Development of a Curcumin-Loaded Lecithin/Chitosan Nanoparticle Utilizing a Box-Behnken Design of Experiment: Formulation Design and Influence of Process Parameters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Curcumin Loaded Lecithin-Chitosan Nanoparticles
2.3. Optimization of CUR/LCSNPs
2.3.1. Particle Size and Zeta Potential Measurements
2.3.2. Determination of Encapsulation Efficiency
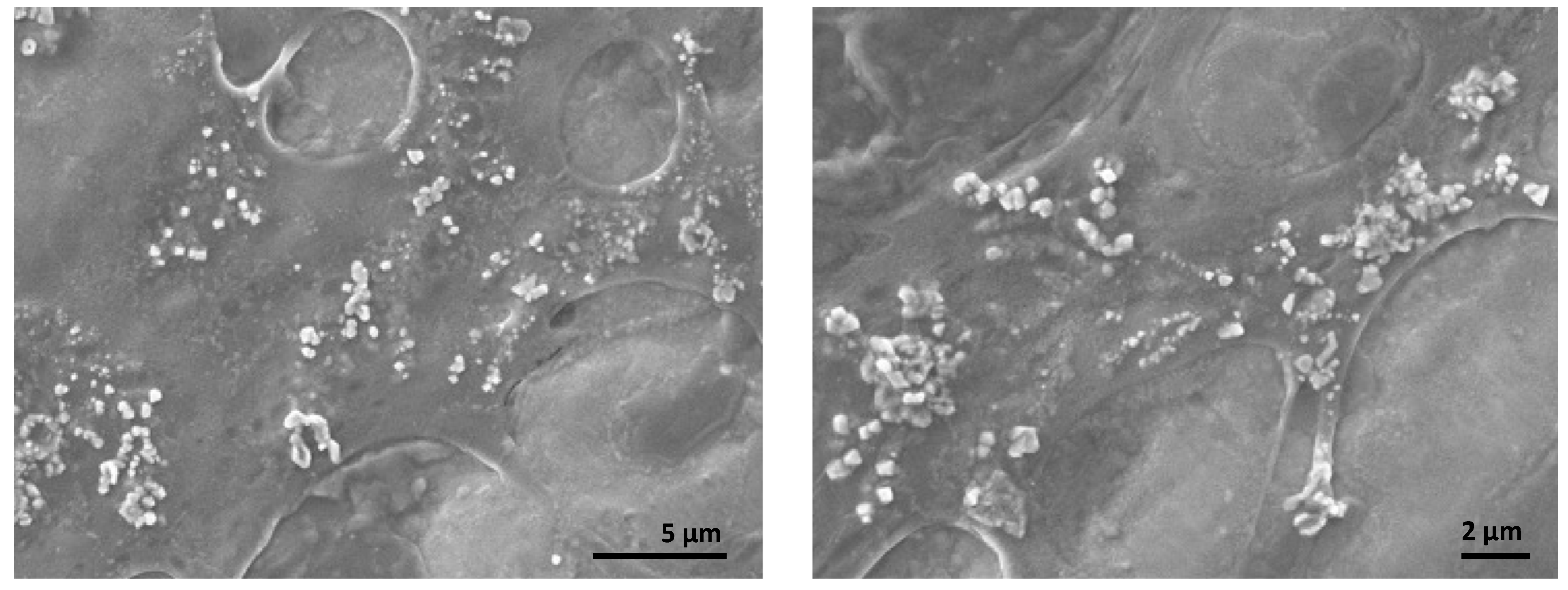
2.3.3. Scanning Electron Microscopy (SEM)
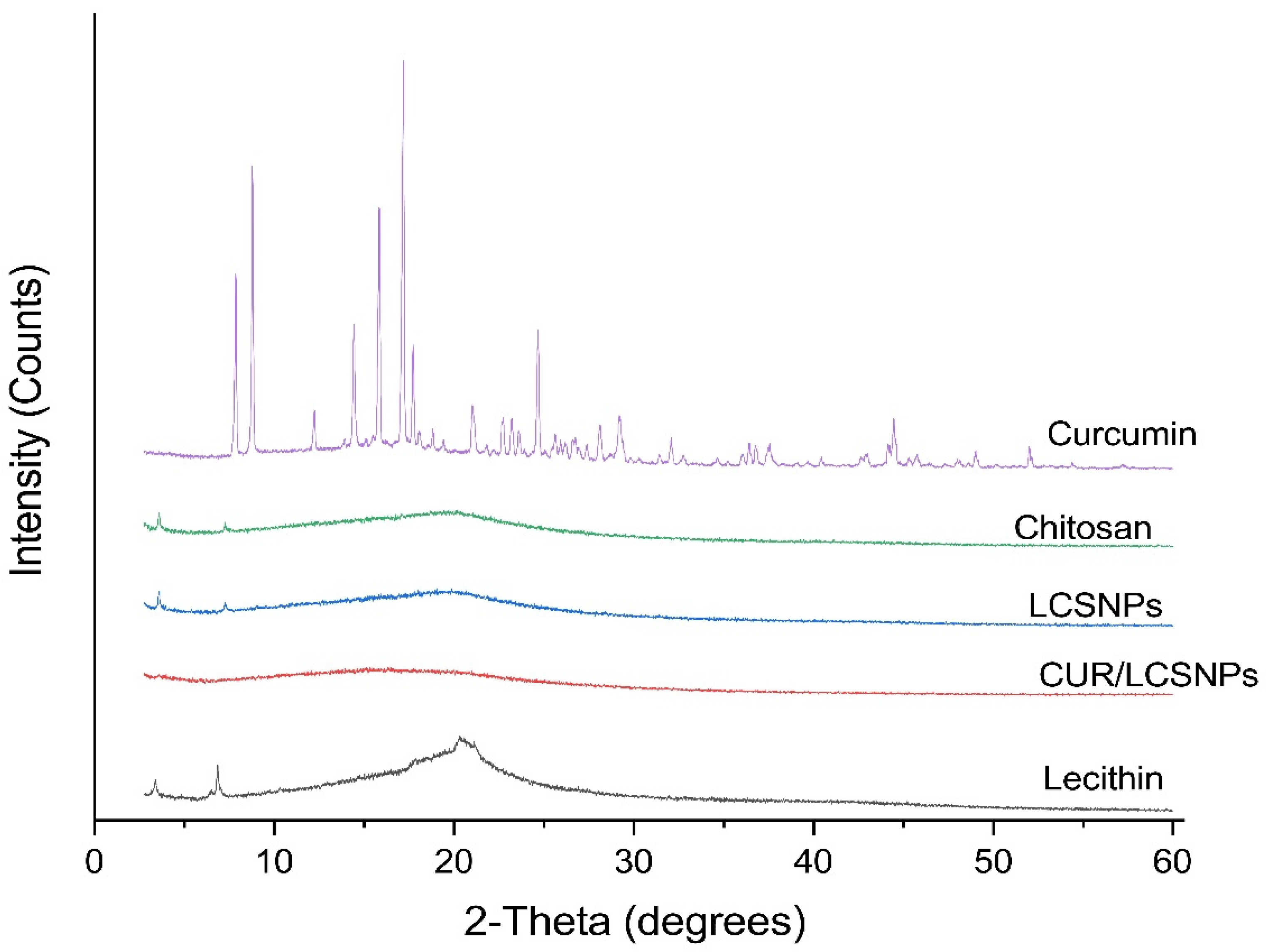
2.3.4. X-ray Diffraction (XRD) Analysis
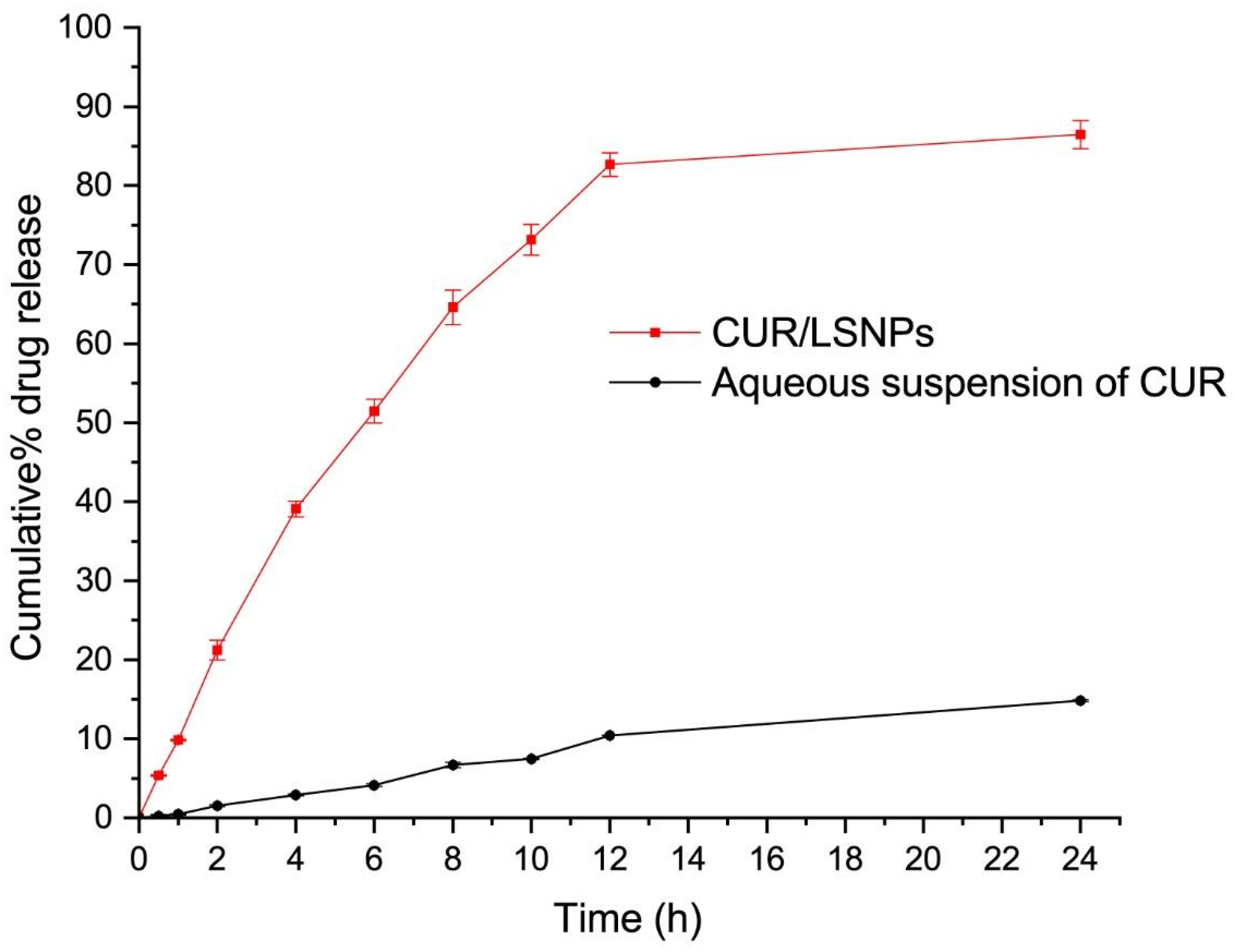
2.4. In Vitro Drug Release and Kinetics
2.5. Physical Stability Study
3. Results and Discussion
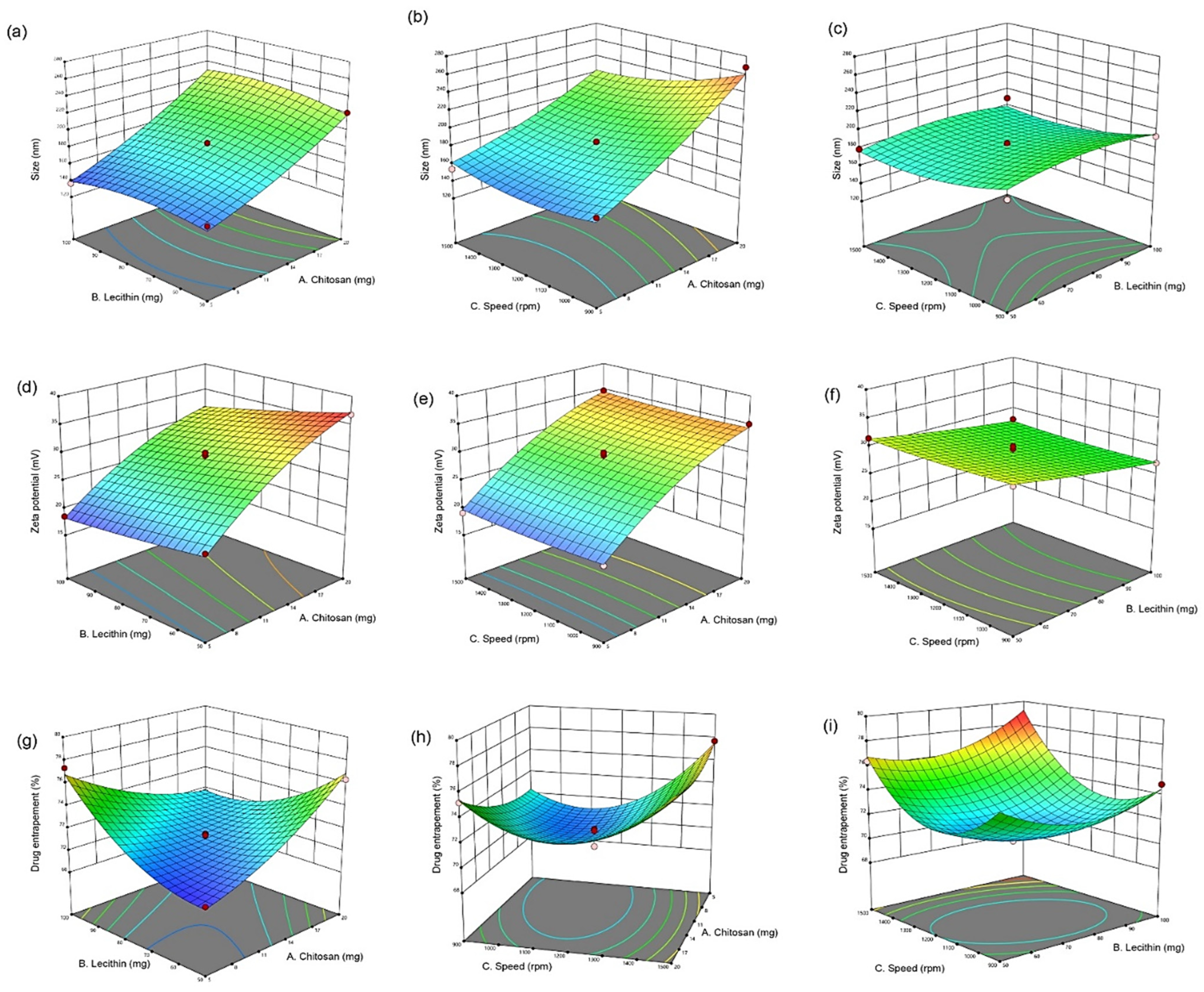
3.1. Optimization of CUR/LCSNPs Utilizing a Box-Behnken Statistical Design
3.1.1. Fitting of the Data on the Model Suggested by a Box-Behnken Statistical Design
3.1.2. Interaction Effect of Independent Variables in the Responses
3.1.3. Applied Model Validation
3.2. SEM and XRD Analysis
3.3. In Vitro Release Kinetics of CUR/LCSNPs
