Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation of Bacteria
2.2. Screening the Isolates for PHB Production
Screening by the Sudan Black B Method
2.3. Screening by Nile Blue A Method
2.4. Molecular Identification of PHB-Accumulating Bacteria by 16S rRNA Gene Analysis
2.5. Amplification of phbB Gene in Potent Isolates
2.6. Production, Detection, and Extraction of PHB
2.7. Quantitative Analysis of PHB
2.8. Characterization of PHB
2.9. Optimization of PHB Production
2.10. Optimization of PHB Production by Response Surface Methodology
2.11. Statistical Analysis
3. Results
3.1. Isolation, Screening, and Selection of PHB-Producing Bacteria
3.2. Identification of High-PHB-Producing Bacterial Isolate Using the 16S rRNA Gene and Its Phylogenetic Analysis
3.3. Amplification of phbB Gene
3.4. Characterization of PHB
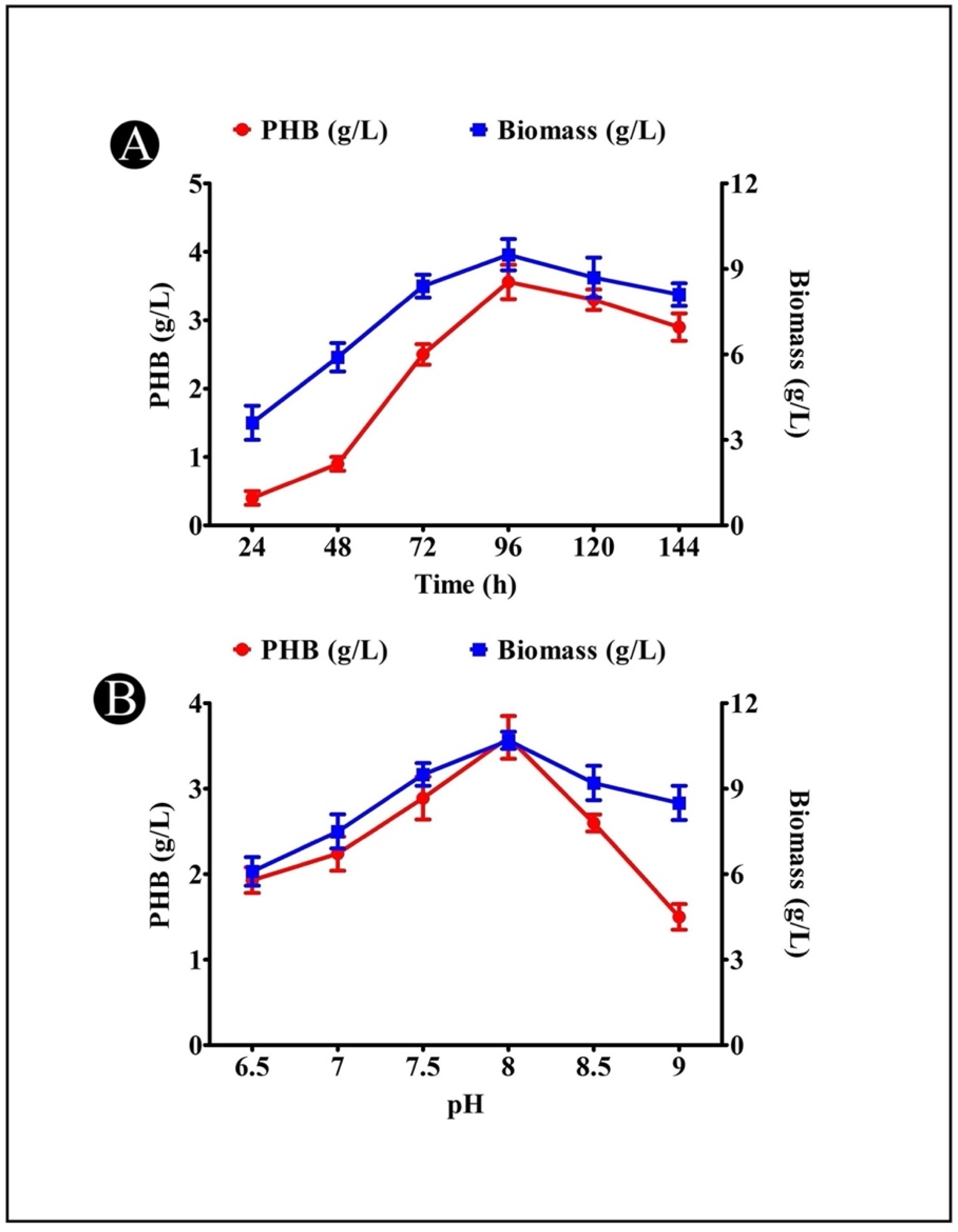
3.5. Effect of Incubation Time on PHB Production
3.6. Effect of Medium pH on PHB Production
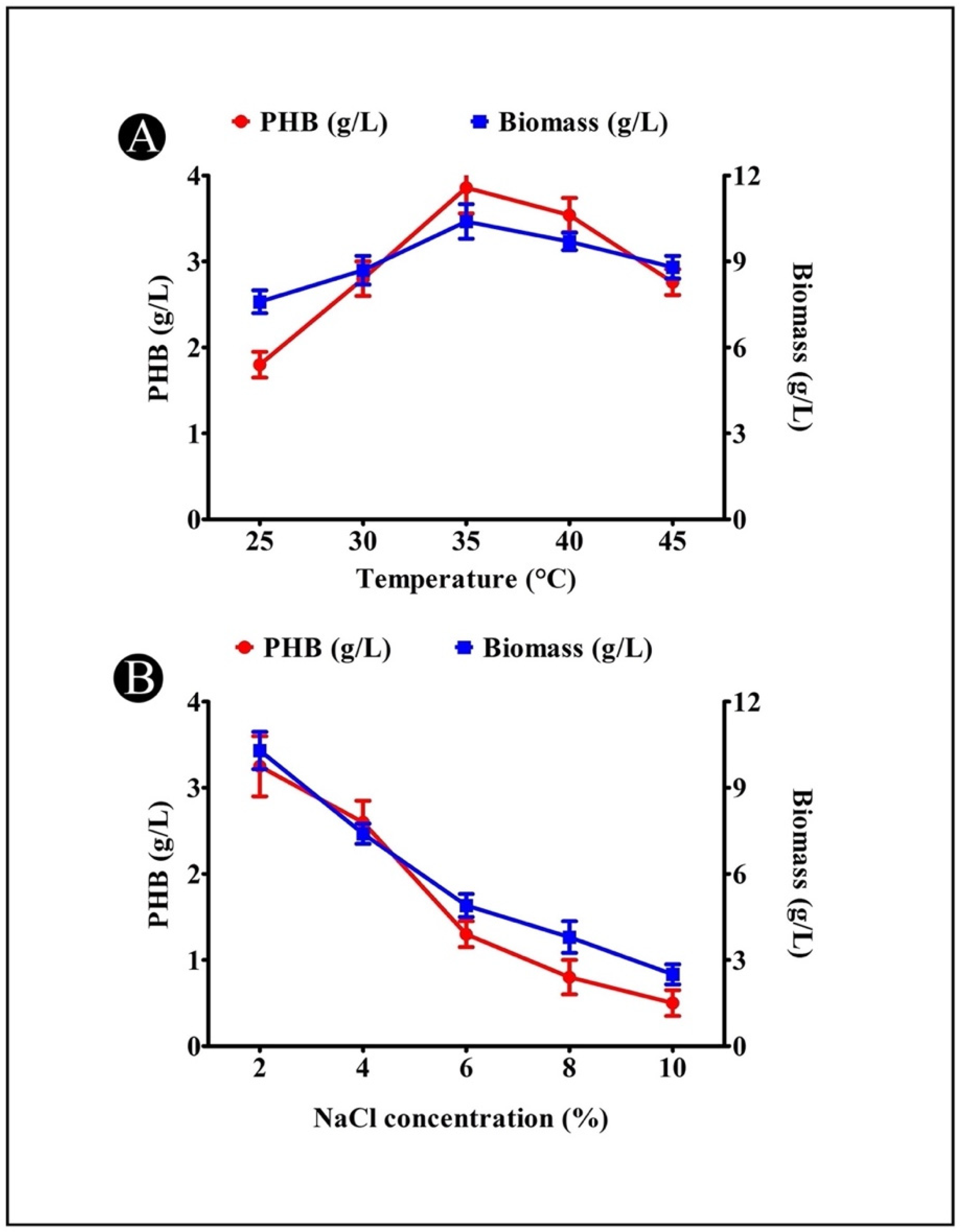
3.7. Effect of Temperature on PHB Production
3.8. Effect of NaCl Concentration on PHB Production
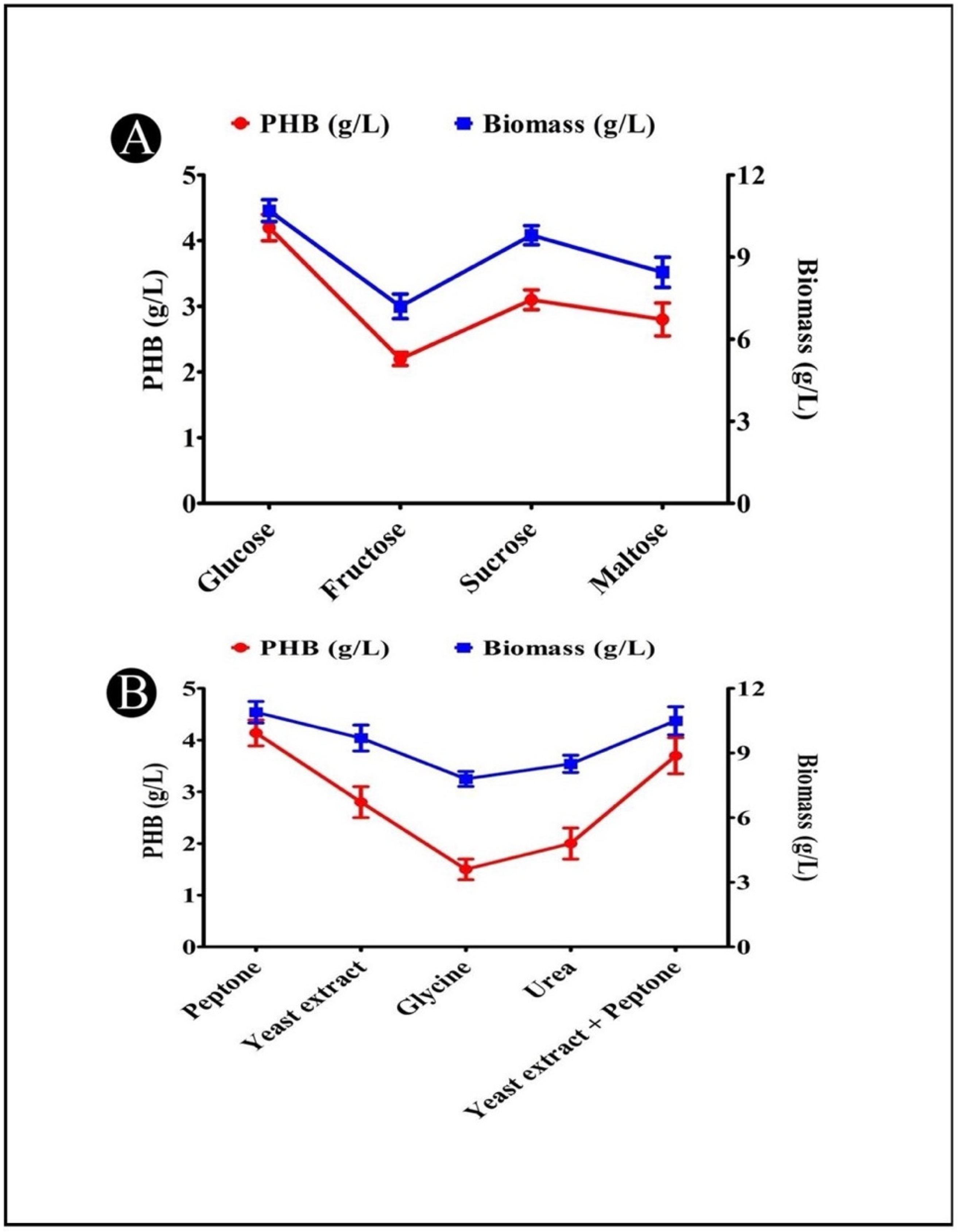
3.9. Effects of Different Carbon Sources on PHB Production
3.10. Effects of Different Nitrogen Sources on PHB Production
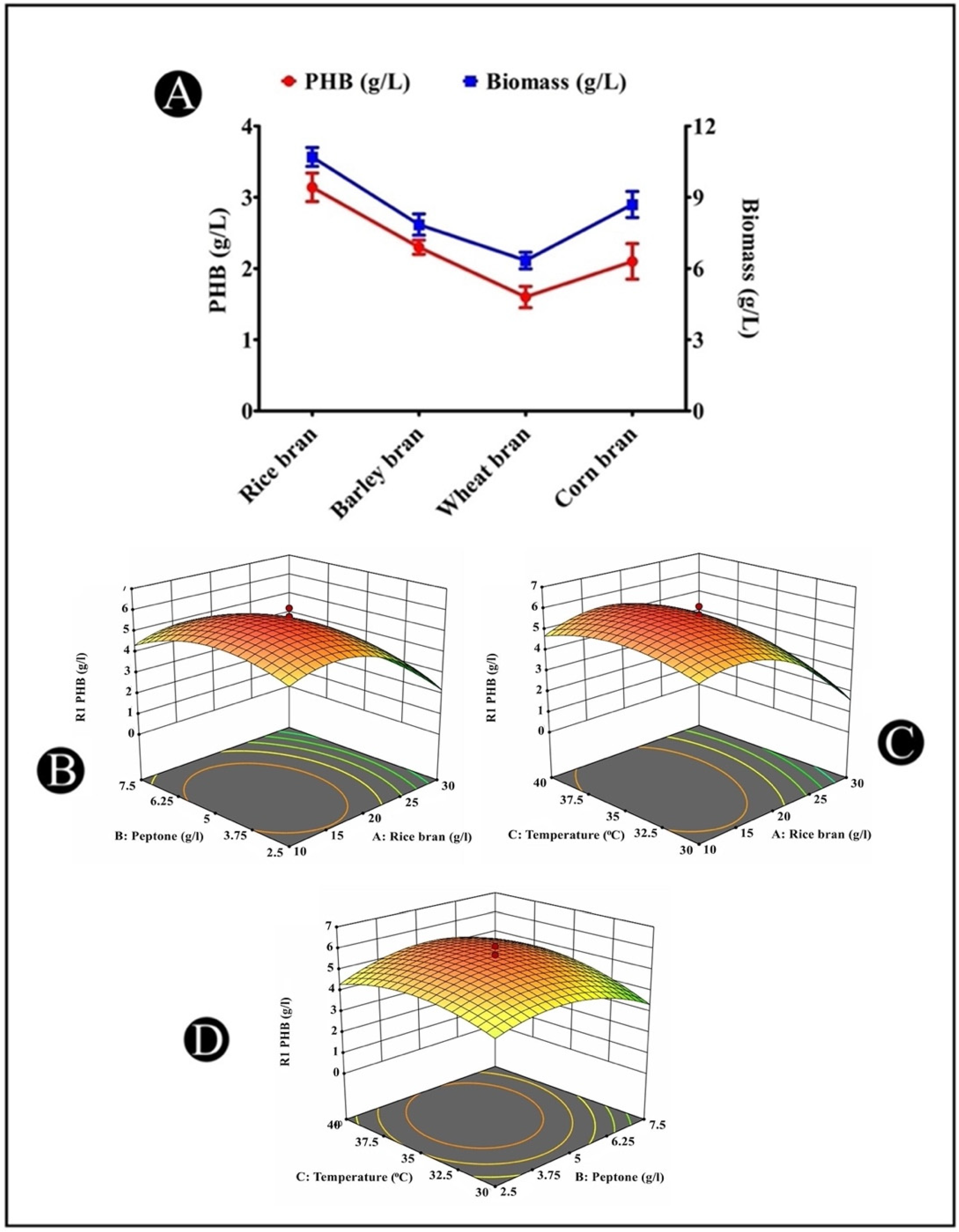
3.11. Production of PHB from Different Agricultural Wastes
3.12. Optimization of PHB Accumulation by Response Surface Methodology
4. Discussion
5. Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Horton, A.A. Plastic Pollution: When Do We Know Enough? J. Hazard. Mater. 2022, 422, 126885. [Google Scholar] [CrossRef]
- Getachew, A.; Woldesenbet, F. Production of biodegradable plastic by polyhydroxybutyrate (PHB) accumulating bacteria using low cost agricultural waste material. BMC Res. Notes 2016, 9, 509. [Google Scholar] [CrossRef]
- Wu, C.-S. Characterization and biodegradability of polyester bioplastic-based green renewable composites from agricultural residues. Polym. Degrad. Stab. 2012, 97, 64–71. [Google Scholar] [CrossRef]
- Agustin, M.; Ahmmad, B.; Alonzo, S.M.M.; Patriana, F.M. Bioplastic based on starch and cellulose nanocrystals from rice straw. J. Reinf. Plast. Compos. 2014, 33, 2205–2213. [Google Scholar] [CrossRef]
- Timmis, K.N.; McGenity, T.; Van Der Meer, J.R.; de Lorenzo, V. Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin, Germany, 2010; ISBN 3540775870. [Google Scholar]
