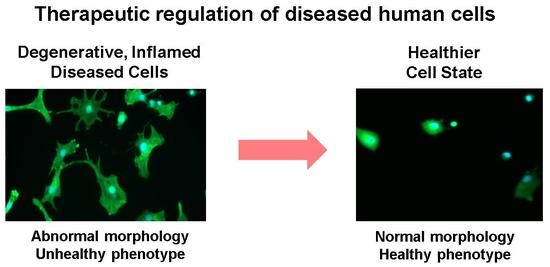
Therapeutic Modulation of Cell Morphology and Phenotype of Diseased Human Cells towards a Healthier Cell State Using Lignin
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of OSL
2.2. Isolation of Human OA Chondrocytes from Articular Cartilage and Treatment with OSL
2.3. Droplet Digital PCR for Absolute Quantification of Gene Expression
2.4. Cell Staining and Microscopy
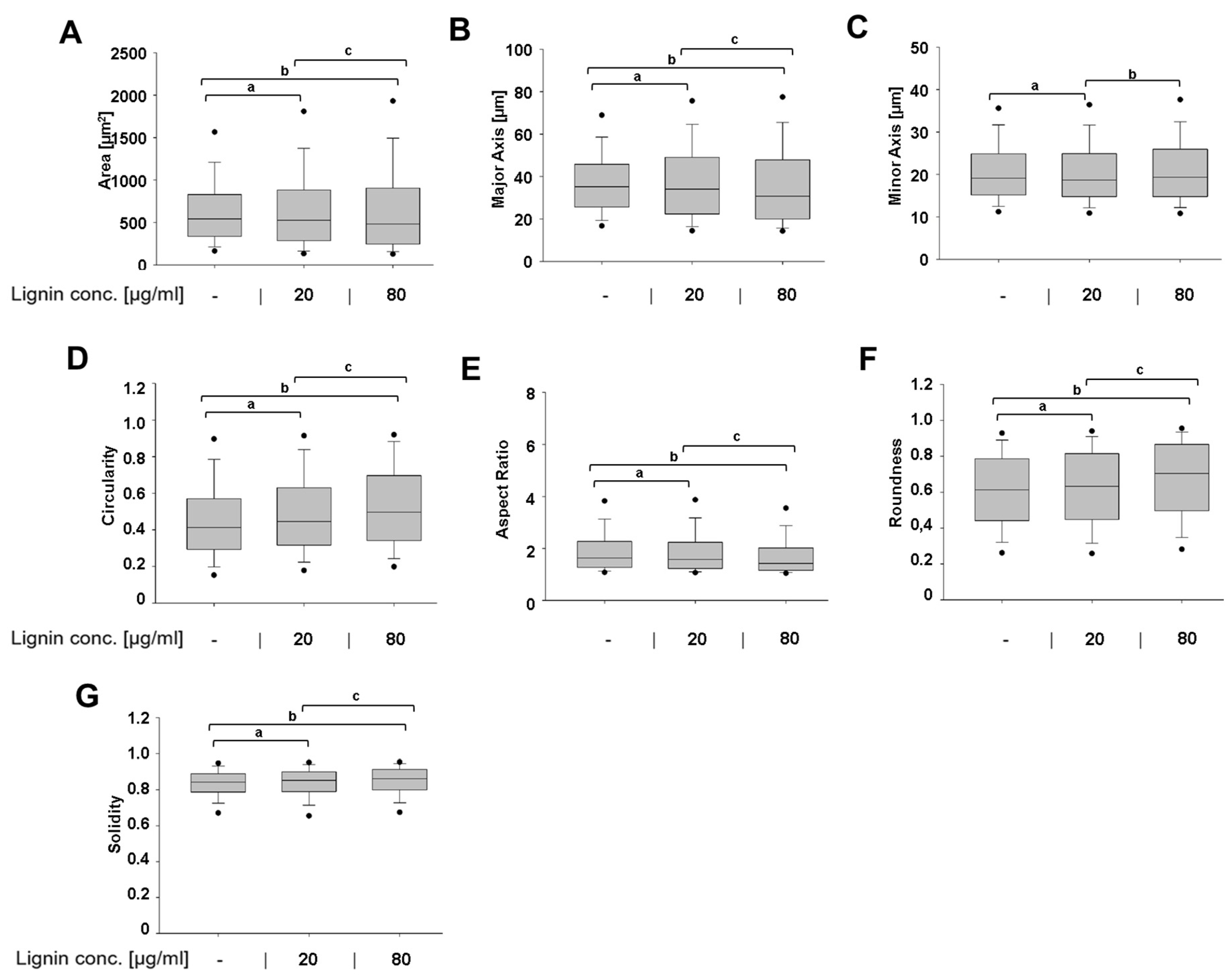
2.5. High-Throughput Quantitative Cell Morphometric Measurements Using a Panel of Cell Shape Descriptors
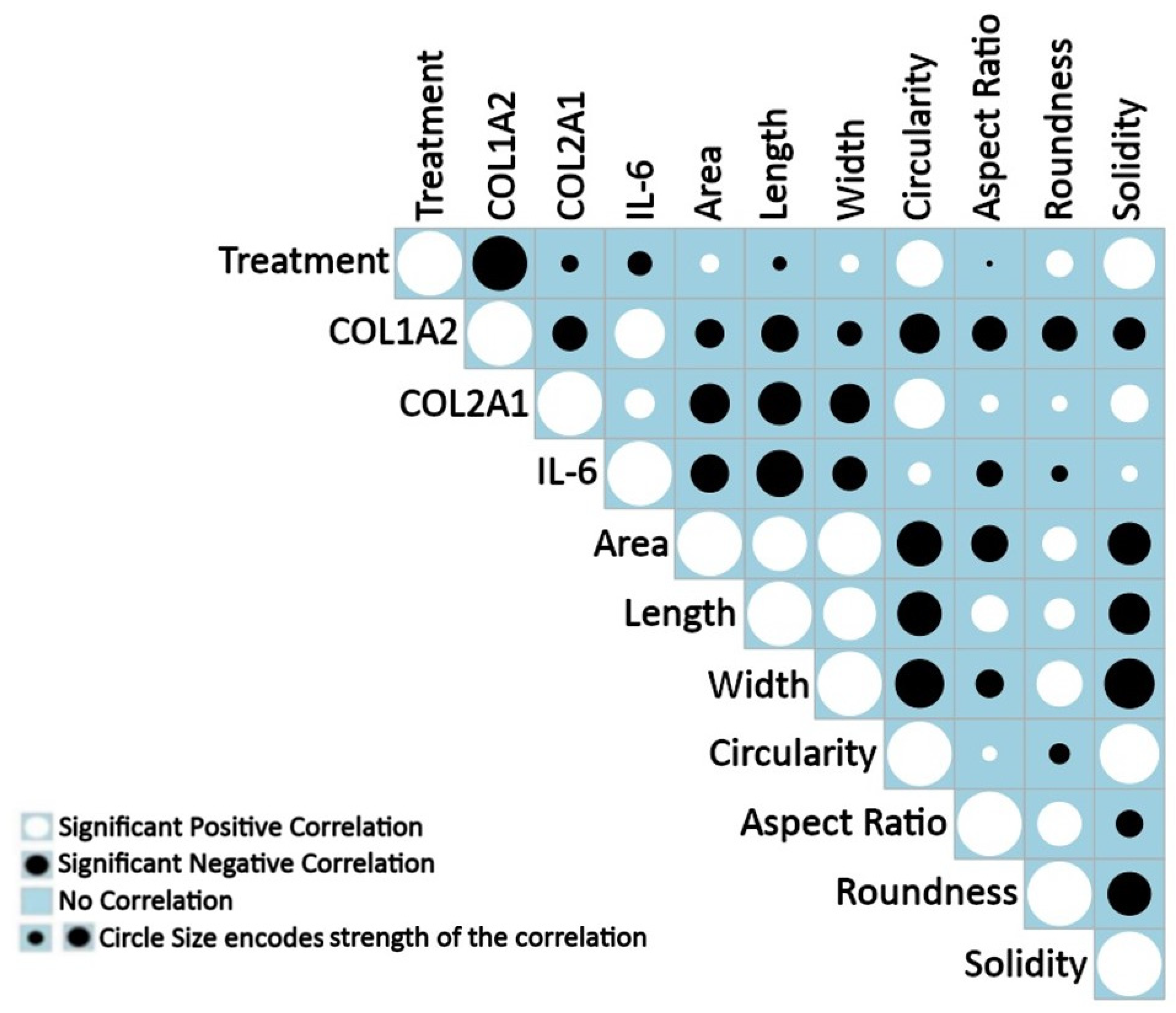
2.6. Correlation Analysis
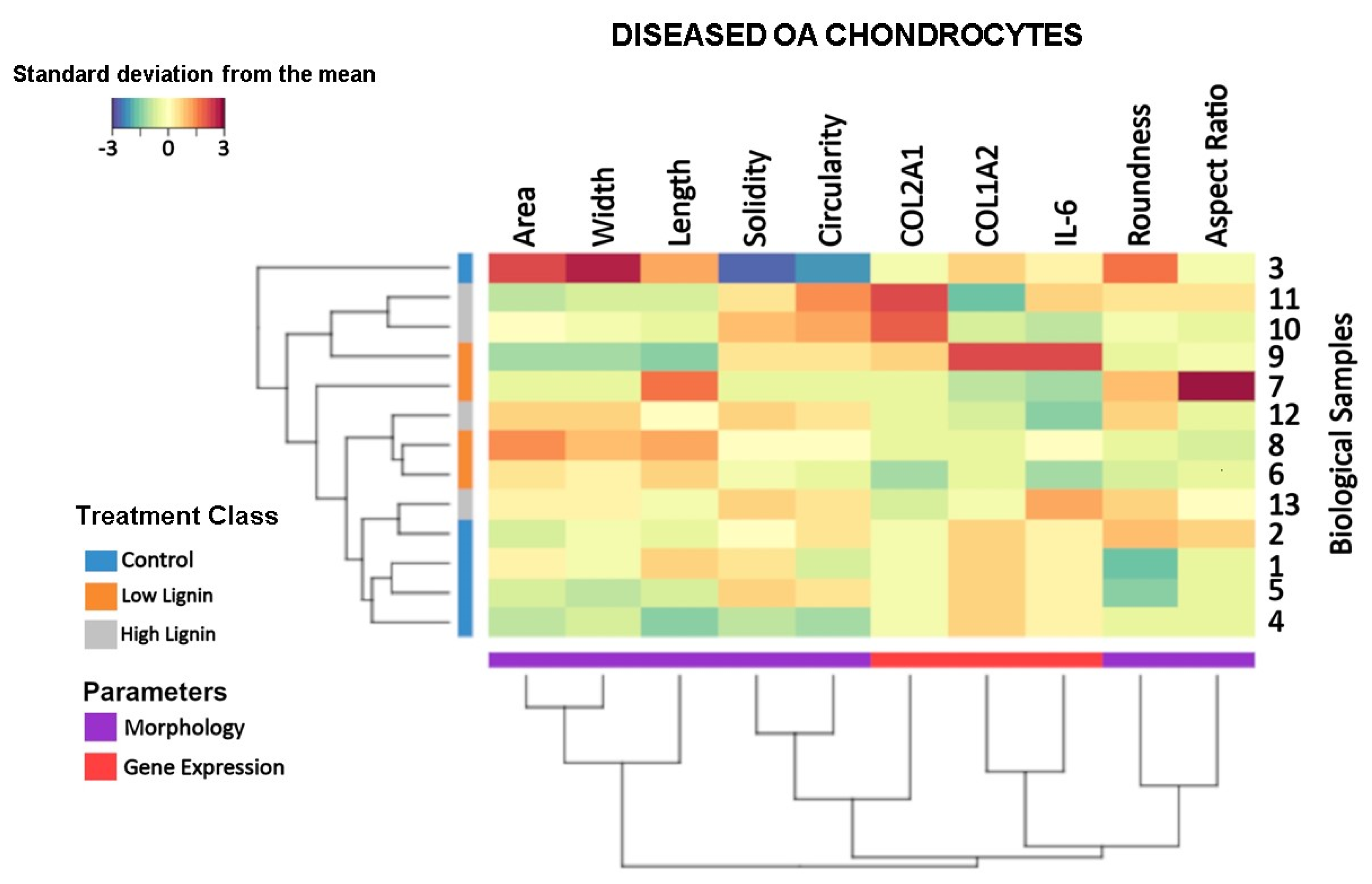
2.7. Clustered Image Map (CIM) Allowing Multi-Level Analyses
2.8. Statistical Analysis
3. Results
3.1. Lignin-Mediated Modulation of Gene Expression in Human Diseased Chondrocytes
3.2. Lignin-Mediated Modulation of a Diseased Cell Shape into a Healthier Cell Shape
3.3. Positive and Negative Correlations between Measured Features under Treatment
3.4. Lignin-Mediated Modulation of the Gene Expression and Cell Morphology at the Sample Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bajwaa, D.S.; Pourhashem, G.; Ullah, A.H.; Bajwac, S.G. A concise review of current lignin production, applications, products and their environmental impact. Indus. Crops Prod. 2019, 139, 111526. [Google Scholar] [CrossRef]
- Melro, E.; Filipe, A.; Sousa, D.; Medronho, B.; Romano, A. Revisiting lignin: A tour through its structural features, characterization methods and applications. New J. Chem. 2021, 45, 6986–7013. [Google Scholar] [CrossRef]
- Sugiarto, S.; Leow, Y.; Tan, C.L.; Wang, G.; Kai, D. How far is Lignin from being a biomedical material? Bioact. Mater. 2022, 8, 71–94. [Google Scholar] [CrossRef]
