Novel Immobilized Biocatalysts Based on Cysteine Proteases Bound to 2-(4-Acetamido-2-sulfanilamide) Chitosan and Research on Their Structural Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. 2-(4-Acetamido-2-sulfanilamide) Chitosan Synthesis and Characterization
2.3. Molecular Docking
2.4. Enzyme Immobilization
2.5. Protein Content Measurement
2.6. Proteolytic Activity Evaluation of the Immobilized Enzymes
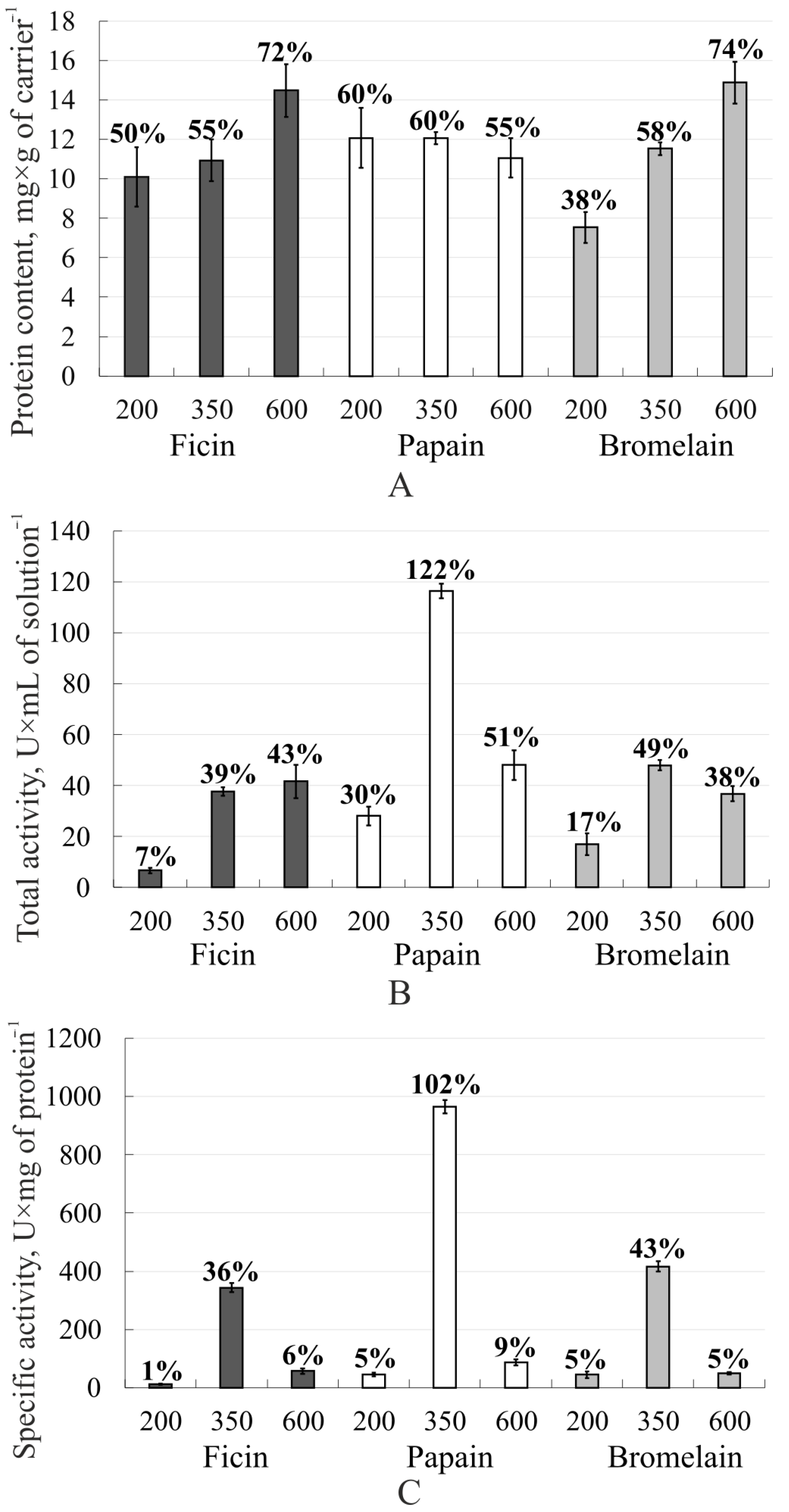
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Meng, Q.; Zhong, S.; Xu, L.; Wang, J.; Zhang, Z.; Gao, Y.; Cui, X. Review on design strategies and considerations of polysaccharide-based smart drug delivery systems for cancer therapy. Carbohyd. Polym. 2022, 279, 119013. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, K.; Peng, X.; Zhang, L. Chitosan-based drug delivery systems: Current strategic design and potential application in human hard tissue repair. Eur. Polym. J. 2022, 166, 110979. [Google Scholar] [CrossRef]
- Hadji, H.; Bouchemal, K. Advances in the treatment of inflammatory bowel disease: Focus on polysaccharide nanoparticulate drug delivery systems. Adv. Drug Deliv. Rev. 2022, 181, 114101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, Y.; Li, R.; Chen, Z.; Fan, X. Advanced drug delivery system against ischemic stroke. J. Control. Release 2022, 344, 173–201. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S. Understanding the burst release phenomenon: Toward designing effective nanoparticulate drug-delivery systems. Ther. Deliv. 2020, 12, 21–36. [Google Scholar] [CrossRef]
- Huang, X.; Brazel, C.S. On the importance and mechanisms of burst release in matrix-controlled drug delivery systems. J. Control. Release 2001, 73, 121–136. [Google Scholar] [CrossRef]
