Abstract
Infective endocarditis (IE) is a heart disease caused by the infection of heart valves, majorly caused by Staphilococcus aureus. IE is initiated by bacteria entering the blood circulation in favouring conditions (e.g., during invasive procedures). So far, the conventional antimicrobial strategies based on the usage of antibiotics remain the major intervention for treating IE. Nevertheless, the therapeutic efficacy of antibiotics in IE is limited not only by the bacterial drug resistance, but also by the formation of biofilms, which resist the penetration of antibiotics into bacterial cells. To overcome these drawbacks, the development of anti-biofilm treatments that can expose bacteria and make them more susceptible to the action of antibiotics, therefore resulting in reduced antimicrobial resistance, is urgently required. A series of anti-biofilm strategies have been developed, and this review will focus in particular on the development of anti-biofilm antibodies. Based on the results previously reported in the literature, several potential anti-biofilm targets are discussed, such as bacterial adhesins, biofilm matrix and bacterial toxins, covering their antigenic properties (with the identification of potential promising epitopes), functional mechanisms, as well as the antibodies already developed against these targets and, where feasible, their clinical translation.
1. Introduction
1.1. The Background of IE and Its Modern Epidemiology
Infective endocarditis (IE) is an infectious heart disease, with an incidence of three to ten episodes per 100,000 per year in the population [1,2,3]. Over the years, the mortality of IE changed with advances in medicine: IE was indeed a highly fatal disease up to the 1940s, when the introduction of penicillin greatly reduced its mortality [4,5]. Later on, the development of valvular surgery sparked new modes of intervention, defining the beginning of the early surgery era [6]. Although the majority of IE cases can be treated with antibiotics, about 25–30% of patients develop severe valvular damage or exhibit ineffectiveness in response to antibiotic treatments, and therefore surgery is required during the early-acute phase of infection. Another 20 to 40% of these patients require surgery later [7].
Concurrently, the ongoing development of antibiotics and surgical interventions renews our understanding of the epidemiology of IE. The changes in antibiotics have altered the patterns of infection and caused resistance worldwide [8,9,10]. On the other hand, in comparison with the pre- and early antibiotic age in which rheumatic heart disease was a leading cause of IE, nowadays, other risk factors are taken into consideration, such as the use of intracardiac devices, prosthetic valve replacement, venous catheters, or haemodialysis. Moreover, the average age of IE patients and the prevalence of comorbidities among them have increased [11,12,13].
At the time of writing, IE is still a serious disease for its high mortality, since 20% of patients die in hospital, and its one-year mortality rate is about 30% [1,2]. Despite the development of trends toward earlier diagnosis and early surgery, the mortality of IE has barely improved in the last four decades, and its one-year mortality has not improved in over two decades [7,14]. So far, the current management of IE is still challenged by difficulties in both prognosis and treatment.
1.2. The Clinical Pathology of IE
As an infectious heart disease, IE is initiated by the bacterial infection of heart valves. Bacteria can enter the bloodstream via the skin, the mouth, gastrointestinal and urinary tracts, through venous catheters or after an invasive medical or surgical procedure. After that, bacteria can reach the heart via body circulation and then adhere to the valves. This will lead to the formation of holes on the valve, or scarring of the valve tissue, which consequently will (or can) result in valve leakage [7,15]. The common signs and symptoms of IE include: chest pain, fatigue, flu-like symptoms and abnormal heart murmurs [15]. The characteristic lesions of IE are vegetations consisting of microorganisms, inflammatory cells, platelets, fibrin and leaflet disruptions. IE might be extended by local spread of the infection, which leads to other complications (Figure 1) [16].
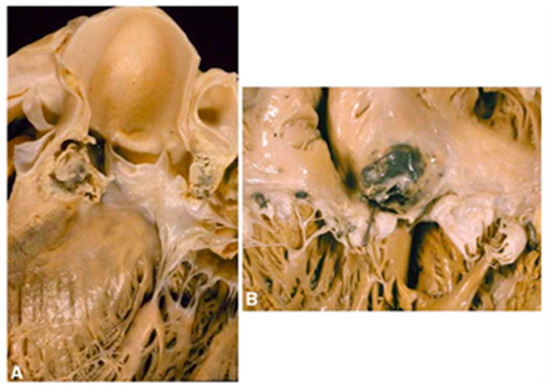
Figure 1.
The local spreading of IE infection. IE originates from the aortic valve and can spread to the ventricular wall (A) and atrium wall (B). Adapted with permission from Thiene and Basso [16]. 2006, Elsevier.
Since IE can occur on both original and prosthetic valves, it is further classified as native valve endocarditis (NVE) and prosthetic valve endocarditis (PVE). In the case of NVE, deformed or damaged endothelium is more accessible for bacterial infection than the healthy one, and IE could develop following injury or thrombosis. Indeed, non-bacterial thrombotic endocarditis (NBTE), which is distinguished from IE for its non-infective pathogenesis, has now been identified as a crucial factor underlying the development of IE. Sterile thrombotic vegetations caused by NBTE are predisposed for bacterial adhesion on valve surfaces; therefore, the entry of microorganisms into the mainstream circulation could convert NBTE into IE. Further, injury to the endocardium and vascular endothelium may generate predilection sites of infection even in the absence of sterile thrombotic vegetations [17].
PVE is initiated by microorganisms invading the prosthesis during surgery or following haematogenic dissemination after surgery (a rare and serious complication of valve replacement). Similar to the pathology of NVE, thrombosis also acts as an underlying inducement for PVE. Thrombus formation on the prosthetic valve counts on a range of factors, including endothelisation, haemodynamics and haemostasis [18]. First of all, the endothelisation of a prosthetic valve takes several weeks to be completed. Therefore, in the early post-operative period, the absence of endothelium could raise the thrombotic risk. Secondly, the altered haemodynamics after valve replacement can contribute to generating turbulent flow in localised regions, which can then lead to stasis and thrombosis on the prosthetic valve. Additionally, the clotting on the valve caused by blood stasis is critical to thrombus formation, and therefore anticoagulant treatment is required for patients after surgery. Eventually, the thrombus composed of fibrin and thrombocytes can serve as a predisposing focus of infection. Taking all of these factors into consideration, although NVE is distinguished from PVE, there are a number of similarities in their pathologies.
1.3. The Challenges in Antibiotic Treatment and Prospective Anti-Biofilm Strategies
IE can be caused by a range of micro-organisms, but Staphylococci are now considered the most frequent causative organisms, since they represent the major pathogens for hospital-acquired infections [4,15,19,20,21,22,23]. Therefore, Staphylococci have become the main targets for antibiotic therapy for this condition. However, the therapeutic efficiency of antibiotics for IE has been challenged by the emergence of resistant Staphylococci since their early introduction. As an example, only a few years after the introduction of penicillin, resistant staphylococcal strains expressing β-lactamase were reported [24]. The introduction of methicillin (a synthetic β-lactamase-resistant penicillin) facilitated the development of Methicillin-resistant Staphylococcus aureus (MRSA) strains [25,26,27]. To compete with the insurgence of resistant bacterial strains, new antibiotics and different combinations of synergistic antibiotics have been tested to improve IE treatment. Importantly, in addition to the acquired drug resistance, virulence factors also critically contribute to the modulation of antibiotic susceptibility in Staphylococci; indeed, IE is particularly difficult to treat due to the ability of MRSA to hinder the action of antibiotics via the formation of biofilms [28,29].
A biofilm can be described as a sessile community of micro-organisms in which cells embed together by attaching to others and a surface in a protective extracellular polymeric matrix, also known as extracellular polymeric substance (EPS). The extracellular matrix of biofilms can effectively increase tolerance to antibiotics via multiple mechanisms [30]. The EPS in staphylococcal biofilms is composed of polysaccharides, extracellular DNA (eDNA) and/or proteins [17,18,31,32,33]. The formation of a biofilm can be generally summarised into three main stages: attachment, maturation and dispersion (Figure 2).
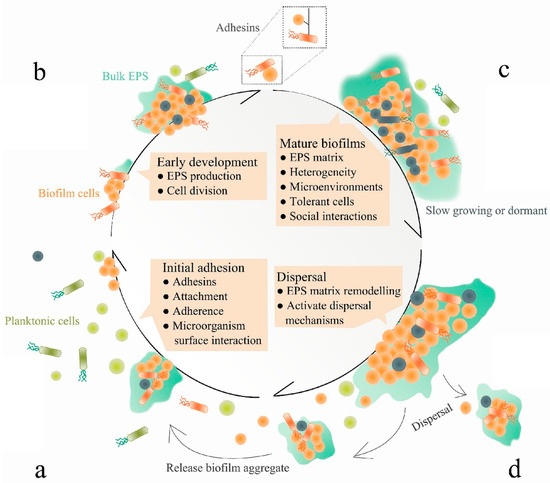
Figure 2.
The molecular mechanisms of biofilm formation. Attachment (a), biofilm formation (b), maturation (c) and dispersion (d). Adapted from Jiang et al. [34]. The different colours and shapes model the different bacterial cells composing the biofilm.
Firstly, attachment is initiated once an individual planktonic bacterial cell reversibly associates with a surface, and such association will become irreversible if the cell remains in contact for a sufficient period of time. Bacterial attachment is mediated by surface proteins such as microbial surface components recognising adhesive matrix molecules (MSCRAMMs) [35]. However, the importance and involvement of these proteins varies largely between strains, and many of these factors function in both attachment and accumulation [34]. Following attachment, biofilm maturation is progressed by bacterial division and their production of the extracellular polymeric matrix, enabling the transition of bacteria from a planktonic to a sessile state. Finally, bacteria within the biofilm can return to the planktonic state; hence, they can disassemble to undergo dispersion [36].