- Galia, M.B. Isolation and Analysis of Storage Compounds. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin, Germany, 2010; ISBN 3540775846. [Google Scholar]
- Verlinden, R.A.J.; Hill, D.J.; Kenward, M.A.; Williams, C.D.; Radecka, I. Bacterial Synthesis of Biodegradable Polyhydroxyal-kanoates. J. Appl. Microbiol. 2007, 102, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Gurieff, N.; Lant, P. Comparative life cycle assessment and financial analysis of mixed culture polyhydroxyalkanoate production. Bioresour. Technol. 2007, 98, 3393–3403. [Google Scholar] [CrossRef]
- Yu, P.H.; Huang, A.L.; Lo, W.; Chua, H.; Chen, G.Q. Conversion of Food Industrial Wastes into Bioplastics. Appl. Biochem. Biotechnol. 1998, 70, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.A.M.; Serafim, L.S.; Lemos, P.C.; Ramos, A.M.; Aguiar, F.R.; Van Loosdrecht, M.C.M. Production of Polyhydroxyal-kanoates by Mixed Microbial Cultures. Bioprocess Biosyst. Eng. 2003, 25, 377–385. [Google Scholar] [CrossRef]
- Shrivastav, A.; Mishra, S.K.; Shethia, B.; Pancha, I.; Jain, D.; Mishra, S. Isolation of promising bacterial strains from soil and marine environment for polyhydroxyalkanoates (PHAs) production utilizing Jatropha biodiesel byproduct. Int. J. Biol. Macromol. 2010, 47, 283–287. [Google Scholar] [CrossRef]
- Mahitha, G.; Madhuri, R. Purification and Characterization of Polyhydroxybutyrate Produced from Marine Bacteria. Int. J. Sci. Eng. Res. 2015, 6, 71–75. [Google Scholar]
- Bhagowati, P.; Pradhan, S.; Dash, H.R.; Das, S. Production, Optimization and Characterization of Polyhydroxybutyrate, a Biodegradable Plastic by Bacillus spp. Biosci. Biotechnol. Biochem. 2015, 79, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbour, NY, USA, 1989; Volume 9, pp. 31–57. [Google Scholar]
- Patel, M.; Siddiqui, A.J.; Hamadou, W.S.; Surti, M.; Awadelkareem, A.M.; Ashraf, S.A.; Alreshidi, M.; Snoussi, M.; Rizvi, S.M.D.; Bardakci, F.; et al. Inhibition of Bacterial Adhesion and Antibiofilm Activities of a Glycolipid Biosurfactant from Lactobacillus rhamnosus with Its Physicochemical and Functional Properties. Antibiotics 2021, 10, 1546. [Google Scholar] [CrossRef]
- Galehdari, H.; Alaee, S.; Mirzaee, M. Cloning of Poly (3-Hydroxybutyrate) Synthesis Genes from Azotobacter Vinelandii into Escherichia Coli. Jundishapur J. Microbiol. 2009, 2, 31–35. [Google Scholar]