- Baidamshina, D.R.; Koroleva, V.A.; Trizna, E.Y.; Pankova, S.M.; Agafonova, M.N.; Chirkova, M.N.; Vasileva, O.S.; Akhmetov, N.; Shubina, V.V.; Porfiryev, A.G.; et al. Anti-biofilm and wound-healing activity of chitosan-immobilized Ficin. Int. J. Biol. Macromol. 2020, 164, 4205–4217. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, W.; Zhang, M.; Zhang, L.; Shen, Y.; Huang, S.; Li, M.; Qiu, W.; Pan, Y.; Zhou, L.; et al. The Inhibitory Effects of Ficin on Streptococcus mutans Biofilm Formation. BioMed Res. Int. 2021, 2021, 6692328. [Google Scholar] [CrossRef]
- Asanarong, O.; Quan, V.M.; Boonrungsiman, S.; Sukyai, P. Bioactive wound dressing using bacterial cellulose loaded with papain composite: Morphology, loading/release and antibacterial properties. Eur. Polym. J. 2021, 143, 110224. [Google Scholar] [CrossRef]
- Hikisz, P.; Bernasinska-Slomczewska, J. Beneficial Properties of Bromelain. Nutrients 2021, 13, 4313. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lucas, J.; Castaneda, D.; Hormingo, D. New trends for a classical enzyme: Papain, a biotechnological success story in the food industry. Trends Food Sci. Technol. 2017, 68, 91–101. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, B.; Lv, K.; Kumissay, L.; Wu, B.; Chu, J.; He, B. Specific immobilization of lipase on functionalized 3D printing scaffolds via enhanced hydrophobic interaction for efficient resolution of racemic 1-indanol. Biochem. Biophys. Res. Commun. 2021, 546, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.; Khoshfetrat, A.B.; Khatami, N.; Ahmadian, M.; Rahbarghzi, R. Comparative study of collagen and gelatin in chitosan-based hydrogels for effective wound dressing: Physical properties and fibroblastic cell behavior. Biochem. Biophys. Res. Commun. 2019, 518, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, X.; Wang, J.; Wang, Y.; Shi, B.; Huang, C.; Yang, X.; Liu, T. Novel chitosan/collagen scaffold containing transforming growth factor-β1 DNA for periodontal tissue engineering, Biochem. Biophys. Res. Commun. 2006, 344, 362–369. [Google Scholar] [CrossRef]
- Ol’shannikova, S.S.; Red’ko, Y.A.; Lavlinskaya, M.S.; Sorokin, A.V.; Holyavka, M.G.; Artyukhov, V.G. Preparation of Papain Complexes with Chitosan Microparticles and Evaluation of Their Stability Using the Enzyme Activity Level. Pharm. Chem. J. 2022, 55, 1240–1244. [Google Scholar] [CrossRef]