All stages of biofilm formation can be targeted for anti-biofilm therapeutic purposes. For example, the initial attachment can be prevented by targeting staphylococcal adhesions [37]; biofilm maturation can be disturbed by blocking surface proteins involved in cell-to-cell adhesion [38]; and finally, the pre-existing biofilm can be decomposed by using dispersal agents, such as cis-2-decenoic acid (C2DA), dispersin B and DNase I [35].
Since anti-biofilm strategies target the molecular pathways involved in the biofilm’s formation and maturation, which is different from the conventional antimicrobial routes, this translates into a significantly reduced selection pressure, which in turn mitigates the potential development of resistance. Further, since factors involved in staphylococcal biofilm formation are highly species-specific (compared to targets for conventional antibiotics), anti-biofilm strategies may allow for the development of narrow-spectrum precision agents, which will have low or no influence on other microbiota [34]. To date, a wide range of molecular targets involved in biofilm formation are being investigated, and the combination of antibiotics and anti-biofilm therapies is likely to be more effective than a single treatment.
1.4. Antibodies as a Promising Approach for Anti-Biofilm IE Treatment
In addition to the molecular agents that could inhibit staphylococcal biofilm formation, different alternative strategies can be used, such as monoclonal antibodies (mAbs) treatment. When staphylococcal infections occur, the immune system generates antibodies against a wide range of antigens, including surface proteins, toxins and cell wall proteins [36,39,40,41]. These antibodies can disrupt the biofilm’s formation via different immunological mechanisms (Figure 3).
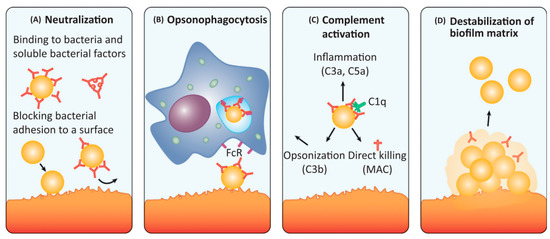
Figure 3.
The antibodies against biofilm can disrupt ongoing biofilm formation and/or disperse established biofilms by different mechanisms: (A) neutralisation, (B) opsonophagocytosis, (C) complement activation and (D) destabilisation of biofilm matrix. Adapted with permission from Raafat et al. [42]. 2019, Elsevier.
A neutralising antibody is an antibody that can block the infectious and pathogenic effects of microbes. High-affinity IgA and IgG antibodies can neutralise the action of secreted staphylococcal proteins, such as immune evasion molecules, exoenzymes, toxins and surface proteins, which are potential therapeutic targets. Further, neutralising antibodies can in turn bind to bacterial adhesins (e.g., MSCRAMMs) and cell wall components to inhibit initial attachment to host matrixes and subsequent initiation of biofilm formation (Figure 3A). Another type of mechanism, known as opsonophagocytosis (i.e., opsonophagocytic-killing, OPK), entails antibodies that can mediate microbial clearances by phagocytes. These surface-bound antibodies (mainly IgG) can trigger OPK by neutrophils and macrophages expressing FcR on their surface. Once antibodies bind to the antigens, their Fc regions are able to activate phagocytes to engulf pathogens (Figure 3B). Similarly, microbial clearance can also be mediated via activating the complement system. The classical complement pathway is activated once the surface-bound antibodies (IgM and IgG) bind to the C1q subunit on the C1 complex, and then the following cascade leads to the formation of the C3 convertase, which cleaves C3 (the central component of all complement pathways) into C3a and C3b. Finally, C3b acts as an opsonin to enable phagocytosis via C3b receptor-expressed phagocytes ingesting C3b-coated pathogens; the soluble C3a (also known as C5a) acts as chemoattractant to recruit immune cells to initiate inflammation. C3 activation also causes the formation of the membrane attack complex (MAC) that can mediate lysis of certain pathogens. These all contribute to the killing of pathogens (Figure 3C). Additionally, antibodies targeting different components of the biofilm matrix can directly destabilise the biofilm structure and thus promote bacterial dispersal (Figure 3D).
The antibodies naturally produced by the host are highly specific, therefore highlighting the potential of antibody therapy as a feasible option for narrow-spectrum anti-biofilm treatment. So far, the feasibility of antibody therapies disrupting staphylococcal biofilms has been shown by some studies, in which different antigens have been targeted, such as surface proteins [43,44,45,46,47], toxins [45], cell wall enzymes [48] and poly-N-acetyl-β-(1,6)-glucosamine (PNAG) [47]. Importantly, the major disadvantage of antibodies therapy, as in the case of antibiotics, is related to the poor penetration into the deepest layers of a biofilm, which would result in incomplete disruption. Moreover, development of resistance is possible, although less likely than with antibiotics. Another hurdle faced by the development of antibodies for anti-biofilm IE treatment is that different Staphylococci strains express different antigens; therefore, finding a universal target is difficult, which renders the antibody development quite challenging [49].
This review, therefore, aims to identify the most promising targets for anti-biofilm IE treatment, with particular emphasis on antibody-based therapy. We will do so by summarising and evaluating the potential targets for anti-biofilm IE treatment, highlighting the most promising ones for antibody development and discussing the challenges in antibody design and development for IE treatment.
2. Molecular Targets for Monoclonal Antibodies Targeting Staphylococcus Biofilms
So far, the natural immunological reactions triggered by biofilm-associated infection are not well understood. Importantly, host antibodies stimulated by S. aureus antigens show reduced efficiency in preventing a reinfection with this pathogen [50]. As mentioned above, the process of biofilm formation can be summarised into three stages in which a wide range of functional proteins are involved. Given the importance of biofilm formation in the pathology of IE, current research on antibodies targeting S. aureus infections has included anti-biofilm strategies. So far, some bacterial proteins contributing to S. aureus biofilm formation have been considered as candidates for developing IE treatments, such as adhesins, biofilm matrix components, cell wall-modifying enzymes, surface glycopolymers and toxins (Figure 4).
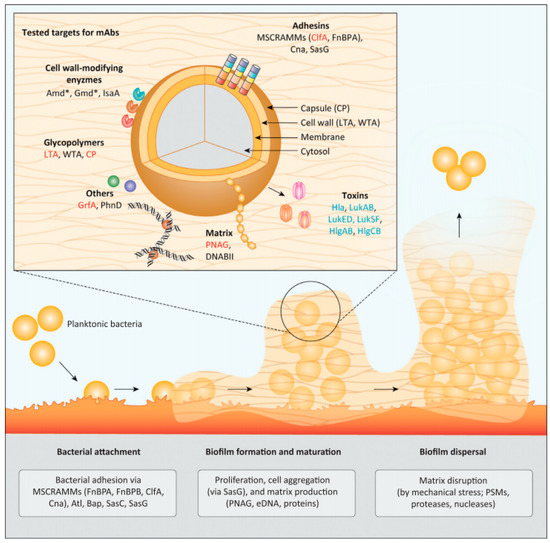
Figure 4.
Potential molecular targets involved in Staphylococcus biofilm formation. The biofilm could be disrupted by generating antibodies against functional proteins involved in each stage of biofilm formation. Adapted with permission from Raafat et al. [42]. 2019, Elsevier.
The functions of these candidates as well as their clinical studies are summarised in Table 1.

Table 1.
Studies and clinical trials on potential anti-biofilm targets and antibodies. Adapted with permission from Raafat et al. [42]. 2019, Elsevier.
3. Anti-Biofilm Strategies
3.1. Inhibition of Bacterial Attachment
The first stage of biofilm formation, bacterial attachment, relies on a number of staphylococcal surface-binding proteins, including MSCRAMMs and other cell wall-associated proteins [e.g., biofilm-associated protein (Bap), S. aureus surface proteins C (SasC) and G (SasG)] (Figure 4) [92]. The initial attachment in vivo is mainly driven by the interaction between MSCRAMMs and the human extracellular matrix (Figure 4) [93]. Therefore, MSCRAMMs are taken as candidates for IE antibody-based therapies and, ideally, their antibodies are supposed to function via a mechanism that prevents the initial bacterial adherence to both abiotic and biotic surfaces by neutralising adhesins and/or promoting OPK [50,84,94].
Among the family of MSCRAMMs, ClfA, ClfB and the fibronectin-binding proteins (FnBPA and FnBPB), are widely found among the S. aureus strains, and other proteins such as collagen-binding protein (Cna) only distribute in a subset of strains [95]. Importantly, however, in comparison to MSCRAMM ClfA, ClfB presents a relatively high frequency of pseudogenes (i.e., DNA sequences that resemble a gene but have been mutated into an inactive form over the course of evolution). This reveals its decreasing importance in the MSCRAMMs family, therefore indicating that ClfB is less suitable as a target for IE antibodies [96].
3.1.1. ClfA: Past Failure of Anti-ClfA Antibodies Enlightens Further Research
The MSCRAMM ClfA is a virulence factor that critically contributes to the colonisation of S. aureus on protein-coated biomaterials and damaged endothelial surfaces by binding to blood plasma protein fibrinogen (Fg). Further, ClfA is found to predominantly promote staphylococcal adhesion under high shear stress [97]. To understand the properties of ClfA that render it an effective target for IE, it is critical to consider its functional mechanism and protein structure, as well as its genetic variations.