- Mercan, N.; Aslim, B.; Yüksekdağ, Z.N.U.R.; Beyatli, Y. Production of Poly-b-Hydroxybutyrate (PHB) by Some Rhizobium Bacteria. Turk. J. Biol. 2002, 26, 215–219. [Google Scholar]
- Paul, S.; Sasikumar, S.C.; Balakumaran, M.D. Optimization, Purification and Characterization of Polyhydroxybutyrate (PHB) Produced by Bacillus Cereus Isolated from Sewage. Int. J. Chem. Technol. Res. 2017, 10, 884–904. [Google Scholar]
- Bhuwal, A.K.; Singh, G.; Aggarwal, N.K.; Goyal, V.; Yadav, A. Isolation and Screening of Polyhydroxyalkanoates Producing Bacteria from Pulp, Paper, and Cardboard Industry Wastes. Int. J. Biomater. 2013, 2013, 752821. [Google Scholar] [CrossRef]
- Mostafa, Y.S.; Alrumman, S.A.; Alamri, S.A.; Otaif, K.A.; Mostafa, M.S.; Alfaify, A.M. Bioplastic (Poly-3-Hydroxybutyrate) Production by the Marine Bacterium Pseudodonghicola Xiamenensis through Date Syrup Valorization and Structural As-sessment of the Biopolymer. Sci. Rep. 2020, 10, 8815. [Google Scholar] [CrossRef]
- Mostafa, Y.S.; Alrumman, S.A.; Otaif, K.A.; Alamri, S.A.; Mostafa, M.S.; Sahlabji, T. Production and Characterization of Bioplastic by Polyhydroxybutyrate Accumulating Erythrobacter aquimaris Isolated from Mangrove Rhizosphere. Molecules 2020, 25, 179. [Google Scholar] [CrossRef]
- Varghese, S.; Dhanraj, N.; Rebello, S.; Sindhu, R.; Binod, P.; Pandey, A.; Jisha, M.; Awasthi, M.K. Leads and hurdles to sustainable microbial bioplastic production. Chemosphere 2022, 305, 135390. [Google Scholar] [CrossRef] [PubMed]
- Rosenboom, J.-G.; Langer, R.; Traverso, G. Bioplastics for a circular economy. Nat. Rev. Mater. 2022, 7, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Doi, Y. Biosynthesis of Polyhydroxyalkanaotes by a Novel Facultatively Anaerobic Vibrio sp. under Marine Conditions. Mar. Biotechnol. 2012, 14, 323–331. [Google Scholar] [CrossRef]
- Palit, K.; Rath, S.; Chatterjee, S.; Das, S. Microbial diversity and ecological interactions of microorganisms in the mangrove ecosystem: Threats, vulnerability, and adaptations. Environ. Sci. Pollut. Res. 2022, 29, 32467–32512. [Google Scholar] [CrossRef] [PubMed]
- Kalaivani, R.; Sukumaran, V. Isolation and Identification of New Strains to Enhance the Production of Biopolymers from Marine Sample in Karankura, Tamil Nadu. Eur. J. Exp. Biol. 2013, 3, 56–64. [Google Scholar]