- Witzler, M.; Alzagameem, A.; Bergs, M.; Khaldi-Hansen, B.E.; Klein, S.E.; Hielscher, D.; Kamm, B.; Kreyenschmidt, J.; Tobiasch, E.; Schulze, M. Lignin-Derived Biomaterials for Drug Release and Tissue Engineering. Molecules 2018, 23, 1885. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Jin, K.; Buehler, M.J. Understanding Plant Biomass via Computational Modeling. Adv. Mater. 2021, 33, e2003206. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Ruggeberg, M.; Gierlinger, N.; Cesarino, I.; Vanholme, R.; Boerjan, W.; Burgert, I. Unravelling the impact of lignin on cell wall mechanics: A comprehensive study on young poplar trees downregulated for Cinnamyl Alcohol Dehydrogenase (CAD). Plant J. 2017, 91, 480–490. [Google Scholar] [CrossRef]
- Ozparpucu, M.; Gierlinger, N.; Cesarino, I.; Burgert, I.; Boerjan, W.; Ruggeberg, M. Significant influence of lignin on axial elastic modulus of poplar wood at low microfibril angles under wet conditions. J. Exp. Bot. 2019, 70, 4039–4047. [Google Scholar] [CrossRef]
- Domínguez-Roblesa, J.; Cárcamo-Martínez, A.; Stewart, S.A.; Donnelly, R.F.; Larrañeta, E.; Borrega, M. Lignin for pharmaceutical and biomedical applications—Could this become a reality? Sustain. Chem. Pharm. 2020, 18, 100320. [Google Scholar] [CrossRef]
- Vinardell, M.P.; Mitjans, M. Lignins and Their Derivatives with Beneficial Effects on Human Health. Int. J. Mol. Sci. 2017, 18, 1219. [Google Scholar] [CrossRef]
- Liu, R.; Dai, L.; Xu, C.; Wang, K.; Zheng, C.; Si, C. Lignin-Based Micro-and Nanomaterials and their Composites in Biomedical Applications. ChemSusChem 2020, 13, 4266–4283. [Google Scholar] [CrossRef]
- Hart, M.L.; Lauer, J.C.; Selig, M.; Hanak, M.; Walters, B.; Rolauffs, B. Shaping the Cell and the Future: Recent Advancements in Biophysical Aspects Relevant to Regenerative Medicine. J. Funct. Morphol. Kinesiol. 2018, 3, 2. [Google Scholar] [CrossRef]
- Lauer, J.C.; Selig, M.; Hart, M.L.; Kurz, B.; Rolauffs, B. Articular chondrocyte phenotype regulation through the cytoskeleton and the signaling processes that originate from or converge on the cytoskeleton: Towards a novel understanding of the intersection between actin dynamics and chondrogenic function. Int. J. Mol. Sci. 2021, 22, 3279. [Google Scholar] [CrossRef]