As a member of MSCRAMMs, ClfA shares a similar domain organisation and structure. Starting from the N-terminus, ClfA contains a signal sequence followed by a ligand-binding N-terminal A region (amino acids 40 to 559) subdivided into independently folded N1, N2 and N3 subdomains, a region consisting of repeated serine-aspartate residues, and a C-terminal region containing an LPXTG motif (Figure 5) [98,99,100].
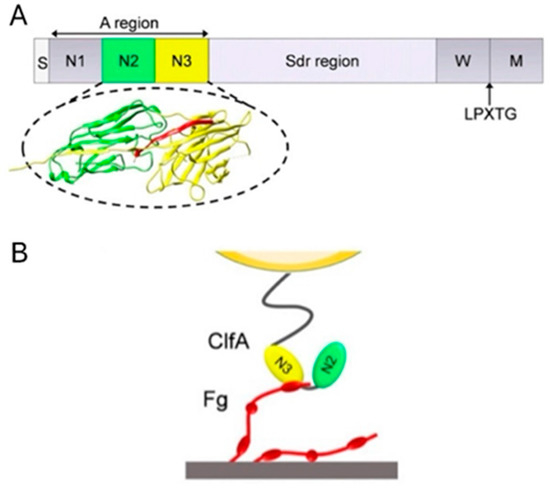
Figure 5.
The protein structure of ClfA and its functional mechanism. (A) From the N- to C- terminus, ClfA is composed of signal sequence (S), A region with subdomains N1, N2 and N3, repeated serine-aspartate residues (Sdr) region, wall spanning (W) region, LPXTG motif (LPXTG) and sorting sequence (M). The binding site of Fg is between N2 (green) and N3 (yellow). (B) The γ-chain of Fg (red) binds to the trench between N2 and N3. Adapted from Herman-Bausier et al. [97].
These components are responsible for its structural function and mechanism. The junction between N2 and N3 is found as a binding trench where the ligand inserts and binds with ClfA. The Sdr region links region A to the C-terminal wall-spanning region and the sorting sequence. The LPXTG motif allows anchoring of the protein to cell wall peptidoglycan by sortase A. The ClfA—Fg interaction occurs via ClfA binding its minimal ligand-binding segment N2 and N3 to the carboxy-terminus of the γ-chain of Fg through variations of a dynamic mechanism termed “Dock, Lock and Latch” (DLL): Firstly, the carboxy-terminus of the γ-chain of Fg docks in a ligand-binding trench located between subdomains N2 and N3 (amino acids 221 to 559). Once the ligand peptide docks into the trench, it is subsequently covered by a flexible C-terminal extension of the N3 domain and thus “locked” in place [101]. After that, the C-terminal part of this extension interacts with the N2 domain and forms an extra β-strand, which complements the pre-existing β-sheet in the N2 domain, and together they serve as a latch to stabilise the MSCRAMM—ligand complex.
The diversity of ClfA has been explored by studies on its variants. The results obtained from structural mapping of CflA subdomains reveal a minimum pairwise identity of 86%, which indicates a low level of structural differences among variants. Therefore, ClfA is considered to be well-conserved, and also one single isolate is predicted to be able to generate cross-reactive antibody responses against a wide range of variants [96]. However, this predication is questioned by a further study, which has shown that ClfA strains present lower binding affinities to heterologous antibodies elicited by their variants, with only 10% variation in aminoacid sequences [102]. This result highlights that small differences in composition across S. aureus strains could result in large effects on antigenicity.
Interestingly, as a member of the MSCRAMM family, S. aureus ClfA shows force sensitivity, and its binding to Fg is significantly enhanced by mechanical force, which shows that the ClfA—Fg binding is increased 15-fold in the presence of mechanical tension [97]. According to the same study, ClfA interacts with Fg via two distinct binding sites, and the stronger binding site is favoured by high shear stress [97]. This mechanical sensitivity of ClfA shows high biological significance to PVE. As mentioned in Section 1.2, the pathology of PVE is critically related to the changed haemodynamics after aortic valve replacement, which causes turbulent flow that can lead to elevated shear stress levels in the ascending aorta [103]. The elevated shear stress caused by the implantation of the prosthetic valve has been proved by a series of past studies, and a maximal stress of 500 N/m2 has been indicated [103,104]. This can probably benefit S. aureus attachment on the prosthetic valve through enhancement in ClfA—Fg binding. However, the confirmation of a potential association between the mechanical sensitivity of ClfA and the pathology of PVE would require deeper investigations.
For the reasons mentioned above, antibody treatments targeting MSCRAMM ClfA could possibly prevent the incidence of IE and especially PVE, and indeed, the therapeutic potential of anti-ClfA antibodies has been evidenced by many studies [44,50,51,84,105]. The application of tefibazumab, a humanised anti-ClfA mAb, contributed to the prevention of IE in a rabbit model [44]. However, tefibazumab has failed to achieve statistically significant improvement in a phase II human clinical trial (ClinicalTrials.gov Identifier: NCT00198289). Surprisingly, the tefibazumab epitope is shown to be located on top of the N3 domain instead of the trench discussed above [106]. Strikingly, this is consistent with the dual mechanisms model of ClfA under mechanical tension. Under low tensile force, Fg binds to the top of the ClfA N3 domain via weak bonds, whereas under high mechanical tension, extension and conformational changes in the ClfA molecule trigger the ultra-strong DLL interaction by the N2 and N3 subdomains, and the γ-chain peptide of Fg inhibits high forces but not low forces binding site model [97]. Therefore, it can be hypothesised that tefibazumab inhibits the function of ClfA by disrupting its low tension-dependent mechanism instead of the DLL mechanism that responds to a high shear stress environment. This hypothesis, to some extent, could also explain the failure of the phase II clinical trial, as S. aureus is exposed to many different levels of shear depending on its location and the type of infectious disease in patients. Notably, although tefibazumab shows therapeutic efficiency in an IE rabbit model, the medical scenario in patients might be much more complex due vascular ageing, calcification and the accompaniment of other cardiovascular diseases [107]. Studies also show that tefibazumab inhibits binding of Fg to ClfA rather than the Fg γ-chain, and residues P467A, Y512A and W518A in the N3 domain are shown to be critically involved in this binding. This provides additional target sites for the future design of effective inhibitors of the ClfA/Fg interaction. Further, as mentioned above, the failure of tefibazumab could be partially explained by its weakened binding affinity when confronting CflA variants from different strains of S. aureus [102,106]. A new anti-ClfA mAb (11h10) was identified by the group of Tkaczyk et al., and its combination with anti-toxin mAb shows more improved efficacy than single-neutralising mAb in responding to S. aureus biofilm infection [50,84]. The same research group also reported another anti-ClfA mAb (SAR114) with >100-fold increased affinity, as well as its combined construction with anti-toxin mAb, in the form of a bispecific antibody (BisAb) [45]. However, to the best of our knowledge, these two anti-ClfA mAbs have not been further investigated since their last publication in 2017. Importantly, though, functional antibodies generated using recombinant ClfA antigens can sufficiently alter the ligand-binding activity of ClfA [108].
3.1.2. FnBP: The Possibility of Developing FnBP Antibody Is Waiting to Be Addressed
FnBP plays multivalent roles in biofilm-associated S. aureus infection, as it not only contributes to bacterial adhesion by binding to human plasma proteins, but also promotes bacterial invasion, intercellular accumulation and biofilm maturation [54,93]. In accordance with MSCRAMM ClfA, FnBP shares a similar protein composition, and it also follows the DLL mechanism mediated by the A domain binding site when interacting with Fg (Figure 6).
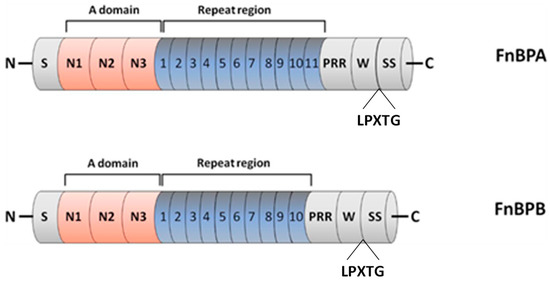
Figure 6.
Domain organisation of FnBPA and FnBPB. From the N- to C- terminus, the FnBPs are composed of signal sequence (S), A domain with subdomains N1, N2 and N3, repeat region that is an unstructured region consisting of tandemly arranged motifs (11 in FnBPA and 10 in FnBPB), proline repeated region (PRR), wall-spanning (W) region, LPXTG motif (LPXTG) and sorting sequence (SS). Adapted with permission from O’Neill et al. [54]. 2008, American Society for Microbiology.
FnBPA and FnBPB proteins also contain a signal sequence at the N-terminus and an A domain composed of three separately folded subdomains, termed N1, N2 and N3, as well as a wall-spanning region and a sorting signal at the C-terminus. Interestingly, an extra binding site of Fg on the top of the N3 subdomain is also identified in FnBP [106]. In contrast to ClfA, it contains a binding site of fibronectin (Fn) in the repeat region, and it has another binding site of plasminogen (Plg) in the A domain, as a single subdomain was required for Plg binding to FnBPs: subdomain N2 for FnBPA and subdomain N3 for FnBPB [109]. Additionally, it exhibits a much greater level of diversity in its subdomains in comparison to ClfA [110,111]. Noticeably, FnBP can promote biofilm formation, but the underlying multivalent mechanisms are still unclear. This has been firstly explained by a low affinity binding between the A domains of FnBP on adjacent bacteria, which can contribute to the aggregation of bacteria [54]. However, a recent study shows that FnBP can contribute to biofilm formation through a previously unknown mechanism that is distinct from its ligand-binding ability as a member of the MSCRAMMs family [112]. Considering its multivalent functions, FnBP is suggested to be a potent candidate to prevent S. aureus infection.