- Burdon, K.L. Fatty Material in Bacteria and Fungi Revealed by Staining Dried, Fixed Slide Preparations. J. Bacteriol. 1946, 52, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Lorch, E.; Rentsch, G. A simple method for staining and counting isolated adipose tissue fat cells. Diabetologia 1969, 5, 356–357. [Google Scholar] [CrossRef][Green Version]
- Thompson, L.R.; Williams, G.J.; Haroon, M.F.; Shibl, A.; Larsen, P.; Shorenstein, J.; Knight, R.; Stingl, U. Metagenomic Co-variation along Densely Sampled Environmental Gradients in the Red Sea. ISME J. 2017, 11, 138–151. [Google Scholar] [CrossRef]
- Aarthi, N.; Ramana, K.V. Identification and Characterization of Polyhydroxybutyrate Producing Bacillus Cereus and Bacillus Mycoides Strains. Int. J. Environ. Sci. 2011, 1, 744. [Google Scholar]
- Alarfaj, A.A.; Arshad, M.; Sholkamy, E.N.; Munusamy, M.A. Extraction and Characterization of Polyhydroxybutyrates (PHB) from Bacillus thuringiensisKSADL127 Isolated from Mangrove Environments of Saudi Arabia. Braz. Arch. Biol. Technol. 2015, 58, 781–788. [Google Scholar] [CrossRef]
- Peoples, O.P.; Sinskey, A.J. Poly-β-Hydroxybutyrate (PHB) Biosynthesis in Alcaligenes Eutrophus H16: Identification and Characterization of the PHB Polymerase Gene (PhbC). J. Biol. Chem. 1989, 264, 15298–15303. [Google Scholar] [CrossRef]
- Yang, M.K.; Lin, Y.C.; Shen, C.H. Identification of two gene loci involved in poly-beta-hydroxybutyrate production in Rhodobacter sphaeroides FJ1. J. Microbiol. Immunol. Infect. 2006, 39, 18–27. [Google Scholar]
- Pettinari, M.J.; Vázquez, G.J.; Silberschmidt, D.; Rehm, B.; Steinbuchel, A.; Méndez, B.S. Poly(3-Hydroxybutyrate) Synthesis Genes in Azotobacter Sp. Strain FA8. Appl. Environ. Microbiol. 2001, 67, 5331–5334. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.; Burdman, S.; Jurkevitch, E.; Okon, Y. Identification and Isolation of Genes Involved in Poly(β-Hydroxybutyrate) Biosynthesis in Azospirillum brasilense and Characterization of a phbC Mutant. Appl. Environ. Microbiol. 2002, 68, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Aneja, P.; Dai, M.; Lacorre, D.A.; Pillon, B.; Charles, T.C. Heterologous complementation of the exopolysaccharide synthesis and carbon utilization phenotypes of Sinorhizobium meliloti Rm1021 polyhydroxyalkanoate synthesis mutants. FEMS Microbiol. Lett. 2004, 239, 277–283. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Production of polyhydroxyalkanoates (PHAs) by Bacillus subtilis and Escherichia coli grown on cane molasses fortified with ethanol. Braz. Arch. Biol. Technol. 2014, 57, 145–154. [Google Scholar] [CrossRef]