- Gleuwitz, F.R.; Sivasankarapillai, G.; Chen, Y.; Friedrich, C.; Laborie, M.G. Lignin-Assisted Stabilization of an Oriented Liquid Crystalline Cellulosic Mesophase, Part B: Toward the Molecular Origin and Mechanism. Biomacromolecules 2020, 21, 2276–2284. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Wang, G.; Sun, H.; Su, W.; Si, C. Lignin fractionation: Effective strategy to reduce molecule weight dependent heterogeneity. Indus. Crops Prod. 2021, 165, 113442. [Google Scholar] [CrossRef]
- Izaguirre, N.; Robles, E.; Llano-Ponte, R.; Labidi, J.; Erdociac, X. Fine-tune of lignin properties by its fractionation with a sequential organic solvent extraction. Indus. Crops Prod. 2022, 175, 114251. [Google Scholar] [CrossRef]
- Menima-Medzogo, J.A.; Walz, K.; Lauer, J.C.; Sivasankarapillai, G.; Gleuwitz, F.R.; Rolauffs, B.; Laborie, M.P.; Hart, M.L. Characterization and In Vitro Cytotoxicity Safety Screening of Fractionated Organosolv Lignin on Diverse Primary Human Cell Types Commonly Used in Tissue Engineering. Biology 2022, 11, 696. [Google Scholar] [CrossRef]
- Saluja, B.; Thakkar, J.N.; Li, H.; Desai, U.R.; Sakagami, M. Novel low molecular weight lignins as potential anti-emphysema agents: In vitro triple inhibitory activity against elastase, oxidation and inflammation. Pulm. Pharmacol. Ther. 2013, 26, 296–304. [Google Scholar] [CrossRef]
- Brown, T.D.; Johnston, R.C.; Saltzman, C.L.; Marsh, J.L.; Buckwalter, J.A. Posttraumatic osteoarthritis: A first estimate of incidence, prevalence, and burden of disease. J. Orthop. Trauma 2006, 20, 739–744. [Google Scholar] [CrossRef]
- Herzog, W.; Federico, S. Considerations on joint and articular cartilage mechanics. Biomech. Model. Mechanobiol. 2006, 5, 64–81. [Google Scholar] [CrossRef]
- Khella, C.M.; Asgarian, R.; Horvath, J.M.; Rolauffs, B.; Hart, M.L. An evidence-based systematic review of human knee post-traumatic osteoarthritis (PTOA): Timeline of clinical presentation and disease markers, comparison of knee joint PTOA models and early disease implications. Int. J. Mol. Sci. 2021, 22, 1996. [Google Scholar] [CrossRef]
- Armiento, A.R.; Alini, M.; Stoddart, M.J. Articular fibrocartilage—Why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019, 146, 289–305. [Google Scholar] [CrossRef]