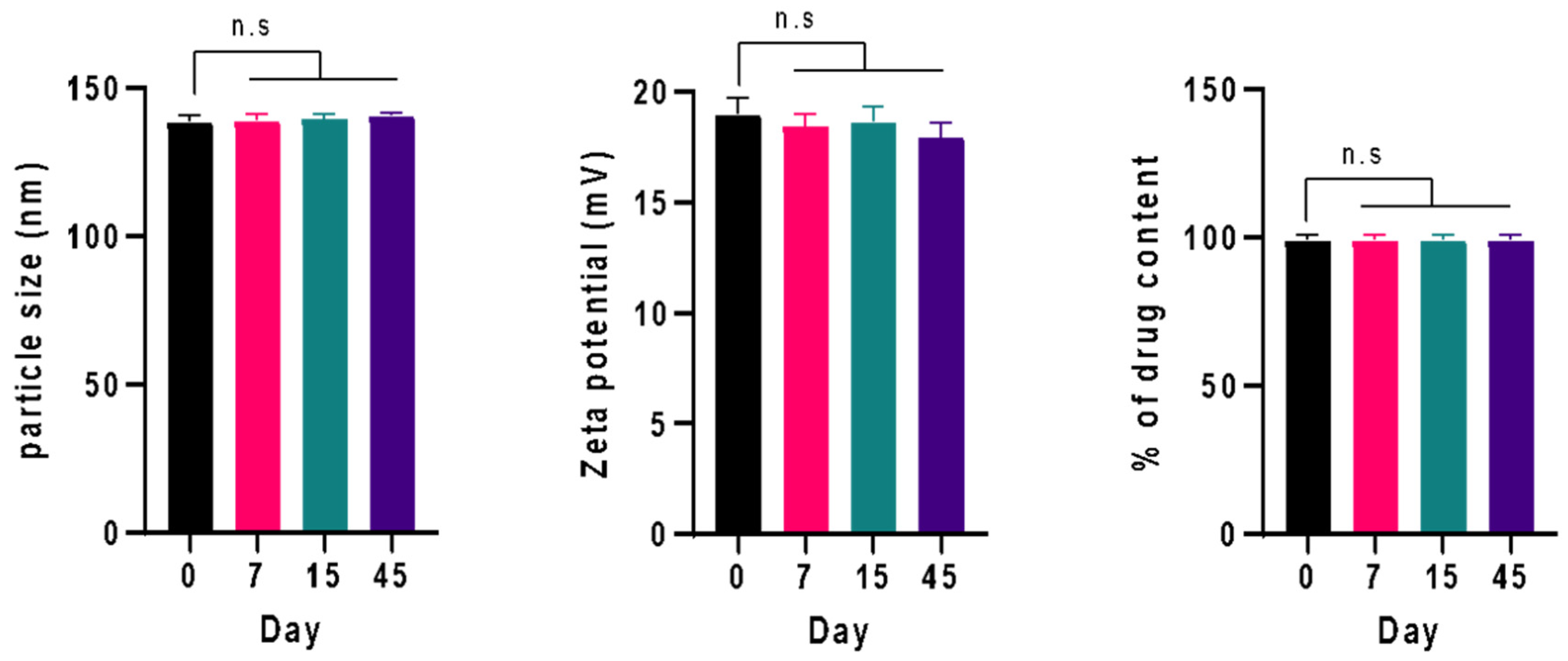
3.4. Physical Stability Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, M.Z.; Alkahtani, S.A.; Akhter, S.; Ahmad, F.J.; Ahmad, J.; Akhtar, M.S.; Mohsin, N.; Abdel-Wahab, B.A. Progress in nanotechnology-based drug carrier in designing of curcumin nanomedicines for cancer therapy: Current state-of-the-art. J. Drug Target. 2016, 24, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.Z.; Akhter, S.; Mohsin, N.; Abdel-Wahab, B.A.; Ahmad, J.; Warsi, M.H.; Rahman, M.; Mallick, N.; Ahmad, F.J. Transformation of curcumin from food additive to multifunctional medicine: Nanotechnology bridging the gap. Curr. Drug Discov. Technol. 2014, 11, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Loo, C.Y.; Young, P.M.; Traini, D.; Mason, R.S.; Rohanizadeh, R. Recent advances in curcumin nanoformulation for cancer therapy. Expert Opin. Drug Deliv. 2014, 11, 1183–1201. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.L.; Chacko, K.M.; Kuruvilla, B.T. Systematic and comprehensive investigation of the toxicity of curcuminoid-essential oil complex: A bioavailable turmeric formulation. Mol. Med. Rep. 2016, 13, 592–604. [Google Scholar] [CrossRef]