- Belal, E.B. Production of Poly-β-Hydroxybutyric Acid (PHB) by Rhizobium Elti and Pseudomonas Stutzeri. Curr. Res. J. Biol. Sci. 2013, 5, 228–273. [Google Scholar]
- Lathwal, P.; Nehra, K.; Singh, M.; Jamdagni, P.; Rana, J.S. Optimization of Culture Parameters for Maximum Polyhydroxy-butyrate Production by Selected Bacterial Strains Isolated from Rhizospheric Soils. Pol. J. Microbiol. 2015, 64, 7. [Google Scholar] [CrossRef]
- Sathiyanarayanan, G.; Saibaba, G.; Kiran, G.S.; Selvin, J. A statistical approach for optimization of polyhydroxybutyrate production by marine Bacillus subtilis MSBN17. Int. J. Biol. Macromol. 2013, 59, 170–177. [Google Scholar] [CrossRef]
- Dhangdhariya, J.H.; Dubey, S.; Trivedi, H.B.; Pancha, I.; Bhatt, J.K.; Dave, B.P.; Mishra, S. Polyhydroxyalkanoate from marine Bacillus megaterium using CSMCRI’s Dry Sea Mix as a novel growth medium. Int. J. Biol. Macromol. 2015, 76, 254–261. [Google Scholar] [CrossRef]
- Omar, S.; Rayes, A.; Eqaab, A.; Voß, I.; Steinbüchel, A. Optimization of Cell Growth and Poly (3-Hydroxybutyrate) Accumu-lation on Date Syrup by a Bacillus Megaterium Strain. Biotechnol. Lett. 2001, 23, 1119–1123. [Google Scholar] [CrossRef]
- Mohandas, S.P.; Balan, L.; Lekshmi, N.; Cubelio, S.S.; Philip, R.; Bright Singh, I.S. Production and Characterization of Poly-hydroxybutyrate from Vibrio Harveyi MCCB 284 Utilizing Glycerol as Carbon Source. J. Appl. Microbiol. 2017, 122, 698–707. [Google Scholar]
- Sasidharan, R.S.; Bhat, S.G.; Chandrasekaran, M. Biocompatible polyhydroxybutyrate (PHB) production by marine Vibrio azureus BTKB33 under submerged fermentation. Ann. Microbiol. 2015, 65, 455–465. [Google Scholar] [CrossRef]
- Hong, J.-W.; Song, H.-S.; Moon, Y.-M.; Hong, Y.-G.; Bhatia, S.K.; Jung, H.-R.; Choi, T.-R.; Yang, S.-Y.; Park, H.-Y.; Choi, Y.-K.; et al. Polyhydroxybutyrate production in halophilic marine bacteria Vibrio proteolyticus isolated from the Korean peninsula. Bioprocess Biosyst. Eng. 2019, 42, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Al-Battashi, H.; Annamalai, N.; Al-Kindi, S.; Nair, A.S.; Al-Bahry, S.; Verma, J.P.; Sivakumar, N. Production of Bioplastic (Poly-3-Hydroxybutyrate) Using Waste Paper as a Feedstock: Optimization of Enzymatic Hydrolysis and Fermentation Em-ploying Burkholderia Sacchari. J. Clean. Prod. 2019, 214, 236–247. [Google Scholar] [CrossRef]