- Hunziker, E.B. Articular cartilage repair: Are the intrinsic biological constraints undermining this process insuperable? Osteoarthr. Cartil. 1999, 7, 15–28. [Google Scholar] [CrossRef]
- Selig, M.; Azizi, S.; Walz, K.; Lauer, J.C.; Rolauffs, B.; Hart, M.L. Cell morphology as a biological fingerprint of chondrocyte phenotype in control and inflammatory conditions. Front. Immunol. 2023, 14, 1102912. [Google Scholar] [CrossRef] [PubMed]
- Kubo, S.; Uraki, Y.; Sano, Y. Preparation of carbon fibers from softwood lignin by atmospheric acetic acid pulping. Carbon 1998, 36, 1119–1124. [Google Scholar] [CrossRef]
- Felka, T.; Rothdiener, M.; Bast, S.; Uynuk-Ool, T.; Zouhair, S.; Ochs, B.G.; De Zwart, P.; Stoeckle, U.; Aicher, W.K.; Hart, M.L.; et al. Loss of spatial organization and destruction of the pericellular matrix in early osteoarthritis in vivo and in a novel in vitro methodology. Osteoarthr. Cartil. 2016, 24, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Rolauffs, B.; Williams, J.M.; Aurich, M.; Grodzinsky, A.J.; Kuettner, K.E.; Cole, A.A. Proliferative remodeling of the spatial organization of human superficial chondrocytes distant from focal early osteoarthritis. Arthritis Rheum. 2010, 62, 489–498. [Google Scholar] [CrossRef]
- Aurich, M.; Hofmann, G.O.; Best, N.; Rolauffs, B. Induced Redifferentiation of Human Chondrocytes from Articular Cartilage Lesion in Alginate Bead Culture After Monolayer Dedifferentiation: An Alternative Cell Source for Cell-Based Therapies? Tissue Eng. Part A 2018, 24, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Barisic, D.; Erb, M.; Follo, M.; Al-Mudaris, D.; Rolauffs, B.; Hart, M.L. Lack of a skeletal muscle phenotype in adult human bone marrow stromal cells following xenogeneic-free expansion. Stem Cell Res. Ther. 2020, 11, 79. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.J. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.J. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H.J.B. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef]