Early studies have highlighted the potential of FnBP antibody therapies, as well as its OPK activity [108,113]. Later, the efficacy of FnBP antibodies was investigated in vivo. An anti-FnBPB mAb (15E11) has been identified in a murine model, with a successful inhibition of bacterial attachment by 70% [114]. The mAb 15E11 binds to an epitope shared by the repeated regions in both FnBPA and FnBPB, which is proximal to (though distinct from) the Fn binding site. Furthermore, the sequence KYEQ(H)GGNIV(I)D in the epitope is thought to be crucial. The authors suggested that the steric hindrance or a conformational change elicited by 15E11 reduces the accessibility of Fn to its binding site. This also provides an extra pathway of interference with the action of FnBP, although further studies are required to clarify its underlying mechanism. In addition, an extra binding site of the Fg ligand on the top of the N3 subdomain is also observed in FnBP, and a force-induced binding mechanism similar to that of ClfA has been suggested [115]. So far, though, no FnBP antibodies are being investigated in clinical trials.
3.2. Decomposition of Biofilm Matrix
After successful attachment to the surface, bacteria start to proliferate and build biofilm by producing a series of EPSs, including polysaccharides (e.g., PNAG), nucleic acids [e.g., environmental DNA (eDNA)], proteins, lipids and other biomolecules (Figure 4). These matrix proteins support the structural integrity of the biofilm by developing a three-dimensional architecture, which in turn enhances biofilm tolerance to both antimicrobial agents and immune cells. So far, two EPSs, PNAG and DNABII, have been extensively considered as potential candidates for IE antibody-based treatment against biofilm-associated infections. They are currently evaluated as candidates for broad-spectrum antimicrobial therapeutics. This is partly due to the critical roles they play in biofilm composition, as well as their wide distributions and conserved properties observed among a variety of microbes, including bacteria, fungi and protozoa [33,113,116,117].
3.2.1. PNAG: The Antibody against PNAG/dPNAG Shows Optimal Anti-Biofilm Effect
PNAG is a major component of the biofilm EPS, also known as polysaccharide intercellular adhesin. PNAG critically mediates intercellular adhesion, thereby leading to the accumulation of bacterial cells, which eventually promote the establishment of biofilms. PNAG not only contributes to the biofilm matrix architecture on the implanted material surface, but also slows down the host defensive responses [118]. The chemical structure of PNAG is shown in Figure 7.
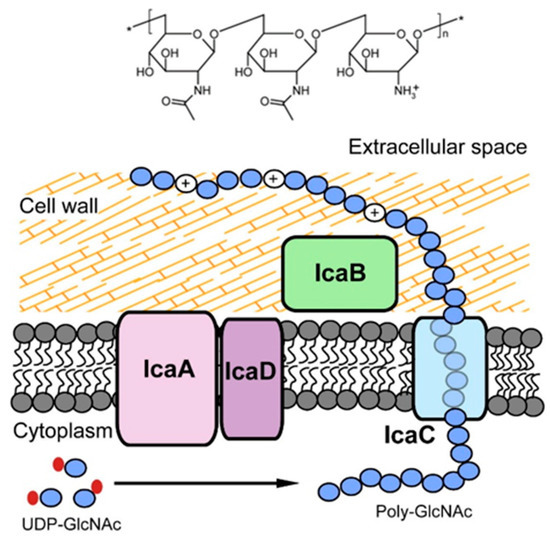
Figure 7.
Chemical structure of PNAG (top) and the Ica system (bottom), which is responsible for the synthesis, translocation and deacetylation of PNAG. Adapted with permission from Nguyen et al. [119]. 2020, Elsevier.
In the case of Gram-positive bacteria, the PNAG synthesis is mediated by the icaADBC locus consisting of four genes, which express four proteins assembling the intercellular adhesion system (Ica) including IcaA, IcaB, IcaC and IcaD (Figure 7). IcaA and IcaD exert primary roles in the exopolysaccharide synthesis. IcaA is a transmembrane enzyme with N-acetylglucosaminyl transferase activity, necessary for the synthesis of the PNAG polymer. However, the enzymatic activity of the product of the icaA gene becomes significant, and oligomers longer than 20 residues are synthesised only when co-expressed with the product of the icaD gene. IcaC translocates the PNAG polymer to the bacterial cell surface, and IcaB operates the deacetylation of the molecule.
Deacetylation of PNAG enables its fixation to the outer bacterial surface, promoting the structural development of exopolysaccharide-based biofilm. Importantly, although PNAG is certainly a critical element of biofilm formation in S. aureus, the existence of PNAG-independent biofilms has also been confirmed. Furthermore, it has been shown that a minor proportion of S. aureus strains can form biofilms even in the absence of the ica locus, which further suggests the existence of ica-independent PNAG-synthesis pathways [120]. As a carbohydrate antigen, the epitope of PNAG is expected to encompass parts of more than one repeating unit, and is often located at the ends of polysaccharide chains [121].
The feasibility of targeting PNAG has been illustrated by many studies [113,122]. Further, different from antibodies against PNAG that aim to decompose biofilm, the antibodies against dPNAG (deacetylated poly-N-β-(1-6)-acetyl-glucosamine) present marked efficacy in the opsonisation and killing of S. aureus [81,123,124]. Thus, dPNAG is so far considered a significant target for the development of antibodies aimed at treating IE. However, PNAGs naturally expressed on bacterial surfaces are highly acetylated (>90%) [125]. Structural studies of PNAG indicate that the acetates are on the polymer’s surface and sticking outwards into environment, exposing themselves as the primarily accessible antigens and thus enabling the immunodominance of the acetate-dependent epitopes [126].
As further shown by recent studies, the conjugation of anti-PNAG and anti-dPNAG antibodies achieving significant OPK and protective effects in vitro and in vivo shows the potential of combined antibodies therapy [117,127]. A human IgG1 mAb (F598, formerly SAR 279356) targeting both PNAG and dPNAG is undergoing preclinical and clinical assessments as a broad-spectrum antimicrobial therapy, since it triggers superior OPK and protective activities against a wide range of microbial pathogens compared to two mAbs that bound optimally to PNAG and minimally to dPNAG (mAbs F628 and F630) [79]. This phenomenon has been explained on the basis of the mAbs’ conformational significance. Once the two Fc regions from two antigen-bound antibodies bind to a C1 complex, the classical pathway of complement cascade is initiated, which is the main killing pathway for many Gram-positive bacterial species (Figure 3C). In the case of PNAG antibodies, as mentioned before, due to the outward positions of their acetate epitopes, the distances between the two Fc regions might not be adequate to bind to a C1 complex and thus fail to activate the complement cascade. However, the binding between the Fc regions of antibodies against both PNAG and dPNAG to the C1 complex is less restricted since their epitopes are located more randomly [126].
F598 recognises PNAG through a large groove-shaped binding site accommodating five N-acetyl-D-glucosamine (GlcNAc) residues as a penta-saccharide epitope, and their interaction is stabilised by two hydrogen bonds linking Asp-109H of F598 to the O3 and O4 atoms of the core GlcNAc (Figure 8).
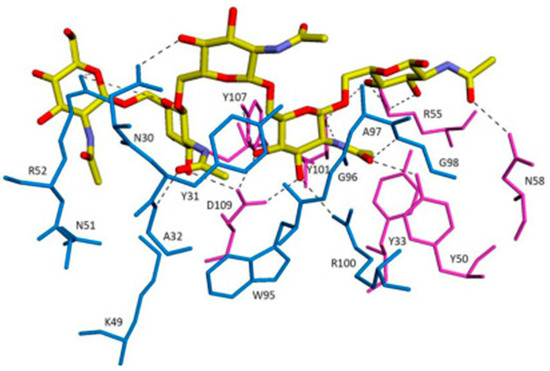
Figure 8.
The overall interactions between F598 antigen-binding fragment (Fab)–GlcNAc and Fab–9NAc crystal structures. Fabs are depicted as thin sticks for the L (blue) and H (magenta) chains. Carbohydrate ligands are shown as thicker sticks with carbon atoms of GlcNAc in green and 9NAc in yellow. Adapted with permission from Soliman et al. [128]. 2014, IUCr.
The Fab arms can span at least 40 GlcNAc residues on an extended PNAG chain [128]. Unfortunately, the clinical trial of F598/SAR 279356 was terminated due to difficulty in patient recruitment (ClinicalTrials.gov Identifier: NCT01389700). Recently, though, the advantage of the anti-biofilm strategy mentioned above has been proved by two studies showing that antibodies against PNAG do not perturb host microbial diversity [129,130]. The current research trends are aimed to develop antibodies against PNAG towards bacterial clearance instead of neutralisation, and both acetate-dependent and -independent epitopes are involved. This, however, brings difficulty to the antibody production process, since the ideal products are proposed to bind both the antigen and C1 complex.