- Aljuraifani, A.A.; Berekaa, M.M.; Ghazwani, A.A. Bacterial biopolymer (polyhydroxyalkanoate) production from low-cost sustainable sources. Microbiologyopen 2019, 8, e00755. [Google Scholar] [CrossRef]
- Santhanam, A.; Sasidharan, S. Microbial Production of Polyhydroxy Alkanotes (PHA) from Alcaligens Spp. and Pseudomonas Oleovorans Using Different Carbon Sources. Afr. J. Biotechnol. 2010, 9, 3144–3150. [Google Scholar]
- Hungund, B.; Vs, S.; Patwardhan, P.; Saleh, A.M. Production of Polyhydroxyalkanoate from Paenibacillus durus BV-1 Isolated from Oil Mill Soil. J. Microb. Biochem. Technol. 2013, 5, 13–17. [Google Scholar] [CrossRef]
- Quillaguamán, J.; Guzmán, H.; Van-Thuoc, D.; Hatti-Kaul, R. Synthesis and Production of Polyhydroxyalkanoates by Halo-philes: Current Potential and Future Prospects. Appl. Microbiol. Biotechnol. 2010, 85, 1687–1696. [Google Scholar] [CrossRef]
- Maheshwari, D.K.; Saraf, M. Halophiles; Springer: Berlin, Germany, 2015. [Google Scholar]
- Patel, K.; Dudhagara, P. Optimization of Xylanase Production by Bacillus Tequilensis Strain UD-3 Using Economical Agri-cultural Substrate and Its Application in Rice Straw Pulp Bleaching. Biocatal. Agric. Biotechnol. 2020, 30, 101846. [Google Scholar] [CrossRef]
- Danial, A.W.; Hamdy, S.M.; Alrumman, S.A.; El-Rab, S.M.F.G.; Shoreit, A.A.M.; Hesham, A.E.-L. Bioplastic Production by Bacillus wiedmannii AS-02 OK576278 Using Different Agricultural Wastes. Microorganisms 2021, 9, 2395. [Google Scholar] [CrossRef]
- Trakunjae, C.; Boondaeng, A.; Apiwatanapiwat, W.; Kosugi, A.; Arai, T.; Sudesh, K.; Vaithanomsat, P. Enhanced Polyhy-droxybutyrate (PHB) Production by Newly Isolated Rare Actinomycetes Rhodococcus sp. Strain BSRT1-1 Using Response Surface Methodology. Sci. Rep. 2021, 11, 1896. [Google Scholar] [CrossRef]