- Mohanty, C.; Das, M.; Sahoo, S.K. Emerging role of nanocarriers to increase the solubility and bioavailability of curcumin. Expert Opin. Drug Deliv. 2012, 9, 1347–1364. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Sharma, R.A.; Gescher, A.J.; Steward, W.P. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef]
- Duan, J.; Zhang, Y.; Han, S.; Chen, Y.; Li, B.; Liao, M.; Chen, W.; Deng, X.; Zhao, J.; Huang, B. Synthesis and in vitro/in vivo anti-cancer evaluation of curcumin-loaded chitosan/poly(butyl cyanoacrylate) nanoparticles. Int. J. Pharm. 2010, 400, 211–220. [Google Scholar] [CrossRef]
- Algahtani, M.S.; Ahmad, M.Z.; Nourein, I.H.; Albarqi, H.A.; Alyami, H.S.; Alyami, M.H.; Alqahtani, A.A.; Alasiri, A.; Algahtani, T.S.; Mohammed, A.A.; et al. Preparation and Characterization of Curcumin Nanoemulgel Utilizing Ultrasonication Technique for Wound Healing: In Vitro, Ex Vivo, and In Vivo Evaluation. Gels 2021, 7, 213. [Google Scholar] [CrossRef]
- Feng, T.; Wei, Y.; Lee, R.J.; Zhao, L. Liposomal curcumin and its application in cancer. Int. J. Nanomed. 2017, 12, 6027–6044. [Google Scholar] [CrossRef] [Green Version]
- Ipar, V.S.; Dsouza, A.; Devarajan, P.V. Enhancing Curcumin Oral Bioavailability Through Nanoformulations. Eur. J. Drug Metab. Pharmacokinet. 2019, 44, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, N.; He, H.; Tang, X. Pharmaceutical strategies of improving oral systemic bioavailability of curcumin for clinical application. J. Control. Release 2019, 316, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Guo, A.; Liu, Z.; Zhang, L.; Liao, W.; Liu, J.; Mao, L.; Yuan, F.; Gao, Y. Development of curcumin loaded core-shell zein microparticles stabilized by cellulose nanocrystals and whey protein microgels through interparticle interactions. Food Funct. 2021, 12, 6936–6949. [Google Scholar] [CrossRef] [PubMed]
- Nano Water Soluble Curcumin Offers Bioavailability Solution. Available online: https://www.vitafoodsinsights.com/ingredients/nano-water-soluble-curcumin-offers-bioavailability-solution (accessed on 17 February 2022).