- Walters, B.; Turner, P.A.; Rolauffs, B.; Hart, M.L.; Stegemann, J.P. Controlled Growth Factor Delivery and Cyclic Stretch Induces a Smooth Muscle Cell-like Phenotype in Adipose-Derived Stem Cells. Cells 2021, 10, 3123. [Google Scholar] [CrossRef]
- Uynuk-Ool, T.; Rothdiener, M.; Walters, B.; Hegemann, M.; Palm, J.; Nguyen, P.; Seeger, T.; Stockle, U.; Stegemann, J.P.; Aicher, W.K.; et al. The geometrical shape of mesenchymal stromal cells measured by quantitative shape descriptors is determined by the stiffness of the biomaterial and by cyclic tensile forces. J. Tissue Eng. Regen. Med. 2017, 11, 3508–3522. [Google Scholar] [CrossRef]
- Rabel, K.; Kohal, R.J.; Steinberg, T.; Tomakidi, P.; Rolauffs, B.; Adolfsson, E.; Palmero, P.; Furderer, T.; Altmann, B. Controlling osteoblast morphology and proliferation via surface micro-topographies of implant biomaterials. Sci. Rep. 2020, 10, 12810. [Google Scholar] [CrossRef]
- Walters, B.; Uynuk-Ool, T.; Rothdiener, M.; Palm, J.; Hart, M.L.; Stegemann, J.P.; Rolauffs, B. Engineering the geometrical shape of mesenchymal stromal cells through defined cyclic stretch regimens. Sci. Rep. 2017, 7, 6640. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013; pp. 275–286. [Google Scholar]
- Harrell, F.E., Jr.; Harrell, M.F.E.J.C., Jr. Package ‘hmisc’. R Package Version 4.2-0. CRAN2018 2019, 2019, 235–236. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J.J.S. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). Statistician 2017, 56, e24. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Welham, Z.; Dejean, S.; Le Cao, K.A. Multivariate Analysis with the R Package mixOmics. Methods Mol. Biol. 2023, 2426, 333–359. [Google Scholar] [CrossRef]
- Hall, A.C. The Role of Chondrocyte Morphology and Volume in Controlling Phenotype-Implications for Osteoarthritis, Cartilage Repair, and Cartilage Engineering. Curr. Rheumatol. Rep. 2019, 21, 38. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, M.; Zhang, Y.; Cao, Q.; Wang, X.; Han, Y.; Sun, G.; Li, Y.; Zhou, J. Novel lignin-chitosan-PVA composite hydrogel for wound dressing. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 104, 110002. [Google Scholar] [CrossRef]