- Biglari, N.; Dashti, M.G.; Abdeshahian, P.; Orita, I.; Fukui, T.; Sudesh, K. Enhancement of bioplastic polyhydroxybutyrate P(3HB) production from glucose by newly engineered strain Cupriavidus necator NSDG-GG using response surface methodology. 3 Biotech 2018, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, H.; Li, K.; Yin, H. Two-stage Fermentation Optimization for Poly-3-hydroxybutyrate Production from Methanol by a New Methylobacterium Isolate from Oil Fields. J. Appl. Microbiol. 2020, 128, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Penkhrue, W.; Jendrossek, D.; Khanongnuch, C.; Pathom-Aree, W.; Aizawa, T.; Behrens, R.L.; Lumyong, S. Response surface method for polyhydroxybutyrate (PHB) bioplastic accumulation in Bacillus drentensis BP17 using pineapple peel. PLoS ONE 2020, 15, e0230443. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Bakhiet, E.K.; Hussein, H.R.; Ali, S.G. Statistical Optimization Studies for Polyhydroxybutyrate (PHB) Produc-tion by Novel Bacillus Subtilis Using Agricultural and Industrial Wastes. Int. J. Environ. Sci. Technol. 2019, 16, 3497–3512. [Google Scholar] [CrossRef]
- Ojha, N.; Das, N. A Statistical approach to optimize the production of Polyhydroxyalkanoates from Wickerhamomyces anomalus VIT-NN01 using Response Surface Methodology. Int. J. Biol. Macromol. 2018, 107, 2157–2170. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Lui, W.-B.; Peng, J. Response Surface Methodology Approach for Optimization of Extrusion Process of Production of Poly (Hydroxyl Butyrate-Co-Hydroxyvalerate)/Tapioca Starch Blends. Int. J. Food Eng. 2017, 13, 20160209. [Google Scholar] [CrossRef]
- Prajapati, J.; Dudhagara, P.; Patel, K. Production of thermal and acid-stable pectinase from Bacillus subtilis strain BK-3: Optimization, characterization, and application for fruit juice clarification. Biocatal. Agric. Biotechnol. 2021, 35, 102063. [Google Scholar] [CrossRef]


| Name of Variable with Code | Unit | Range and Levels | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Rice bran (A) | g/L | 10 | 20 | 30 |
| Peptone (B) | g/L | 2.5 | 5 | 7.5 |
| Temperature (C) | °C | 30 | 35 | 40 |
| Run | A | B | C | PHB Production (g/L) ± SD | |
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| 1 | 1 | −1 | 1 | 1.58 ± 0.08 | 1.99 |
| 2 | 1 | −1 | −1 | 0.55 ± 0.06 | 1.11 |
| 3 | 0 | −1.6817 | 0 | 4.17 ± 0.15 | 3.46 |
| 4 | −1 | −1 | 1 | 3.52 ± 0.08 | 3.78 |
| 5 | 1.6817 | 0 | 0 | 0.01 ± 0.01 | −0.4603 |
| 6 | 0 | 0 | 0 | 5.57 ± 0.15 | 5.65 |
| 7 | 0 | 0 | 1.6817 | 4.44 ± 0.09 | 4.33 |
| 8 | 0 | 0 | −1.6817 | 3.31 ± 0.1 | 3.21 |
| 9 | −1 | 1 | 1 | 4.33 ± 0.1 | 3.91 |
| 10 | −1.6817 | 0 | 0 | 3.35 ± 0.18 | 3.62 |
| 11 | −1 | −1 | −1 | 4.35 ± 0.07 | 4.44 |