- Pankova, S.M.; Sakibaev, F.A.; Holyavka, M.G.; Vyshkvorkina, Y.M.; Lukin, A.N.; Artyukhov, V.G. Studies of the Processes of the Trypsin Interactions with Ion Exchange Fibers and Chitosan. Rus. J. Bioorg. Chem. 2021, 47, 765–776. [Google Scholar] [CrossRef]
- Baidamshina, D.R.; Koroleva, V.A.; Olshannikova, S.S.; Trizna, E.Y.; Bogachev, M.I.; Artyukhov, V.G.; Holyavka, M.G.; Kayumov, A.R. Biochemical Properties and Anti-Biofilm Activity of Chitosan-Immobilized Papain. Mar. Drugs. 2021, 19, 197. [Google Scholar] [CrossRef]
- Holyavka, M.G.; Makin, S.M.; Kondratyev, M.S.; Abdullatypov, A.V.; Kovaleva, T.A.; Artyukhov, V.G. Supramolecular Organization of Inulinases from Aspergillus awamori, Aspergillus ficuum and Kluyveromyces marxianus: A Comparative Aspect. Biophysics 2018, 63, 866–875. [Google Scholar] [CrossRef]
- Patila, M.; Chalmpes, N.; Dounousi, E.; Stamatis, H.; Gournis, D. Use of functionalized carbon nanotubes for the development of robust nanobiocatalysts. Methods Enzymol. 2020, 630, 263–301. [Google Scholar] [CrossRef]
- Chalmpes, N.; Patila, M.; Kouloumpis, A.; Alatzoglou, C.; Spyrou, K.; Subrati, M.; Polydera, A.C.; Bourlinos, A.B.; Stamatis, H.; Gournis, D. Graphene Oxide–Cytochrome c Multilayered Structures for Biocatalytic Applications: Decrypting the Role of Surfactant in Langmuir–Schaefer Layer Deposition. ACS Appl. Mater. Interfaces 2022, 14, 26204–26215. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhuang, S. Antibacterial activity of chitosan and its derivatives and their interaction mechanism with bacteria: Current state and perspectives. Eur. Polym. J. 2020, 138, 109984. [Google Scholar] [CrossRef]
- Kumar, M.; Muzzarelli, R.A.A.; Muzzarelli, C.; Sashiwa, H.; Domb, A.J. Chitosan chemistry and pharmaceutical perspectives. Chem. Rev. 2004, 104, 6017–6084. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Du, Y.M.; Xu, Y.M. Adsorption and complexation of chitosan wet-end additives in papermaking systems. J. Appl. Polym. Sci. 2004, 91, 2642–2648. [Google Scholar] [CrossRef]