- Ahmad, M.Z.; Rizwanullah, M.; Ahmad, J.; Alasmary, M.Y.; Akhter, M.H.; Abdel-Wahab, B.A.; Warsi, M.H.; Haque, A. Progress in nanomedicine-based drug delivery in designing of chitosan nanoparticles for cancer therapy. Int. J. Polym. Mater. Polym. Biomater. 2021, 71, 602–623. [Google Scholar] [CrossRef]
- Naruphontjirakul, P.; Viravaidya-Pasuwat, K. Development of anti-HER2-targeted doxorubicin-core-shell chitosan nanoparticles for the treatment of human breast cancer. Int. J. Nanomed. 2019, 14, 4105–4121. [Google Scholar] [CrossRef]
- Yoncheva, K.; Merino, M.; Shenol, A.; Daskalov, N.T.; Petkov, P.S.; Vayssilov, G.N.; Garrido, M.J. Optimization and in-vitro/in-vivo evaluation of doxorubicin-loaded chitosan-alginate nanoparticles using a melanoma mouse model. Int. J. Pharm. 2019, 556, 1–8. [Google Scholar] [CrossRef]
- Lopes, V.F.; Giongo, C.N.; de Almeida Campos, L.; Abraham, W.R.; Mainardes, R.M.; Khalil, N.M. Chitosan Nanoparticles Potentiate the in vitro and in vivo Effects of Curcumin and other Natural Compounds. Curr. Med. Chem. 2021, 28, 4935–4953. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, W.; He, M.; Lu, Y.; Lou, K.; Gao, F. Preparation and characterization of β-cyclodextrin grafted N-maleoyl chitosan nanoparticles for drug delivery. Asian J. Pharm. Sci. 2017, 12, 558–568. [Google Scholar] [CrossRef]
- Yemisci, M.; Caban, S.; Fernandez-Megia, E.; Capan, Y.; Couvreur, P.; Dalkara, T. Preparation and Characterization of Biocompatible Chitosan Nanoparticles for Targeted Brain Delivery of Peptides. Methods Mol. Biol. 2018, 1727, 443–454. [Google Scholar] [CrossRef]
- Alkholief, M. Optimization of Lecithin-Chitosan nanoparticles for simultaneous encapsulation of doxorubicin and piperine. J. Drug Deliv. Sci. Technol. 2019, 52, 204–214. [Google Scholar] [CrossRef]
- Lopes Rocha Correa, V.; Assis Martins, J.; Ribeiro de Souza, T.; de Castro Nunes Rincon, G.; Pacheco Miguel, M.; Borges de Menezes, L.; Correa Amaral, A. Melatonin loaded lecithin-chitosan nanoparticles improved the wound healing in diabetic rats. Int. J. Biol. Macromol. 2020, 162, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.; Ravi, P.R.; Kathuria, H.; Vats, R. Self-assembled lecithin-chitosan nanoparticles improve the oral bioavailability and alter the pharmacokinetics of raloxifene. Int. J. Pharm. 2020, 588, 119731. [Google Scholar] [CrossRef]
- Panda, D.S.; Eid, H.M.; Elkomy, M.H.; Khames, A.; Hassan, R.M.; Abo El-Ela, F.I.; Yassin, H.A. Berberine Encapsulated Lecithin-Chitosan Nanoparticles as Innovative Wound Healing Agent in Type II Diabetes. Pharmaceutics 2021, 13, 1197. [Google Scholar] [CrossRef] [PubMed]
- Negi, P.; Sharma, G.; Verma, C.; Garg, P.; Rathore, C.; Kulshrestha, S.; Lal, U.R.; Gupta, B.; Pathania, D. Novel thymoquinone loaded chitosan-lecithin micelles for effective wound healing: Development, characterization, and preclinical evaluation. Carbohydr. Polym. 2020, 230, 115659. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ruiz, A.G.; Ganem, A.; Olivares-Corichi, I.M.; García-Sánchez, J.R. Lecithin–chitosan–TPGS nanoparticles as nanocarriers of (−)-epicatechin enhanced its anticancer activity in breast cancer cells. RSC Adv. 2018, 8, 34773–34782. [Google Scholar] [CrossRef] [PubMed]
- Ilk, S.; Saglam, N.; Özgen, M. Kaempferol loaded lecithin/chitosan nanoparticles: Preparation, characterization, and their potential applications as a sustainable antifungal agent. Artif. Cells Nanomed. Biotechnol. 2017, 45, 907–916. [Google Scholar] [CrossRef]
- Sonvico, F.; Cagnani, A.; Rossi, A.; Motta, S.; Di Bari, M.T.; Cavatorta, F.; Alonso, M.J.; Deriu, A.; Colombo, P. Formation of self-organized nanoparticles by lecithin/chitosan ionic interaction. Int. J. Pharm. 2006, 324, 67–73. [Google Scholar] [CrossRef]