| 12 | 0 | 0 | 0 | 5.3 ± 0.05 | 5.65 |
| 13 | 0 | 0 | 0 | 6.11 ± 0.16 | 5.65 |
| 14 | 0 | 0 | 0 | 5.7 ± 0.1 | 5.65 |
| 15 | 1 | 1 | −1 | 0.52 ± 0.05 | 0.4064 |
| 16 | 0 | 0 | 0 | 5.47 ± 0.160 | 5.65 |
| 17 | 0 | 1.6817 | 0 | 2.47 ± 0.05 | 2.98 |
| 18 | 0 | 0 | 0 | 5.72 ± 0.07 | 5.65 |
| 19 | −1 | 1 | −1 | 3.72 ± 0.08 | 3.46 |
| 20 | 1 | 1 | 1 | 2.34 ± 0.12 | 2.39 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value Probe > F |
|---|---|---|---|---|---|
| Model | 64.1 | 9 | 7.12 | 31.37 | <0.0001 * |
| A: Rice bran | 20.05 | 1 | 20.05 | 88.3 | <0.0001 * |
| B: Peptone | 0.2782 | 1 | 0.2782 | 1.23 | 0.2943 |
| C: Temperature | 1.5 | 1 | 1.5 | 6.62 | 0.0278 * |
| AB | 0.0378 | 1 | 0.0378 | 0.1665 | 0.6918 |
| AC | 1.18 | 1 | 1.18 | 5.19 | 0.046 * |
| BC | 0.6216 | 1 | 0.6216 | 2.74 | 0.129 |
| A² | 29.89 | 1 | 29.89 | 131.64 | <0.0001 * |
| B² | 10.67 | 1 | 10.67 | 46.98 | <0.0001 * |
| C² | 6.36 | 1 | 6.36 | 27.99 | 0.0004 * |
| Residual | 2.27 | 10 | 0.2271 | ||
| Lack of Fit | 1.89 | 5 | 0.3781 | 4.97 | 0.0515 ns |
| Pure Error | 0.3801 | 5 | 0.076 | ||
| Cor Total | 66.37 | 19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan, M.; Siddiqui, A.J.; Ashraf, S.A.; Snoussi, M.; Badraoui, R.; Alreshidi, M.; Elasbali, A.M.; Al-Soud, W.A.; Alharethi, S.H.; Sachidanandan, M.; et al. Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization. Polymers 2022, 14, 3982. https://doi.org/10.3390/polym14193982
Adnan M, Siddiqui AJ, Ashraf SA, Snoussi M, Badraoui R, Alreshidi M, Elasbali AM, Al-Soud WA, Alharethi SH, Sachidanandan M, et al. Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization. Polymers. 2022; 14(19):3982. https://doi.org/10.3390/polym14193982
Chicago/Turabian StyleAdnan, Mohd, Arif Jamal Siddiqui, Syed Amir Ashraf, Mejdi Snoussi, Riadh Badraoui, Mousa Alreshidi, Abdelbaset Mohamed Elasbali, Waleed Abu Al-Soud, Salem Hussain Alharethi, Manojkumar Sachidanandan, and et al. 2022. "Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization" Polymers 14, no. 19: 3982. https://doi.org/10.3390/polym14193982
APA StyleAdnan, M., Siddiqui, A. J., Ashraf, S. A., Snoussi, M., Badraoui, R., Alreshidi, M., Elasbali, A. M., Al-Soud, W. A., Alharethi, S. H., Sachidanandan, M., & Patel, M. (2022). Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization. Polymers, 14(19), 3982. https://doi.org/10.3390/polym14193982