3.2.2. DNABII: A Promising Antibody Target for Anti-Biofilm Treatment
Similar to PNAG, eDNA is also widely distributed among various microbes and critically involved in the construction of biofilm matrix as a part of EPS. The eDNA potentially acts as an electrostatic polymer that anchors cells to a surface via the negative charge it carries [129]. The structural integrity of those eDNAs in the biofilm matrix is conversely supported by bacterial DNA-binding proteins (DNABII family). The DNABII family includes HU proteins and integration host factor (IHF), condensing bacterial DNA and also acting as regulators in many cellular processes. HU and IHF have conserved homologs in a wide variety of bacterial species, and they share structural features and the key activity of DNA-bending [131].
The members of the HU family are typically small bacterial proteins (16~20 KDa) and exist as homodimers in Gram-positive bacteria such as S. aureus. The S. aureus HU (SHU) consists of a hydrophobic core composed of two α-helices and two negatively charged β-sheets arms, and therefore can be divided into two portions: the α-helical “body” and the β-ribbon “arms”. HU acts similarly to a histone through binding and supercoiling DNA into a circular structure; in addition, it also critically acts as a molecular glue, packing eDNA and stabilising the bacterial biofilm by similar patterns [132,133]. This glue-like ability for eDNA is supported by functional characteristics of HU, including a non-specific DNA binding profile [134], high abundance [135] and its ability to migrate into the extracellular medium through multiple mechanisms [124,136,137]. Another DNABII member, IHF, is also known as a DNA-“bending” protein that can create kinks in DNA strands. The DNA-binding events of IHF are more sequence-specific and are only observed in Gram-negative bacteria [138]. Overall, HU can be considered a potential target to decompose S. aureus biofilms, and the study of its interactions with DNA can assist the development of anti-DNABII antibodies.
The structure and the DNA-binding mechanisms of SHU have been established (PDB ID: 4QJU). The high flexibility of β-arm is observed in the binding of DNA, and the residue Arg55 positioned in the hinge region of the β-arm exhibits a critical role in their flexible nature. The C atom of Arg55 from chain D is used as the vertex of the angle (Figure 9A,B) [139].
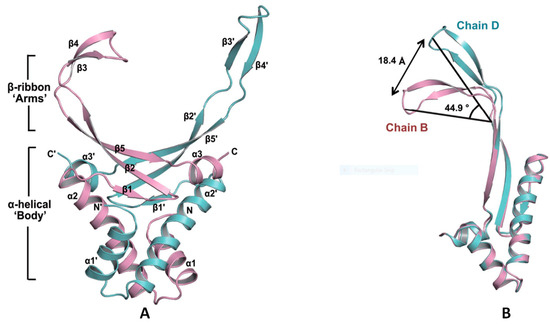
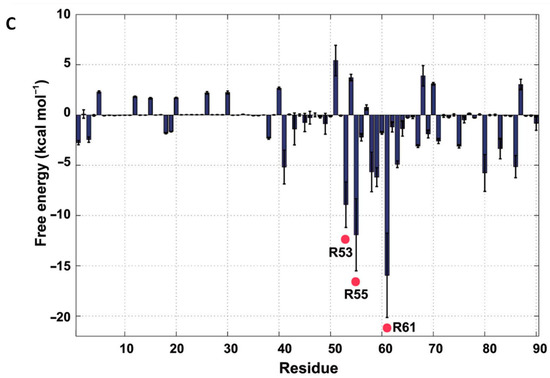
Figure 9.
HU structure and the arginine residues involved in its DNA-binding site. (A) Ribbon diagram of the SHU homodimer. The α–helices, β–sheets and the N– and C–termini are labelled. The residue Arg55 is labelled in a circle. (B) The flexible β–ribbon arms deviate with a distance of 18.4 Å and with a bending angle of approximately 44.9°. (C) The binding free energies in the SHU–DNA complex, as determined by MD simulation. The three Arg residues (Arg53, Arg55 and Arg61) that show low free energies are marked with red points. Adapted from Kim et al. [139].
Further, as shown by molecular dynamics analysis on the SHU—DNA complex, the β-arms residues Arg53, Arg55 and Arg61 present low free energy, especially for the Arg61, which suggests their essential role in recognising and binding DNA (Figure 9C). Based on these results, it can be summarised that these three arginine residues on the β-arms are essential for β-arm flexibility, which affects the DNA binding as well as the biological function. Additionally, the involvement of the arginine residues on the β-arms is universal in HU homologues. Interestingly, though, the two essential residues, Arg55 and Arg61, are not completely conserved, whereas Arg58 is conserved in all HU homologues even though it exhibits a relatively small affinity contribution to DNA binding. Further understanding the mechanism of these essential residues might provide us with helpful indications for the design of anti-biofilm agents including but not limited to anti-DNABII antibodies.
Based on its abundance and promising eDNA binding function among different bacteria, DNABII is considered a potent candidate for the development of antibodies. Neutralising of DNABII by specific antibodies has been shown to decompose the biofilm and thus promote the clearance of bacteria by antibiotics and immune cells [140]. Further, targeting DNABII attains high therapeutic efficiency in a wide range of biofilm-associated infections in vivo, which significantly indicates its therapeutic potential for polymicrobial IE in a clinical scenario [123,130,132,140]. So far, a native human mAb (TRL1068) generated by Estellés et al. shows anti-biofilm efficacy in two biofilm-associated infectious models that are in the setting of medical devices invasion [81,85]. The epitope of TRL1068 is part of the β-sheet capped with a β-turn in HU, and a specific binding sequence of AARKGRNPQTGKEID within the DNA-binding domain of HU has been identified [81]. This is consistent with the functional importance of β-arm mentioned before. Currently, TRL1068 is undergoing clinical trials (ClinicalTrials.gov Identifier: NCT04763759).
3.3. Targeting S. aureus Toxins as Supplemental Therapy
The formation of biofilms is partly assisted by various bacterial toxins, such as pore-forming toxins (e.g., α-toxin (AT), Leukocidin A/B (LukAB) and γ-hemolysin (HlgAB and HlgCB)), and some even produced higher amounts than that by planktonic cells (Figure 4) [85]. Since the pore-forming toxins could counter anti-biofilm immune response via mediating lysis of host immune cells, it has been postulated that mAbs neutralising toxins might promote host defences and clearance of planktonic and biofilm cells. AT is the most extensively studied target among these bacterial toxins.
AT Antibody: The Only Type of Antibody Currently Successful in Clinical Trials
S. aureus AT is a molecule of ~33 kDa, which exerts its virulence upon a two-step-mediated mechanism. It first binds to its receptor (ADAM10) on the surface of host immune cells and endothelial cells. After that, AT molecules undergo a conformational change to promote oligomerisation, which in turn results in the formation of membrane pores, followed by cell lysis and tissue damage. The AT heptameric complex is comprised of two cylinders: a wider cylinder that comprises the cap and rim domains, and a narrow cylinder called the “pore-forming region” (Figure 10).
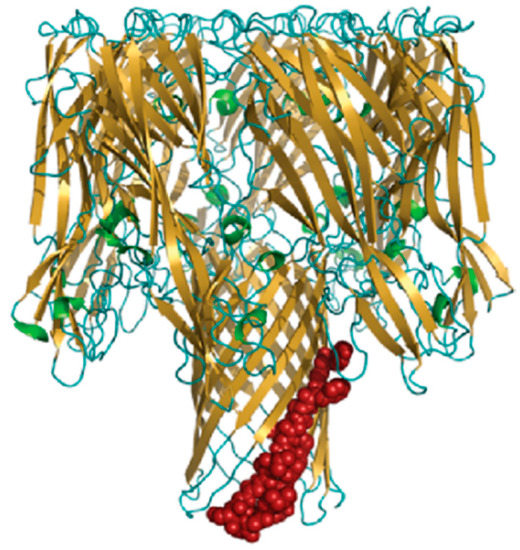
Figure 10.
The structure of AT heptameric complex. Adapted from Oscherwitz et al. [141].
The N-terminal 20 amino acids in the cap region of seven monomers interact with each other to lock in a heptameric conformation, and each monomer contributes two β-strands to assemble a fourteen-stranded anti-parallel β-barrel that forms a pore in the cell membrane. The rim domains appear to be proximal to the membrane, as they are directly involved in AT binding to cell [86]. The important residues involved in its pore-forming mechanism have been identified, including N-terminal Arg66 and Glu70 and C-terminal Arg200, Asp254, Asp255 and Asp276 [142]. Furthermore, a determinative sequence of AT pore forming is identified, which could be referred to as a linear neutralising epitope: it is located in the β-barrel pore-forming region of AT, likely including residues 122 to 137 [141].
In addition to lysing immune and host cells, AT can contribute to biofilm formation by promoting bacterial survival, destroying the host epithelium and facilitating bacterial cell-to-cell interactions. Therefore, the antibody against AT is suggested to be a multi-mechanistic anti-biofilm strategy. This suggestion is strongly supported by studies on human mAb (MEDI4893) [85]. This study shows MEDI4893 could exert its neutralising effect through a dual mechanism, as it not only blocks the binding between AT and its cellular receptor ADAM10, but it also inhibits its heptameric conformational change that enables cell lysis [85]. Furthermore, MEDI4893 has been extensively tested in a series of biofilm-associated infection models [84,85]. So far, a phase II clinical trial of MEDI4893 has been completed, which confirms its efficacy in preventing S. aureus infection (ClinicalTrials.gov Identifier: NCT02296320). In addition, another AT-neutralising antibody named as AR-301 has successfully passed its phase IIa clinical trial on patients with hospital-acquired bacterial pneumonia (ClinicalTrials.gov Identifier: NCT01589185). Overall, AT antibodies show promising effects on reducing S. aureus virulence, which can be used in conjunction with anti-biofilm therapy for achieving higher therapeutic effects.