- Alam, M.S.; Algahtani, M.S.; Ahmad, J.; Kohli, K.; Shafiq-Un-Nabi, S.; Warsi, M.H.; Ahmad, M.Z. Formulation design and evaluation of aceclofenac nanogel for topical application. Ther. Deliv. 2020, 11, 767–778. [Google Scholar] [CrossRef]
- Algahtani, M.S.; Ahmad, M.Z.; Ahmad, J. Nanoemulsion loaded polymeric hydrogel for Topical delivery of curcumin in Psoriasis. J. Drug Deliv. Sci. Technol. 2020, 59, 101847. [Google Scholar] [CrossRef]
- Algahtani, M.S.; Ahmad, M.Z.; Nourein, I.H.; Ahmad, J. Co-delivery of imiquimod and curcumin by nanoemugel for improved topical delivery and reduced psoriasis-like skin lesions. Biomolecules 2020, 10, 968. [Google Scholar] [CrossRef]
- Abul Kalam, M.; Khan, A.A.; Khan, S.; Almalik, A.; Alshamsan, A. Optimizing indomethacin-loaded chitosan nanoparticle size, encapsulation, and release using Box-Behnken experimental design. Int. J. Biol. Macromol. 2016, 87, 329–340. [Google Scholar] [CrossRef]
- Abul Kalam, M.; Sultana, Y.; Ali, A.; Aqil, M.; Mishra, A.K.; Aljuffali, I.A.; Alshamsan, A. Part I: Development and optimization of solid-lipid nanoparticles using Box-Behnken statistical design for ocular delivery of gatifloxacin. J. Biomed. Mater. Res. Part A 2013, 101, 1813–1827. [Google Scholar] [CrossRef] [PubMed]
- Zidan, A.S.; Mokhtar Ibrahim, M.; Megrab, N.A.E. Optimization of methotrexate loaded niosomes by Box-Behnken design: An understanding of solvent effect and formulation variability. Drug Dev. Ind. Pharm. 2017, 43, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Gunda, R.K.; Manchineni, P.R. Statistical Design and Optimization of Sustained Release Formulations of Pravastatin. Turk. J. Pharm. Sci. 2020, 17, 221–227. [Google Scholar] [CrossRef]
- Hosny, K.M.; Alharbi, W.S.; Almehmady, A.M.; Bakhaidar, R.B.; Alkhalidi, H.M.; Sindi, A.M.; Hariri, A.H.; Shadab, M.D.; Zaki, R.M. Preparation and optimization of pravastatin-naringenin nanotransfersomes to enhance bioavailability and reduce hepatic side effects. J. Drug Deliv. Sci. Technol. 2020, 57, 101746. [Google Scholar] [CrossRef]
- Abosabaa, S.A.; ElMeshad, A.N.; Arafa, M.G. Chitosan Nanocarrier Entrapping Hydrophilic Drugs as Advanced Polymeric System for Dual Pharmaceutical and Cosmeceutical Application: A Comprehensive Analysis Using Box-Behnken Design. Polymers 2021, 13, 677. [Google Scholar] [CrossRef]
- Zidan, A.S.; Mokhtar, M. Multivariate Optimization of Formulation Variables Influencing Flurbiprofen Proniosomes Characteristics. J. Pharm. Sci. 2011, 100, 2212–2221. [Google Scholar] [CrossRef]
- Sayyar, Z.; Malmiri, H.J. Photocatalytic and antibacterial activities study of prepared self-cleaning nanostructure surfaces using synthesized and coated ZnO nanoparticles with Curcumin nanodispersion. Z. Für Krist.-Cryst. Mater. 2019, 234, 307–328. [Google Scholar] [CrossRef]
- Asare-Addo, K.; Conway, B.R.; Hajamohaideen, M.J.; Kaialy, W.; Nokhodchi, A.; Larhrib, H. Aqueous and hydro-alcoholic media effects on polyols. Colloids Surf. B Biointerfaces 2013, 111, 24–29. [Google Scholar] [CrossRef]
- Siepmann, J.; Siepmann, F. Mathematical modeling of drug dissolution. Int. J. Pharm. 2013, 453, 12–24. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceutics 2017, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Notario-Pérez, F.; Martín-Illana, A.; Cazorla-Luna, R.; Ruiz-Caro, R.; Bedoya, L.M.; Tamayo, A.; Rubio, J.; Veiga, M.D. Influence of Chitosan Swelling Behaviour on Controlled Release of Tenofovir from Mucoadhesive Vaginal Systems for Prevention of Sexual Transmission of HIV. Mar. Drugs 2017, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Yurtdaş Kirimlioğlu, G.; Öztürk, A.A. Levocetirizine Dihydrochloride-Loaded Chitosan Nanoparticles: Formulation and In Vitro Evaluation. Turk. J. Pharm. Sci. 2020, 17, 27–35. [Google Scholar] [CrossRef] [PubMed]