- Muzzarelli, R.A.A.; Muzzarelli, C. Chitosan Chemistry: Relevance to the Biomedical Sciences. In Polysaccharides I. Advances in Polymer Science; Heinze, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 186. [Google Scholar] [CrossRef]
- Silva, D.F.; Rosa, H.; Carvalho, A.F.A.; Oliva-Neto, P. Immobilization of Papain on Chitin and Chitosan and Recycling of Soluble Enzyme for Deflocculation of Saccharomyces cerevisiaefrom Bioethanol Distilleries. Enzyme Res. 2015, 2015, 573721. [Google Scholar] [CrossRef] [Green Version]
- Zappino, M.; Cacciotti, I.; Benucci, I.; Nanni, F.; Liburdi, K.; Valentini, F.; Esti, M. Bromelain immobilization on microbial and animal source chitosan films, plasticized with glycerol, for application in wine-like medium: Microstructural, mechanical and catalytic characterisations. Food Hydrocoll. 2015, 45, 41–47. [Google Scholar] [CrossRef]
- Jiang, T.; James, R.; Kumbar, S.G.; Laurencin, C.T. Chitosan as a Biomaterial. In Natural and Synthetic Biomedical Polymers, 1st ed.; Kumbar, S.G., Laurencin, C.T., Deng, M., Eds.; Elsevier: Burlington, VT, USA, 2014. [Google Scholar]
- Akpan, E.I.; Gbenebor, O.P.; Adeosun, S.O.; Cletus, O. Solubility, degree of acetylation, and distribution of acetyl groups in chitosan. In Handbook of Chitin and Chitosan. Volume 1: Preparation and Properties; Gopi, S., Thomas, S., Pius, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Niu, X.; Zhu, L.; Xi, L.; Guo, L.; Wang, H. An antimicrobial agent prepared by N-succinyl chitosan immobilized lysozyme and its application in strawberry preservation. Food Control 2019, 108, 106829. [Google Scholar] [CrossRef]
- Zhong, Z.; Chen, R.; Xing, R.; Chen, X.; Liu, S.; Guo, Z.; Ji, X.; Wang, L.; Li, P. Synthesis and antifungal properties of sulfanilamide derivatives of chitosan. Carbohyd. Res. 2007, 342, 2390–2395. [Google Scholar] [CrossRef]