The molecular interaction between AT and MEDI4893 has been studied by Oganesyan et al. [86]. MEDI4893 binds to a novel epitope that is a rim domain of AT, comprised of residues Asn177 to Arg200 and Thr261 to Lys271, and the residues within these regions were further confirmed as both functionally and structurally important for MEDI4893 binding. The attachment between the rim and AT is mediated by several hydrogen bonds (Figure 11).
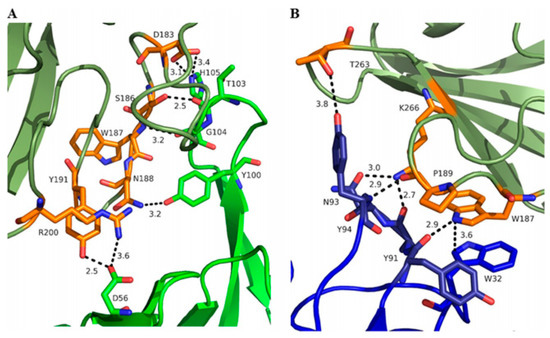
Figure 11.
Interface between MEDI4893 HC (green) and AT (olive) (A) and MEDI4893 LC (blue) and AT (olive) (B). Both HC and LC are in close contact with the rim of AT and create several hydrogen bonds (dotted lines) and one stacking interaction between MEDI4893 Fab Trp32 (LC) and AT Trp187. AT residues in contact with MEDI4893 are shown in orange. Adapted from Oganesyan et al. [86].
Further, this study interpreted the dual mechanism of action of MEDI4893. In addition to its ability to inhibit AT binding to its receptor, MEDI4893 can also bind to the opposite side of the rim domain when compared with another AT mAb named LTM14 (Figure 12).
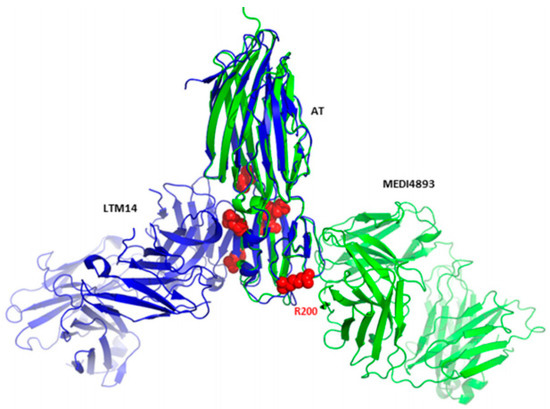
Figure 12.
Superimposition of monomeric AT bound to MEDI4893 Fab (green) with monomeric AT bound to mAb LTM14 (PDB code 4IDJ, blue). Both Fab molecules bind to the same rim domain, though on opposite sides. Residues known to be critical for AT interaction with the cell surface receptor are shown as red spheres. Adapted from Oganesyan et al. [86].
This reveals another plausible mechanism in which MEDI4893 prevents pore formation by preventing AT molecules from occupying neighbouring positions. In particular, the MEDI4893 light chain creates a steric hindrance with the neighbouring AT protomer in the rim region, and its heavy chain restricts two additional AT protomers from extending their stem. According to a previous study, which described the importance of residue Arg200 for AT binding to cell membranes and cell lysis [142], Oganesyan et al. supposed in response that Arg200 is the functional site blocked by MEDI4893. Later, these hypotheses were verified by introducing specific mutations [109]. According to the results, residues K266 and N188 critically contribute to MEDI4893 binding affinity to AT, and residue R200 was important for AT cell lysis, in agreement with the previous study [142].
3.4. Other Targets for Anti-Biofilm Treatment
In addition to the targets and their corresponding antibodies mentioned above, there are other types of anti-biofilm targets, including: (i) the cell wall-modifying enzymes and proteins, (ii) capsule and cell wall components and (iii) ATP-binding cassette (ABC) transporters. The first category includes the S. aureus autolysin (Atl) and immunodominant staphylococcal antigen A (IsaA), which are involved in cell division, growth and biofilm formation [143,144]. In the second category, lipoteichoic acid (LTA) and wall teichoic acid (WTA), as well as the capsular polysaccharides (CP) all contribute to the fundamental aspects of Gram-positive bacterial physiology [145,146]. As for the final category, the ABC transporter is responsible for cellular transport processes and drug resistance [147]. Studies on their antibodies are summarised in Table 1.
4. Conclusions and Future Outlook
This review aimed to illustrate the possibility of generating anti-biofilm antibodies for IE treatment, and a range of potential targets are mentioned and discussed above. According to the current studies, it can be summarised that biofilm formation is one of the major barriers that cause the persistence and drug resistance of IE. The S. aureus biofilm can form on both native and prosthetic valves. It has been shown that surface roughness is an important factor in stabilising cellular attachment to surfaces followed by biofilm formation [17]. In the case of NVE, bacterial infection causes vegetations on valve leaflets and the surrounding areas, which can increase the roughness of cardiac tissue and thus further promote the biofilm formation (see Section 1.2). Further, the most common material used to produce bioprosthetic valves is either bovine or pig pericardium, which are characterised by highly fibrous surfaces [104]. On the other hand, in comparison with antibiotic treatments, the major benefit of the antibody-based anti-biofilm strategy is the lower selection pressure it effects on bacteria, thus preventing further development of drug resistance. Moreover, it can both prevent the incidence of infection and directly intervene in the ongoing development of infections, and it can also be used as a supplementary therapy with antibiotics.
Amongst these candidates, two members of MSCRAMMs (ClfA and FnBP), two components of the biofilm matrix (PNAG and HU) and AT are mainly discussed. The first two candidates from the MSCRAMM family have been considered as the least promising for producing antibodies due to their poorly clarified shear stress-dependent binding mechanism. Furthermore, small differences in their composition can have large effects on antigenicity, shown as ClfA presenting lower binding affinities to antibodies elicited by other ClfA variants. As for the FnBPs, they even exhibit a much greater level of diversity in the A region subdomain to which their ligands Fg and Fn bind.
The three other targets, PNAG, HU and AT, are all considered promising candidates for generating antibodies due to their high accessibility of epitopes and conserved expression amongst different Staphylococci strains, which could also be combined in multimechanistic therapies (e.g., bispecific antibodies neutralising both HU and AT or bi-antibodies targeting both DNAP and dDNAP) [148]. Antibodies against S. aureus AT have been applied as a part of a combination therapy for IE in some previous studies, together with antibiotics and anti-ClfA antibodies [45,50,149]. AT is a reliable target for developing antibodies, with a robust body of evidence built since 1951. Furthermore, two anti-AT antibodies have already successfully completed a phase II clinical trial.
Taking all data together, targeting biofilms using antibodies can be considered an extremely promising treatment option for IE. Nonetheless, the development of natural antibodies is burdened by several limitations, such as short shelf-life, high costs of manufacturing and relatively low stability [150,151,152,153,154]. The ongoing development of nanotechnology tools for medical use might provide a suitable solution. Recently, the use of molecularly imprinted polymeric nanoparticles (MIP NPs) has been considered a reliable alternative strategy to conventional antibodies for their antibody-mimicking properties. Although this technology is still in its infancy for therapeutic applications, there is a growing body of evidence that indicates the potential feasibility of this approach [155,156,157,158,159,160,161,162,163,164].