- Belgodere, J.A.; Zamin, S.A.; Kalinoski, R.M.; Astete, C.E.; Penrod, J.C.; Hamel, K.M.; Lynn, B.C.; Rudra, J.S.; Shi, J.; Jung, J.P. Modulating Mechanical Properties of Collagen-Lignin Composites. ACS Appl. Bio. Mater. 2019, 2, 3562–3572. [Google Scholar] [CrossRef] [PubMed]
- Eivazzadeh-Keihan, R.; Moghim Aliabadi, H.A.; Radinekiyan, F.; Sobhani, M.; Farzane, K.; Maleki, A.; Madanchi, H.; Mahdavi, M.; Shalan, A.E. Investigation of the biological activity, mechanical properties and wound healing application of a novel scaffold based on lignin-agarose hydrogel and silk fibroin embedded zinc chromite nanoparticles. RSC Adv. 2021, 11, 17914–17923. [Google Scholar] [CrossRef]
- Silva-Barroso, A.S.; Cabral, C.S.D.; Ferreira, P.; Moreira, A.F.; Correia, I.J. Lignin-enriched tricalcium phosphate/sodium alginate 3D scaffolds for application in bone tissue regeneration. Int. J. Biol. Macromol. 2023, 239, 124258. [Google Scholar] [CrossRef] [PubMed]
- Polo, C.C.; Pereira, L.; Mazzafera, P.; Flores-Borges, D.N.A.; Mayer, J.L.S.; Guizar-Sicairos, M.; Holler, M.; Barsi-Andreeta, M.; Westfahl, H., Jr.; Meneau, F. Correlations between lignin content and structural robustness in plants revealed by X-ray ptychography. Sci. Rep. 2020, 10, 6023. [Google Scholar] [CrossRef] [PubMed]
- Tschaikowsky, M.; Selig, M.; Brander, S.; Balzer, B.N.; Hugel, T.; Rolauffs, B.J.O. Proof-of-concept for the detection of early osteoarthritis pathology by clinically applicable endomicroscopy and quantitative AI-supported optical biopsy. Osteoarthr. Cartil. 2021, 29, 269–279. [Google Scholar] [CrossRef]
- Selig, M.; Lauer, J.C.; Hart, M.L.; Rolauffs, B.J. Mechanotransduction and stiffness-sensing: Mechanisms and opportunities to control multiple molecular aspects of cell phenotype as a design cornerstone of cell-instructive biomaterials for articular cartilage repair. Int. J. Mol. Sci. 2020, 21, 5399. [Google Scholar] [CrossRef]