- Holyavka, M.; Faizullin, D.; Koroleva, V.; Olshannikova, S.; Zakhartchenko, N.; Zuev, Y.; Kondtatyev, M.; Zakharova, E.; Artyukhov, V. Novel biotechnological formulations of cysteine proteases, immobilized on chitosan. Structure, stability and activity. Int. J. Biol. Macromol. 2021, 180, 161–176. [Google Scholar] [CrossRef]
- Holyavka, M.G.; Kondratyev, M.S.; Samchenko, A.A.; Kabanov, A.V.; Komarov, V.M.; Artyukhov, V.G. In silico design of high-affinity ligands for the immobilization of inulinase. Comput. Biol. Med. 2016, 71, 198–204. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Faar, A.L.; Randall, R.J. Protein measurement with Folin-phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Artyukhov, V.G.; Kovaleva, T.A.; Kholyavka, M.G.; Bityutskaya, L.A.; Grechkina, M.V. Thermal inactivation of free and immobilized inulinase. Appl. Biochem. Microbiol. 2010, 46, 385–389. [Google Scholar] [CrossRef]
- Sabirova, A.R.; Rudakova, N.L.; Balaban, N.P.; Ilyinskaya, O.N.; Demidyuk, I.V.; Kostrov, S.V.; Rudenskaya, G.N.; Sharipova, M.R. A novel secreted metzincin metalloproteinase from Bacillus intermedius. FEBS Lett. 2010, 584, 4419–4425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellamy, L.J. The Infra-Red Spectra of Complex Molecules, 1st ed.; Methuen & Co. Ltd.: London, UK; John Wiley & Sons, Inc.: New York, NY, USA, 1958. [Google Scholar]
- Sorokin, A.; Lavlinskaya, M. Synthesis of the superabsorbents enriched in chitosan derivatives with excellent water absorption properties. Polym. Bull. 2022, 79, 407–427. [Google Scholar] [CrossRef]
- Tantala, J.; Thumanu, K.; Rachtanapun, C. An assessment of antibacterial mode of action of chitosan on Listeria innocua cells using real-time HATR-FTIR spectroscopy. Int. J. Biol. Macromol. 2019, 135, 386–393. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, T.; He, R.; Ding, Y.; Duan, T.; Xiao, B. Understanding the interfacial interactions of bioinspired chitosan-calcite nanocomposites by first principles molecular dynamics simulations and experimental FT-IR spectroscopy. Carbohyd. Polym. 2019, 223, 115054. [Google Scholar] [CrossRef]