| Factors | Level Used | ||
|---|---|---|---|
| Low (−1) | Medium (0) | High (+1) | |
| Independent variables | |||
| A = Chitosan (mg) | 5 | 12.5 | 20 |
| B = Lecithin (mg) | 50 | 75 | 100 |
| C = Speed (rpm) | 900 | 1200 | 1500 |
| Independent Variables | Dependent Variables | |||||
|---|---|---|---|---|---|---|
| A (mg) | B (mg) | C (rpm) | Y1 (nm) | Y2 (mV) | Y3 (%) | |
| F1 | 12.5 | 50 | 900 | 183.74 ± 1.70 | 31.5 ± 1.25 | 75.62 ± 2.60 |
| F2 | 12.5 | 75 | 1200 | 185.33 ± 3.50 | 29.6 ± 1.15 | 70.04 ± 2.40 |
| F3 | 5 | 50 | 1200 | 150.63 ± 4.90 | 21.7 ± 2.10 | 69.84 ± 1.50 |
| F4 | 20 | 75 | 1500 | 218.63 ± 2.70 | 34.8 ± 2.10 | 77.79 ± 2.90 |
| F5 | 12.5 | 75 | 1200 | 183.15 ± 2.70 | 28.8 ± 1.40 | 70.91 ± 1.60 |
| F6 | 20 | 50 | 1200 | 221.22 ± 4.20 | 36.8 ± 3.20 | 76.33 ± 2.10 |
| F7 | 12.5 | 100 | 1500 | 189.40 ± 1.10 | 27.9 ± 2.20 | 78.50 ± 2.10 |
| F8 | 20 | 100 | 1200 | 224.01 ± 4.30 | 31.2 ± 12.0 | 71.59 ± 2.50 |
| F9 | 5 | 100 | 1200 | 136.44 ± 1.70 | 18.5 ± 1.40 | 77.34 ± 2.20 |
| F10 | 5 | 75 | 1500 | 154.10 ± 2.10 | 19.1 ± 1.20 | 77.69 ± 2.70 |
| F11 | 5 | 75 | 900 | 162.43 ± 2.50 | 19.7 ± 1.10 | 71.46 ± 2.30 |
| F12 | 12.5 | 75 | 1200 | 185.48 ± 4.30 | 28.3 ± 2.40 | 71.53 ± 2.70 |
| F13 | 12.5 | 75 | 1200 | 184.32 ± 3.10 | 27.9 ± 2.40 | 71.37 ± 3.0 |
| F14 | 12.5 | 100 | 900 | 193.45 ± 3.50 | 27.1 ± 2.15 | 74.86 ± 2.50 |
| F15 | 12.5 | 50 | 1500 | 179.01 ± 1.90 | 31.5 ± 1.90 | 76.37 ± 2.60 |
| F16 | 12.5 | 75 | 1200 | 185.66 ± 3.10 | 30.1 ± 1.80 | 71.47 ± 2.30 |
| F17 | 20 | 75 | 900 | 267.94 ± 3.70 | 35.1 ± 3.20 | 75.20 ± 2.90 |
| Response | Model | R-Squared (r2) | Adjusted r2 | Predicted r2 | Std. Dev. | %CV | Adequate Precision | Remarks |
|---|---|---|---|---|---|---|---|---|
| Particle size (nm) | Linear | 0.9513 | 0.9401 | 0.9010 | 6.86 | - | - | - |
| Second order (2FI) | 0.9206 | 0.8730 | 0.6111 | 11.19 | - | - | - | |
| Quadratic | 0.9984 | 0.9834 | 0.9389 | 4.57 | 2.44 | 47.4997 | Suggested | |
| Zeta potential (mV) | Linear | 0.9481 | 0.9360 | 0.9028 | 1.41 | - | - | - |
| Second order (2FI) | 0.9513 | 0.9220 | 0.7981 | 1.55 | - | - | - | |
| Quadratic | 0.9896 | 0.9762 | 0.9411 | 0.86 | 3.04 | 28.75 | Suggested | |
| %EE | Linear | 0.8778 | 0.8229 | 0.6338 | 3.08 | - | - | - |
| Second order (2FI) | 0.8643 | 0.8429 | 0.6580 | 2.83 | - | - | - | |
| Quadratic | 0.9837 | 0.9670 | 0.9378 | 0.88 | 1.19 | 14.20 | Suggested |
| Source | Sum of Square | Degree of Freedom | Mean Square | F value | p-Value | Remarks |
|---|---|---|---|---|---|---|
| Y1 (nm) | ||||||
| Model-Quadratic | 15,323.95 | 9 | 1702.66 | 26.81 | 0.0001 | significant |
| A-Chitosan | 13,464.08 | 1 | 13,464.08 | 211.99 | <0.0001 | |
| B-Lecithin | 9.43 | 1 | 9.43 | 0.1485 | 0.7114 | |
| C-Speed | 551.62 | 1 | 551.62 | 8.69 | 0.0215 | |
| AB | 71.99 | 1 | 71.99 | 1.13 | 0.3224 | |
| AC | 419.86 | 1 | 419.86 | 6.61 | 0.0369 | |
| BC | 0.1146 | 1 | 0.1146 | 0.0018 | 0.9673 | |
| A2 | 168.61 | 1 | 168.61 | 2.65 | 0.1473 | |
| B2 | 272.47 | 1 | 272.47 | 4.29 | 0.0771 | |
| C2 | 392.63 | 1 | 392.63 | 6.18 | 0.0418 | |
| Lack of fit | 440.16 | 3 | 146.72 | 1.468 | 0.7217 | Not significant |
| Pure error | 4.43 | 4 | 1.11 | |||
| Total correlation | 15,768.54 | 16 | ||||
| Y2 (mV) | ||||||
| Model-Quadratic | 489.50 | 9 | 54.39 | 73.82 | <0.0001 | significant |
| A-Chitosan | 433.65 | 1 | 433.65 | 588.56 | <0.0001 | |