- Ramirez, F.; Tanaka, S.; Bou-Gharios, G. Transcriptional regulation of the human alpha2(I) collagen gene (COL1A2), an informative model system to study fibrotic diseases. Matrix Biol. J. Int. Soc. Matrix Biol. 2006, 25, 365–372. [Google Scholar] [CrossRef]
- Van Kempen, L.C.; Rijntjes, J.; Mamor-Cornelissen, I.; Vincent-Naulleau, S.; Gerritsen, M.J.; Ruiter, D.J.; van Dijk, M.C.; Geffrotin, C.; van Muijen, G.N. Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int. J. Cancer 2008, 122, 1019–1029. [Google Scholar] [CrossRef]
- Liang, Y.; Diehn, M.; Bollen, A.W.; Israel, M.A.; Gupta, N. Type I collagen is overexpressed in medulloblastoma as a component of tumor microenvironment. J. Neurooncol. 2008, 86, 133–141. [Google Scholar] [CrossRef]
- Li, J.; Ding, Y.; Li, A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J. Surg. Oncol. 2016, 14, 297. [Google Scholar] [CrossRef] [PubMed]
- Gabay, C. Interleukin-6 and chronic inflammation. Arthritis Res. Ther. 2006, 8, S3. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selig, M.; Walz, K.; Lauer, J.C.; Rolauffs, B.; Hart, M.L. Therapeutic Modulation of Cell Morphology and Phenotype of Diseased Human Cells towards a Healthier Cell State Using Lignin. Polymers 2023, 15, 3041. https://doi.org/10.3390/polym15143041
Selig M, Walz K, Lauer JC, Rolauffs B, Hart ML. Therapeutic Modulation of Cell Morphology and Phenotype of Diseased Human Cells towards a Healthier Cell State Using Lignin. Polymers. 2023; 15(14):3041. https://doi.org/10.3390/polym15143041
Chicago/Turabian StyleSelig, Mischa, Kathrin Walz, Jasmin C. Lauer, Bernd Rolauffs, and Melanie L. Hart. 2023. "Therapeutic Modulation of Cell Morphology and Phenotype of Diseased Human Cells towards a Healthier Cell State Using Lignin" Polymers 15, no. 14: 3041. https://doi.org/10.3390/polym15143041
APA StyleSelig, M., Walz, K., Lauer, J. C., Rolauffs, B., & Hart, M. L. (2023). Therapeutic Modulation of Cell Morphology and Phenotype of Diseased Human Cells towards a Healthier Cell State Using Lignin. Polymers, 15(14), 3041. https://doi.org/10.3390/polym15143041