Foisner, R.; Birnstiel, M.L. Transferrin-polycation-DNA complexes: The effect of polycations on the structure of the complex and DNA delivery to cells. Proc. Natl. Acad. Sci. USA 1991, 88, 4255–4259. [Google Scholar] [CrossRef]
- Ferkol, T.; Perales, J.C.; Eckman, E.; Kaetzel, C.S.; Hanson, R.W.; Davis, P.B. Gene transfer into the airway epithelium of animals by targeting the polymeric immunoglobulin receptor. J. Clin. Investig. 1995, 95, 493–502. [Google Scholar] [CrossRef] [Green Version]
- Kimoto, S.; Dick, W.D.; Hunt, B.; Szymanski, W.W.; McMurry, P.H.; Roberts, D.L.; Pui, D.Y.H. Characterization of nanosized silica size standards. Aerosol Sci. Technol. 2017, 51, 936–945. [Google Scholar] [CrossRef] [Green Version]
- Raja Mohd Hafidz, R.N.; Yaakob, C.M.; Amin, I.; Noorfaizan, A. Chemical and functional properties of bovine and porcine skin gelatin. Int. Food Res. J. 2011, 18, 813–817. [Google Scholar]
- Nelson, M.T.; Keith, J.P.; Li, B.-B.; Stocum, D.L.; Li, J. Electrospun composite polycaprolactone scaffolds for optimized tissue regeneration. Proc. Inst. Mech. Eng. Part N J. Nanoeng. Nanosyst. 2012, 226, 111–121. [Google Scholar] [CrossRef]
- Lopresti, F.; Pavia, F.C.; Ceraulo, M.; Capuana, E.; Brucato, V.; Ghersi, G.; Botta, L.; La Carrubba, V. Physical and biological properties of electrospun poly(d,l-lactide)/nanoclay and poly(d,l-lactide)/nanosilica nanofibrous scaffold for bone tissue engineering. J. Biomed. Mater. Res. A 2021, 109, 2120–2136. [Google Scholar] [CrossRef] [PubMed]
- Ulusoy, U. A Review of Particle Shape Effects on Material Properties for Various Engineering Applications: From Macro to Nanoscale. Minerals 2023, 13, 91. [Google Scholar] [CrossRef]
- Talevi, A.; Ruiz, M.E. Drug Release. The ADME Encyclopedia; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–7. ISBN 978-3-030-51519-5. [Google Scholar]
- Fu, Y.; Kao, W.J. Drug release kinetics and transport mechanisms of non-degradable and degradable polymeric delivery systems. Expert Opin. Drug Deliv. 2010, 7, 429–444. [Google Scholar] [CrossRef]
- Chelobanov, B.P.; Laktionov, P.P.; Vlasov, V.V. Proteins involved in binding and cellular uptake of nucleic acids. Biochemistry 2006, 71, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, K.A.; Stepanova, A.O.; Kuznetsov, N.A.; Chernonosova, V.S.; Kharkova, M.V.; Romanova, I.V.; Karpenko, A.A.; Laktionov, P.P. Diclofenac release from polycaprolactone 3D matrices produced by electrospinning: Influence of fiber structure and composition of the surrounding medium. Int. J. Polym. Mater. Polym. Biomater. 2019, 68, 27–33. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, W.; Bartolo, P. Investigation of polycaprolactone for bone tissue engineering scaffolds: In vitro degradation and biological studies. Mater. Des. 2022, 216, 110582. [Google Scholar] [CrossRef]