Author Contributions
Conceptualisation, A.P.; methodology, J.H. and A.P.; formal analysis, J.H.; investigation, J.H.; data curation, J.H. and A.P.; writing—original draft preparation, J.H.; writing—review and editing, J.H. and A.P.; visualisation, J.H.; supervision, A.P.; project administration, A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pant, S.; Patel, N.J.; Deshmukh, A.; Golwala, H.; Patel, N.; Badheka, A.; Hirsch, G.A.; Mehta, J.L. Trends in infective endocarditis incidence, microbiology, and valve replacement in the United States from 2000 to 2011. J. Am. Coll. Cardiol. 2015, 65, 2070–2076. [Google Scholar] [CrossRef]
- Bor, D.H.; Woolhandler, S.; Nardin, R.; Brusch, J.; Himmelstein, D.U. Infective endocarditis in the U.S., 1998–2009: A nationwide study. PLoS ONE 2013, 8, e60033. [Google Scholar] [CrossRef] [PubMed]
- Federspiel, J.J.; Stearns, S.C.; Peppercorn, A.F.; Chu, V.H.; Fowler, V.G., Jr. Increasing US rates of endocarditis with Staphylococcus aureus: 1999–2008. Arch. Intern. Med. 2012, 172, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Loewe, L.; Rosenblatt, P.; Greene, H.J. Combined Penicillin and Heparin Therapy of Subacute Bacterial Endocarditis. Bull. N. Y. Acad. Med. 1946, 22, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Cates, J.E.; Christie, R.V.; Garrod, L.P. Penicillin-resistant subacute bacterial endocarditis treated by a combination of penicillin and streptomycin. Br. Med. J. 1951, 1, 653–656. [Google Scholar] [CrossRef]
- Wallace, A.G.; Young, W.G., Jr.; Osterhout, S. Treatment of Acute Bacterial Endocarditis by Valve Excision and Replacement. Circulation 1965, 31, 450–453. [Google Scholar] [CrossRef]
- Slipczuk, L.; Codolosa, J.N.; Davila, C.D.; Romero-Corral, A.; Yun, J.; Pressman, G.S.; Figueredo, V.M. Infective endocarditis epidemiology over five decades: A systematic review. PLoS ONE 2013, 8, e82665. [Google Scholar] [CrossRef]
- Martin, G.S.; Mannino, D.M.; Eaton, S.; Moss, M. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 2003, 348, 1546–1554. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef]
- Fluit, A.C.; Jones, M.E.; Schmitz, F.J.; Acar, J.; Gupta, R.; Verhoef, J. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin. Infect. Dis. 2000, 30, 454–460. [Google Scholar] [CrossRef]
- Cabell, C.H.; Heidenreich, P.A.; Chu, V.H.; Moore, C.M.; Stryjewski, M.E.; Corey, G.R.; Fowler, V.G., Jr. Increasing rates of cardiac device infections among Medicare beneficiaries: 1990–1999. Am. Heart J. 2004, 147, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Darouiche, R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004, 350, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.T.; Steckelberg, J.M. Infective endocarditis in patients receiving long-term hemodialysis. Mayo Clin. Proc. 2000, 75, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.J.; Baddour, L.M.; Habib, G.; Hoen, B.; Salaun, E.; Pettersson, G.B.; Schafers, H.J.; Prendergast, B.D. Challenges in Infective Endocarditis. J. Am. Coll. Cardiol. 2017, 69, 325–344. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, D.R.; Corey, G.R.; Hoen, B.; Miro, J.M.; Fowler, V.G., Jr.; Bayer, A.S.; Karchmer, A.W.; Olaison, L.; Pappas, P.A.; Moreillon, P.; et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: The International Collaboration on Endocarditis-Prospective Cohort Study. Arch. Intern. Med. 2009, 169, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Thiene, G.; Basso, C. Pathology and pathogenesis of infective endocarditis in native heart valves. Cardiovasc Pathol. 2006, 15, 256–263. [Google Scholar] [CrossRef]
- Freedman, L.R.; Valone, J., Jr. Experimental infective endocarditis. Prog. Cardiovasc Dis. 1979, 22, 169–180. [Google Scholar] [CrossRef]
- Gencbay, M.; Turan, F.; Degertekin, M.; Eksi, N.; Mutlu, B.; Unalp, A. High prevalence of hypercoagulable states in patients with recurrent thrombosis of mechanical heart valves. J. Heart Valve Dis. 1998, 7, 601–609. [Google Scholar]
- Selton-Suty, C.; Celard, M.; Le Moing, V.; Doco-Lecompte, T.; Chirouze, C.; Iung, B.; Strady, C.; Revest, M.; Vandenesch, F.; Bouvet, A.; et al. Preeminence of Staphylococcus aureus in infective endocarditis: A 1-year population-based survey. Clin. Infect. Dis. 2012, 54, 1230–1239. [Google Scholar] [CrossRef]
- Friedrich, A.W. Control of hospital acquired infections and antimicrobial resistance in Europe: The way to go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef]
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Jia, K.; Fang, T.; Wang, X.; Liu, Y.; Sun, W.; Wang, Y.; Ding, T.; Wang, J.; Li, C.; Xu, D.; et al. Antibiotic Resistance Patterns of Staphylococcus aureus Isolates from Retail Foods in Mainland China: A Meta-Analysis. Foodborne Pathog. Dis. 2020, 17, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.L.; Apisarnthanarak, A.; Madriaga, G. The Burden of Healthcare-Associated Infections in Southeast Asia: A Systematic Literature Review and Meta-analysis. Clin. Infect. Dis. 2015, 60, 1690–1699. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci. Prog. 2002, 85, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Kishii, K.; Ito, T.; Watanabe, S.; Okuzumi, K.; Hiramatsu, K. Recurrence of heterogeneous methicillin-resistant Staphylococcus aureus (MRSA) among the MRSA clinical isolates in a Japanese university hospital. J. Antimicrob. Chemother. 2008, 62, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oguri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef]
- Jensen, S.O.; Lyon, B.R. Genetics of antimicrobial resistance in Staphylococcus aureus. Future Microbiol. 2009, 4, 565–582. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Pinto, R.M.; Lopes-de-Campos, D.; Martins, M.C.L.; Van Dijck, P.; Nunes, C.; Reis, S. Impact of nanosystems in Staphylococcus aureus biofilms treatment. FEMS Microbiol. Rev. 2019, 43, 622–641. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.; Foulston, L.; DeFrancesco, A.S.; Losick, R. An Electrostatic Net Model for the Role of Extracellular DNA in Biofilm Formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.N.C.; Yildiz, F.H. Biofilm Matrix Proteins. Microbiol Spectr 2015, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Geng, M.; Bai, L. Targeting Biofilms Therapy: Current Research Strategies and Development Hurdles. Microorganisms 2020, 8, 1222. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011, 19, 449–455. [Google Scholar] [CrossRef]
- Colque-Navarro, P.; Jacobsson, G.; Andersson, R.; Flock, J.I.; Mollby, R. Levels of antibody against 11 Staphylococcus aureus antigens in a healthy population. Clin. Vaccine Immunol. 2010, 17, 1117–1123. [Google Scholar] [CrossRef]
- Feuillie, C.; Formosa-Dague, C.; Hays, L.M.C.; Vervaeck, O.; Derclaye, S.; Brennan, M.P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc. Natl. Acad. Sci. USA 2017, 114, 3738–3743. [Google Scholar] [CrossRef]
- Leonard, A.C.; Petrie, L.E.; Cox, G. Bacterial Anti-adhesives: Inhibition of Staphylococcus aureus Nasal Colonization. ACS Infect. Dis. 2019, 5, 1668–1681. [Google Scholar] [CrossRef]
- Dryla, A.; Prustomersky, S.; Gelbmann, D.; Hanner, M.; Bettinger, E.; Kocsis, B.; Kustos, T.; Henics, T.; Meinke, A.; Nagy, E. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin. Diagn. Lab. Immunol. 2005, 12, 387–398. [Google Scholar] [CrossRef]
- Thomer, L.; Emolo, C.; Thammavongsa, V.; Kim, H.K.; McAdow, M.E.; Yu, W.; Kieffer, M.; Schneewind, O.; Missiakas, D. Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J. Exp. Med. 2016, 213, 293–301. [Google Scholar] [CrossRef]
- Kumar, A.; Ray, P.; Kanwar, M.; Sharma, M.; Varma, S. A comparative analysis of antibody repertoire against Staphylococcus aureus antigens in patients with deep-seated versus superficial staphylococcal infections. Int. J. Med. Sci. 2005, 2, 129–136. [Google Scholar] [CrossRef]
- Raafat, D.; Otto, M.; Reppschlager, K.; Iqbal, J.; Holtfreter, S. Fighting Staphylococcus aureus Biofilms with Monoclonal Antibodies. Trends Microbiol. 2019, 27, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Belyi, Y.; Rybolovlev, I.; Polyakov, N.; Chernikova, A.; Tabakova, I.; Gintsburg, A. Staphylococcus aureus Surface Protein G is An Immunodominant Protein and a Possible Target in An Anti-Biofilm Drug Development. Open Microbiol. J. 2018, 12, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Domanski, P.J.; Patel, P.R.; Bayer, A.S.; Zhang, L.; Hall, A.E.; Syribeys, P.J.; Gorovits, E.L.; Bryant, D.; Vernachio, J.H.; Hutchins, J.T.; et al. Characterization of a humanized monoclonal antibody recognizing clumping factor A expressed by Staphylococcus aureus. Infect. Immun. 2005, 73, 5229–5232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tkaczyk, C.; Kasturirangan, S.; Minola, A.; Jones-Nelson, O.; Gunter, V.; Shi, Y.Y.; Rosenthal, K.; Aleti, V.; Semenova, E.; Warrener, P.; et al. Multimechanistic Monoclonal Antibodies (MAbs) Targeting Staphylococcus aureus Alpha-Toxin and Clumping Factor A: Activity and Efficacy Comparisons of a MAb Combination and an Engineered Bispecific Antibody Approach. Antimicrob. Agents Chemother. 2017, 61, e00629-17. [Google Scholar] [CrossRef]
- Varshney, A.K.; Kuzmicheva, G.A.; Lin, J.; Sunley, K.M.; Bowling, R.A., Jr.; Kwan, T.Y.; Mays, H.R.; Rambhadran, A.; Zhang, Y.; Martin, R.L.; et al. A natural human monoclonal antibody targeting Staphylococcus Protein A protects against Staphylococcus aureus bacteremia. PLoS ONE 2018, 13, e0190537. [Google Scholar] [CrossRef]
- Franca, A.; Vilanova, M.; Cerca, N.; Pier, G.B. Monoclonal antibody raised against PNAG has variable effects on static S. epidermidis biofilm accumulation in vitro. Int. J. Biol. Sci. 2013, 9, 518–520. [Google Scholar] [CrossRef]
- Romero Pastrana, F.; Neef, J.; Koedijk, D.; de Graaf, D.; Duipmans, J.; Jonkman, M.F.; Engelmann, S.; van Dijl, J.M.; Buist, G. Human antibody responses against non-covalently cell wall-bound Staphylococcus aureus proteins. Sci. Rep. 2018, 8, 3234. [Google Scholar] [CrossRef]
- Giersing, B.K.; Dastgheyb, S.S.; Modjarrad, K.; Moorthy, V. Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine 2016, 34, 2962–2966. [Google Scholar] [CrossRef]
- Tkaczyk, C.; Hamilton, M.M.; Sadowska, A.; Shi, Y.; Chang, C.S.; Chowdhury, P.; Buonapane, R.; Xiao, X.; Warrener, P.; Mediavilla, J.; et al. Targeting Alpha Toxin and ClfA with a Multimechanistic Monoclonal-Antibody-Based Approach for Prophylaxis of Serious Staphylococcus aureus Disease. mBio 2016, 7, e00528-16. [Google Scholar] [CrossRef]
- Hall, A.E.; Domanski, P.J.; Patel, P.R.; Vernachio, J.H.; Syribeys, P.J.; Gorovits, E.L.; Johnson, M.A.; Ross, J.M.; Hutchins, J.T.; Patti, J.M. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect. Immun. 2003, 71, 6864–6870. [Google Scholar] [CrossRef] [PubMed]
- Weems, J.J., Jr.; Steinberg, J.P.; Filler, S.; Baddley, J.W.; Corey, G.R.; Sampathkumar, P.; Winston, L.; John, J.F.; Kubin, C.J.; Talwani, R.; et al. Phase II, randomized, double-blind, multicenter study comparing the safety and pharmacokinetics of tefibazumab to placebo for treatment of Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2006, 50, 2751–2755. [Google Scholar] [CrossRef] [PubMed]
- Squibb, B.-M. A Phase IIa Dose Escalation Study to Assess Safety and Pharmacokinetics of Aurexis® in Cystic Fibrosis Subjects Chronically Colonized with Staphylococcus aureus in Their Lungs. Available online: https://clinicaltrials.gov/ct2/show/NCT00198289 (accessed on 21 April 2022).