- Baumann, H.; Faust, V. Concepts for improved regioselective placement of O-sulfo, N-sulfo, N-acetyl, and N-carboxymethyl groups in chitosan derivatives. Carbohyd. Res. 2001, 331, 43–57. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernandez-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef] [Green Version]

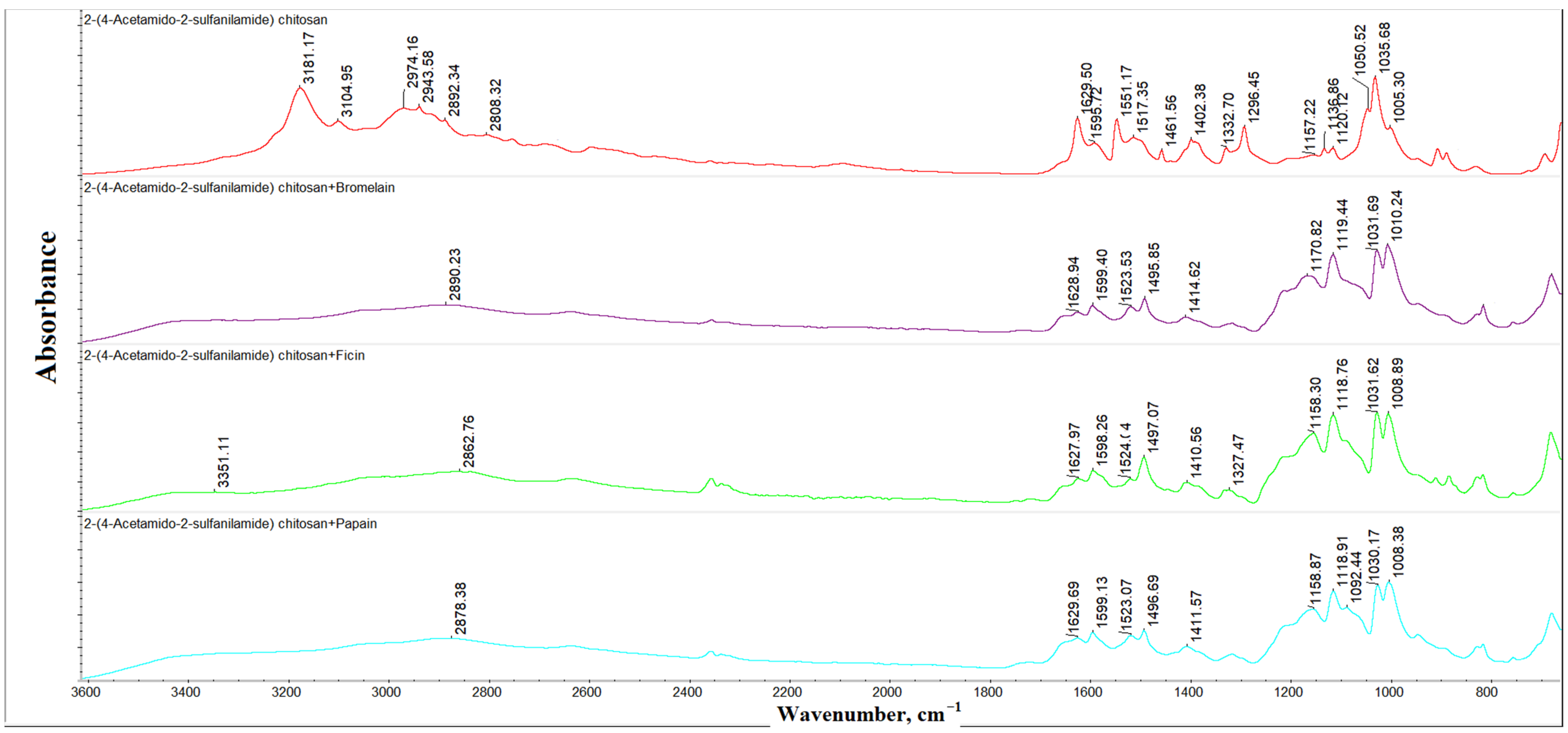
| Wavenumber, cm−1 | Band Description |
|---|---|
| 1005 | Planar deformation vibrations of the C-H bonds of the 1,4-disubstituted aromatic cycles |
| 1035 | Vibrations of the C-O-C bonds in the pyranose cycles |
| 1050 | |
| 1120 | Planar deformation vibrations of the C-H bonds of the 1,4-disubstituted aromatic cycles |
| 1136 | Stretching symmetric vibrations of the SO2 groups |
| 1157 | Stretching vibrations of the 1,4-glycosidic bonds |
| 1296 | Vibrations of the SO2 groups |
| 1332 | Amide III, stretching vibrations of the C-N bonds |
| 1402 | Stretching asymmetric vibrations of the SO2 groups |
| 1461 | Stretching breathing vibrations of the benzene cycles |
| 1517 | |
| 1551 | Amide II, deformation vibrations of the N-H bonds |
| 1595 | Deformation planar vibrations of the primary amine N-H bonds |
| 1629 | Amide I, stretching vibrations of the C=O bonds |
| 2758 | Stretching symmetric vibrations of the methylene groups |
| 2808 | |
| 2892 | |
| 2934 | Stretching asymmetric vibrations of the methylene groups |
| 2974 | Stretching asymmetric vibrations of the methyl groups |
| 3104 | Stretching vibrations of the C-H bonds in aromatic cycles |
| 3181 |
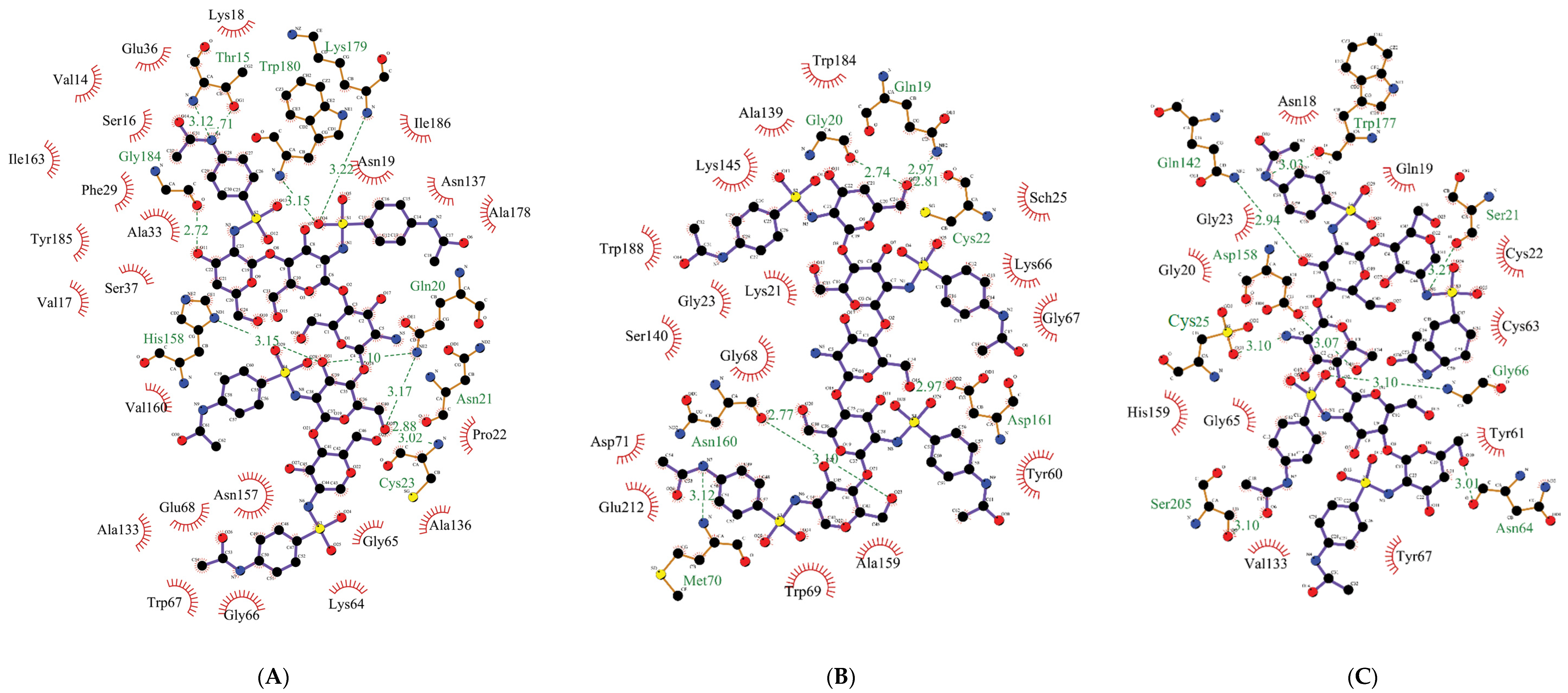
| Affinity, kcal/mol | Amino Acid Residues Forming | ||
|---|---|---|---|
| H-Bonds, Length, Å | Other Interactions | ||
| (1) Bromelain | |||
| −10.0 | Thr15 Gln20 Asn21 Cys23 His158 Lys179 Trp180 Gly184 | 3.12 3.17 2.88 3.02 3.15 3.22 3.15 2.72 | Val14, Ser16, Val17, Lys18, Asn19, Pro22, Phe29, Ala33, Glu36, Ser37, Lys64, Gly65, Gly66, Trp67, Glu68, Ala133, Ala136, Asn137, Asn157, Val160, Ile163, Ala178, Tyr185, Ile186 |