- Dias, J.R.; Sousa, A.; Augusto, A.; Bártolo, P.J.; Granja, P.L. Electrospun Polycaprolactone (PCL) Degradation: An In Vitro and In Vivo Study. Polymers 2022, 14, 3397. [Google Scholar] [CrossRef]
- Wissing, T.B.; Bonito, V.; van Haaften, E.E.; van Doeselaar, M.; Brugmans, M.M.C.P.; Janssen, H.M.; Bouten, C.V.C.; Smits, A.I.P.M. Macrophage-Driven Biomaterial Degradation Depends on Scaffold Microarchitecture. Front. Bioeng. Biotechnol. 2019, 7, 87. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Martín-Montañez, E.; López-Téllez, J.F.; Acevedo, M.J.; Pavía, J.; Khan, Z.U. Efficiency of gene transfection reagents in NG108-15, SH-SY5Y and CHO-K1 cell lines. Methods Find. Exp. Clin. Pharmacol. 2010, 32, 291–297. [Google Scholar] [CrossRef]
- Yamano, S.; Dai, J.; Moursi, A.M. Comparison of transfection efficiency of nonviral gene transfer reagents. Mol. Biotechnol. 2010, 46, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Moro, E.; Pettenazzo, A.; Behr, J.P.; Zacchello, F.; Scarpa, M. ExGen 500 is an efficient vector for gene delivery to lung epithelial cells in vitro and in vivo. Gene Ther. 1997, 4, 1100–1106. [Google Scholar] [CrossRef] [Green Version]
- Mendes, B.B.; Conniot, J.; Avital, A.; Yao, D.; Jiang, X.; Zhou, X.; Sharf-Pauker, N.; Xiao, Y.; Adir, O.; Liang, H.; et al. Nanodelivery of nucleic acids. Nat. Rev. Methods Primers 2022, 2, 24. [Google Scholar] [CrossRef]
- Zhu, Y.; Shen, R.; Vuong, I.; Reynolds, R.A.; Shears, M.J.; Yao, Z.-C.; Hu, Y.; Cho, W.J.; Kong, J.; Reddy, S.K.; et al. Multi-step screening of DNA/lipid nanoparticles and co-delivery with siRNA to enhance and prolong gene expression. Nat. Commun. 2022, 13, 4282. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A. Electrospun Nanofibers as Carriers of Microorganisms, Stem Cells, Proteins, and Nucleic Acids in Therapeutic and Other Applications. Front. Bioeng. Biotechnol. 2020, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Sebe, I.; Szabó, P.; Kállai-Szabó, B.; Zelkó, R. Incorporating small molecules or biologics into nanofibers for optimized drug release: A review. Int. J. Pharm. 2015, 494, 516–530. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourquin, C.; Anz, D.; Zwiorek, K.; Lanz, A.-L.; Fuchs, S.; Weigel, S.; Wurzenberger, C.; von der Borch, P.; Golic, M.; Moder, S.; et al. Targeting CpG Oligonucleotides to the Lymph Node by Nanoparticles Elicits Efficient Antitumoral Immunity. J. Immunol. 2008, 181, 2990–2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–760. [Google Scholar] [CrossRef]
- Verbon, E.H.; Post, J.A.; Boonstra, J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene 2012, 511, 1–6. [Google Scholar] [CrossRef]
- He, S.; Xia, T.; Wang, H.; Wei, L.; Luo, X.; Li, X. Multiple release of polyplexes of plasmids VEGF and bFGF from electrospun fibrous scaffolds towards regeneration of mature blood vessels. Acta Biomater. 2012, 8, 2659–2669. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequences (5′-3′) | PCR Conditions |
|---|---|---|