| B-Lecithin | 35.28 | 1 | 35.28 | 47.88 | 0.0002 | |
| C-Speed | 0.0012 | 1 | 0.0012 | 0.0017 | 0.9683 | |
| AB | 1.44 | 1 | 1.44 | 1.95 | 0.2048 | |
| AC | 0.0225 | 1 | 0.0225 | 0.0305 | 0.8662 | |
| BC | 0.1600 | 1 | 0.1600 | 0.2172 | 0.6554 | |
| A2 | 18.65 | 1 | 18.65 | 25.31 | 0.0015 | |
| B2 | 0.2047 | 1 | 0.2047 | 0.2778 | 0.6144 | |
| C2 | 0.5026 | 1 | 0.5026 | 0.6822 | 0.4361 | |
| Lack of fit | 1.80 | 3 | 0.6008 | 0.7163 | 0.5918 | not significant |
| Pure error | 3.36 | 4 | 0.8388 | |||
| Total correlation | 494.66 | 16 | ||||
| Y3 (%EE) | ||||||
| Model-Quadratic | 144.09 | 9 | 16.01 | 20.64 | 0.0003 | significant |
| A-Chitosan | 2.61 | 1 | 2.61 | 3.37 | 0.1090 | |
| B-Lecithin | 2.11 | 1 | 2.11 | 2.72 | 0.1429 | |
| C-Speed | 21.86 | 1 | 21.86 | 28.18 | 0.0011 | |
| AB | 37.45 | 1 | 37.45 | 48.30 | 0.0002 | |
| AC | 3.30 | 1 | 3.30 | 4.26 | 0.0780 | |
| BC | 2.09 | 1 | 2.09 | 2.69 | 0.1448 | |
| A2 | 3.85 | 1 | 3.85 | 4.96 | 0.0612 | |
| B2 | 13.00 | 1 | 13.00 | 16.76 | 0.0046 | |
| C2 | 52.13 | 1 | 52.13 | 67.22 | <0.0001 | |
| Lack of fit | 3.89 | 3 | 1.30 | 3.37 | 0.1358 | not significant |
| Pure error | 1.54 | 4 | 0.3850 | |||
| Total correlation | 149.52 | 16 | ||||
| Response | Predicted Value | Observed Value | Prediction Error (%) |
|---|---|---|---|
| Particle size | 141.40 | 138.4 ± 2.10 | −2.09% |
| Zeta potential | +18.19 | +18.98 ± 0.72 | 4.36% |
| %EE | 76.78 | 77.40 ± 1.70 | 0.79% |
| Formulation | Zero-Order | First-Order | Higuchi Model | Hixon–Crowell | Korsmeyer–Peppas Model | |
|---|---|---|---|---|---|---|
| R2 | R2 | R2 | R2 | R2 | n | |
| Optimized CUR/LCSNPs | 0.7918 | 0.9794 | 0.9876 | 0.9747 | 0.9937 | 0.343 |
| Aqueous suspension of CUR | 0.8812 | 0.9786 | 0.9875 | 0.9833 | 0.9245 | 0.519 |
| Periods | ||||
|---|---|---|---|---|
| Parameter (Mean ± SD, n = 3) | 0 Days | 7 Days | 15 Days | 45 Days |
| Particle size (nm) | 138.43 ± 2.09 | 138.97 ± 2.05 | 139.56 ± 1.55 | 140.54 ± 0.92 |
| PDI | 0.172 ± 0.031 | 0.175 ± 0.029 | 0.182 ± 0.024 | 0.188 ± 0.025 |
| Zeta potential (mV) | +18.98 ± 0.72 | 18.46 ± 0.53 | 18.63 ± 0.68 | 17.93 ± 0.66 |
| %drug content | 99.19 ± 1.71 | 99.01 ± 1.77 | 98.97 ± 1.88 | 98.95 ± 1.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walbi, I.A.; Ahmad, M.Z.; Ahmad, J.; Algahtani, M.S.; Alali, A.S.; Alsudir, S.A.; Aodah, A.H.; Albarqi, H.A. Development of a Curcumin-Loaded Lecithin/Chitosan Nanoparticle Utilizing a Box-Behnken Design of Experiment: Formulation Design and Influence of Process Parameters. Polymers 2022, 14, 3758. https://doi.org/10.3390/polym14183758
Walbi IA, Ahmad MZ, Ahmad J, Algahtani MS, Alali AS, Alsudir SA, Aodah AH, Albarqi HA. Development of a Curcumin-Loaded Lecithin/Chitosan Nanoparticle Utilizing a Box-Behnken Design of Experiment: Formulation Design and Influence of Process Parameters. Polymers. 2022; 14(18):3758. https://doi.org/10.3390/polym14183758
Chicago/Turabian StyleWalbi, Ismail A., Mohammad Zaki Ahmad, Javed Ahmad, Mohammed S. Algahtani, Amer S. Alali, Samar A. Alsudir, Alhassan H. Aodah, and Hassan A. Albarqi. 2022. "Development of a Curcumin-Loaded Lecithin/Chitosan Nanoparticle Utilizing a Box-Behnken Design of Experiment: Formulation Design and Influence of Process Parameters" Polymers 14, no. 18: 3758. https://doi.org/10.3390/polym14183758
APA StyleWalbi, I. A., Ahmad, M. Z., Ahmad, J., Algahtani, M. S., Alali, A. S., Alsudir, S. A., Aodah, A. H., & Albarqi, H. A. (2022). Development of a Curcumin-Loaded Lecithin/Chitosan Nanoparticle Utilizing a Box-Behnken Design of Experiment: Formulation Design and Influence of Process Parameters. Polymers, 14(18), 3758. https://doi.org/10.3390/polym14183758