| (2) Papain | |||
| −7.4 | Ser21 Cys25 Asn64 Gly66 Gln142 Asp158 Trp177 Ser205 | 3.27 3.10 3.01 3.10 2.94 3.07 3.03 3.10 | Asn18, Gln19, Gly20, Cys22, Gly23, Tyr61, Cys63, Gly65, Tyr67, Val133, His159 |
| (3) Ficin | |||
| −7.9 | Gln19 Gly20 Cys22 Met70 Asn160 Asp161 | 2.97 2.74 2.81 3.12 2.77 2.97 | Lys21, Gly23, Sch25, Tyr60, Lys66, Gly67, Gly68, Trp69, Asp71, Ala139, Ser140, Lys145, Ala159, Trp184, Trp188, Glu212 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olshannikova, S.S.; Malykhina, N.V.; Lavlinskaya, M.S.; Sorokin, A.V.; Yudin, N.E.; Vyshkvorkina, Y.M.; Lukin, A.N.; Holyavka, M.G.; Artyukhov, V.G. Novel Immobilized Biocatalysts Based on Cysteine Proteases Bound to 2-(4-Acetamido-2-sulfanilamide) Chitosan and Research on Their Structural Features. Polymers 2022, 14, 3223. https://doi.org/10.3390/polym14153223
Olshannikova SS, Malykhina NV, Lavlinskaya MS, Sorokin AV, Yudin NE, Vyshkvorkina YM, Lukin AN, Holyavka MG, Artyukhov VG. Novel Immobilized Biocatalysts Based on Cysteine Proteases Bound to 2-(4-Acetamido-2-sulfanilamide) Chitosan and Research on Their Structural Features. Polymers. 2022; 14(15):3223. https://doi.org/10.3390/polym14153223
Chicago/Turabian StyleOlshannikova, Svetlana S., Nataliya V. Malykhina, Maria S. Lavlinskaya, Andrey V. Sorokin, Nikolay E. Yudin, Yulia M. Vyshkvorkina, Anatoliy N. Lukin, Marina G. Holyavka, and Valeriy G. Artyukhov. 2022. "Novel Immobilized Biocatalysts Based on Cysteine Proteases Bound to 2-(4-Acetamido-2-sulfanilamide) Chitosan and Research on Their Structural Features" Polymers 14, no. 15: 3223. https://doi.org/10.3390/polym14153223
APA StyleOlshannikova, S. S., Malykhina, N. V., Lavlinskaya, M. S., Sorokin, A. V., Yudin, N. E., Vyshkvorkina, Y. M., Lukin, A. N., Holyavka, M. G., & Artyukhov, V. G. (2022). Novel Immobilized Biocatalysts Based on Cysteine Proteases Bound to 2-(4-Acetamido-2-sulfanilamide) Chitosan and Research on Their Structural Features. Polymers, 14(15), 3223. https://doi.org/10.3390/polym14153223