| GFP | F: CGTGTCTTCGCCAAGTAC R: CTTCATCATGGTGATGTCGT Probe:(Cy3)-CTCGTGGGAGC-(BHQ2) | 95 °C for 3 min, 35 cycles 95 °C for 40 s, and 60 °C for 60 s. |
| GAPDH | F:TGAAGGTCGGAGTCAACGGATTTGGT R: CATCGCCCCACTTGATTTTGGAGGG | 95 °C for 3 min, 40 cycles 95 °C for 20 s, 57 °C for 20 s, and 72 °C for 40 s. |
| 18S | F: AAACGGCTACCACATCCAAG R: CAATTACAGGGCCTCGAAAG | 95 °C for 3 min, 40 cycles 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 40 s. |
| Dynamic Light Scattering | TEM | p# | |
|---|---|---|---|
| Silica NPs | 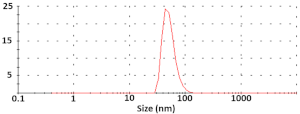 Diameter = 52 ± 5 nm ζ-potential = −2.26 ± 0.02 mV |  Size = 56.3 ± 14.6 nm | 0.138 |
| pDNA-NPs | 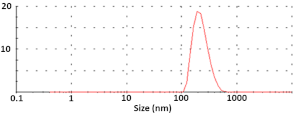 Diameter = 252 ± 45 nm ζ-potential = −10.7 ± 0.2 mV |  Size = 77.9 ± 17.1 nm | 0.008 |
| (pDNA-NPs) + PEI9 |  Diameter = 205 ± 73 nm ζ-potential = −4.55 ± 0.3 mV |  Size = 63.0 ± 9.7 nm | 0.001 |
| p1,2 = 0.006; p1,3 = 0.006; p2,3 = 0.447 | p1,2 < 0.001; p1,3 = 0.159; p2,3 = 0.099 |
| Scaffold | O | N | Si | P |
|---|---|---|---|---|
| M control | 331.1 | 26.3 | - | - |
| M with NPs | 347.2 | 30.1 | 5.4 | - |
| M with pDNA-NPs | 351.0 | 35.1 | 1.4 | - |
| M with (pDNA-NPs) + PEI9 | 344.4 | 38.3 | 3.4 | - |
| Scaffold | Higuchi | Zero-Order | First-Order | Korsmeyer–Peppas |
|---|---|---|---|---|
| M with pDNA-NPs | 0.9536 | 0.9536 | 0.8687 | 0.9715 |
| M with (pDNA-NPs) + PEI3 | 0.9730 | 0.9730 | 0.9330 | 0.9655 |
| M with (pDNA-NPs) + PEI9 | 0.6205 | 0.6205 | 0.4653 | 0.8253 |
| M with (pDNA-NPs) + PEI27 | 0.5705 | 0.5705 | 0.4206 | 0.7261 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernonosova, V.; Khlebnikova, M.; Popova, V.; Starostina, E.; Kiseleva, E.; Chelobanov, B.; Kvon, R.; Dmitrienko, E.; Laktionov, P. Electrospun Scaffolds Enriched with Nanoparticle-Associated DNA: General Properties, DNA Release and Cell Transfection. Polymers 2023, 15, 3202. https://doi.org/10.3390/polym15153202
Chernonosova V, Khlebnikova M, Popova V, Starostina E, Kiseleva E, Chelobanov B, Kvon R, Dmitrienko E, Laktionov P. Electrospun Scaffolds Enriched with Nanoparticle-Associated DNA: General Properties, DNA Release and Cell Transfection. Polymers. 2023; 15(15):3202. https://doi.org/10.3390/polym15153202
Chicago/Turabian StyleChernonosova, Vera, Marianna Khlebnikova, Victoriya Popova, Ekaterina Starostina, Elena Kiseleva, Boris Chelobanov, Ren Kvon, Elena Dmitrienko, and Pavel Laktionov. 2023. "Electrospun Scaffolds Enriched with Nanoparticle-Associated DNA: General Properties, DNA Release and Cell Transfection" Polymers 15, no. 15: 3202. https://doi.org/10.3390/polym15153202
APA StyleChernonosova, V., Khlebnikova, M., Popova, V., Starostina, E., Kiseleva, E., Chelobanov, B., Kvon, R., Dmitrienko, E., & Laktionov, P. (2023). Electrospun Scaffolds Enriched with Nanoparticle-Associated DNA: General Properties, DNA Release and Cell Transfection. Polymers, 15(15), 3202. https://doi.org/10.3390/polym15153202