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- Rennermalm, A.; Li, Y.H.; Bohaufs, L.; Jarstrand, C.; Brauner, A.; Brennan, F.R.; Flock, J.I. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine 2001, 19, 3376–3383. [Google Scholar] [CrossRef]
- Visai, L.; Xu, Y.; Casolini, F.; Rindi, S.; Hook, M.; Speziale, P. Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylococcus aureus from a collagen substrate. J. Biol. Chem. 2000, 275, 39837–39845. [Google Scholar] [CrossRef]
- Pharma, N. A Multi Centre, Double-Blind, Randomised, Placebo Controlled Prospective Study on the Safety and Efficacy of Aurograb in Patients with Severe, Deep-Seated Staphylococcal Infections Receiving Vancomycin. Available online: https://www.clinicaltrials.gov/ct2/show/NCT00217841 (accessed on 21 April 2022).
- Nilsson, I.M.; Patti, J.M.; Bremell, T.; Hook, M.; Tarkowski, A. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J. Clin. Investig. 1998, 101, 2640–2649. [Google Scholar] [CrossRef]
- McCarthy, H.; Waters, E.M.; Bose, J.L.; Foster, S.; Bayles, K.W.; O’Neill, E.; Fey, P.D.; O’Gara, J.P. The major autolysin is redundant for Staphylococcus aureus USA300 LAC JE2 virulence in a murine device-related infection model. FEMS Microbiol. Lett. 2016, 363, fnw087. [Google Scholar] [CrossRef]
- Nair, N.; Vinod, V.; Suresh, M.K.; Vijayrajratnam, S.; Biswas, L.; Peethambaran, R.; Vasudevan, A.K.; Biswas, R. Amidase, a cell wall hydrolase, elicits protective immunity against Staphylococcus aureus and S. epidermidis. Int. J. Biol. Macromol. 2015, 77, 314–321. [Google Scholar] [CrossRef]
- Haghighat, S.; Siadat, S.D.; Sorkhabadi, S.M.; Sepahi, A.A.; Mahdavi, M. Cloning, expression and purification of autolysin from methicillin-resistant Staphylococcus aureus: Potency and challenge study in Balb/c mice. Mol. Immunol. 2017, 82, 10–18. [Google Scholar] [CrossRef]
- Varrone, J.J.; de Mesy Bentley, K.L.; Bello-Irizarry, S.N.; Nishitani, K.; Mack, S.; Hunter, J.G.; Kates, S.L.; Daiss, J.L.; Schwarz, E.M. Passive immunization with anti-glucosaminidase monoclonal antibodies protects mice from implant-associated osteomyelitis by mediating opsonophagocytosis of Staphylococcus aureus megaclusters. J. Orthop. Res. 2014, 32, 1389–1396. [Google Scholar] [CrossRef]
- van den Berg, S.; Bonarius, H.P.; van Kessel, K.P.; Elsinga, G.S.; Kooi, N.; Westra, H.; Bosma, T.; van der Kooi-Pol, M.M.; Koedijk, D.G.; Groen, H.; et al. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int. J. Med. Microbiol. 2015, 305, 55–64. [Google Scholar] [CrossRef]
- Oesterreich, B.; Lorenz, B.; Schmitter, T.; Kontermann, R.; Zenn, M.; Zimmermann, B.; Haake, M.; Lorenz, U.; Ohlsen, K. Characterization of the biological anti-staphylococcal functionality of hUK-66 IgG1, a humanized monoclonal antibody as substantial component for an immunotherapeutic approach. Hum. Vaccin. Immunother. 2014, 10, 926–937. [Google Scholar] [CrossRef] [PubMed]
- Ohlsen, K.; Lorenz, U. Immunotherapeutic strategies to combat staphylococcal infections. Int. J. Med. Microbiol. 2010, 300, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Morisaki, J.H.; et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef]
- Jung, D.-J.; An, J.-H.; Kurokawa, K.; Jung, Y.-C.; Kim, M.-J.; Aoyagi, Y.; Matsushita, M.; Takahashi, S.; Lee, H.-S.; Takahashi, K.; et al. Specific Serum Ig Recognizing Staphylococcal Wall Teichoic Acid Induces Complement-Mediated Opsonophagocytosis against Staphylococcus aureus. J. Immunol. 2012, 189, 4951. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, J.; Lee, J.C. Antibodies to capsular polysaccharides are not protective against experimental Staphylococcus aureus endocarditis. Infect. Immun. 1995, 63, 375–380. [Google Scholar] [CrossRef]
- Zhou, C.; Lehar, S.; Gutierrez, J.; Rosenberger, C.M.; Ljumanovic, N.; Dinoso, J.; Koppada, N.; Hong, K.; Baruch, A.; Carrasco-Triguero, M.; et al. Pharmacokinetics and pharmacodynamics of DSTA4637A: A novel THIOMAB antibody antibiotic conjugate against Staphylococcus aureus in mice. MAbs 2016, 8, 1612–1619. [Google Scholar] [CrossRef]
- Wang-Lin, S.X.; Zhou, C.; Kamath, A.V.; Hong, K.; Koppada, N.; Saad, O.M.; Carrasco-Triguero, M.; Khojasteh, C.; Deng, R. Minimal physiologically-based pharmacokinetic modeling of DSTA4637A, A novel THIOMAB antibody antibiotic conjugate against Staphylococcus aureus, in a mouse model. MAbs 2018, 10, 1131–1143. [Google Scholar] [CrossRef]
- Lee, J.C.; Park, J.S.; Shepherd, S.E.; Carey, V.; Fattom, A. Protective efficacy of antibodies to the Staphylococcus aureus type 5 capsular polysaccharide in a modified model of endocarditis in rats. Infect. Immun. 1997, 65, 4146–4151. [Google Scholar] [CrossRef]
- Liu, B.; Park, S.; Thompson, C.D.; Li, X.; Lee, J.C. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence 2017, 8, 859–874. [Google Scholar] [CrossRef][Green Version]
- Rupp, M.E.; Holley, H.P., Jr.; Lutz, J.; Dicpinigaitis, P.V.; Woods, C.W.; Levine, D.P.; Veney, N.; Fowler, V.G., Jr. Phase II, randomized, multicenter, double-blind, placebo-controlled trial of a polyclonal anti-Staphylococcus aureus capsular polysaccharide immune globulin in treatment of Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2007, 51, 4249–4254. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.K.; Schelonka, R.; White, R.; Holley, H.P.; Bifano, E.; Cummings, J.; Adcock, K.; Kaufman, D.; Puppala, B.; Riedel, P.; et al. A blinded, randomized, multicenter study of an intravenous Staphylococcus aureus immune globulin. J. Perinatol. 2006, 26, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Weisman, L.E. Antibody for the prevention of neonatal noscocomial staphylococcal infection: A review of the literature. Arch. De Pédiatrie 2007, 14, S31–S34. [Google Scholar] [CrossRef]
- Weisman, L.E.; Thackray, H.M.; Steinhorn, R.H.; Walsh, W.F.; Lassiter, H.A.; Dhanireddy, R.; Brozanski, B.S.; Palmer, K.G.; Trautman, M.S.; Escobedo, M.; et al. A randomized study of a monoclonal antibody (pagibaximab) to prevent staphylococcal sepsis. Pediatrics 2011, 128, 271–279. [Google Scholar] [CrossRef]
- Patel, M.; Kaufman, D.A. Anti-lipoteichoic acid monoclonal antibody (pagibaximab) studies for the prevention of staphylococcal bloodstream infections in preterm infants. Expert Opin. Biol. Ther. 2015, 15, 595–600. [Google Scholar] [CrossRef]
- Cerca, N.; Jefferson, K.K.; Maira-Litran, T.; Pier, D.B.; Kelly-Quintos, C.; Goldmann, D.A.; Azeredo, J.; Pier, G.B. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect. Immun. 2007, 75, 3406–3413. [Google Scholar] [CrossRef]
- Kelly-Quintos, C.; Cavacini, L.A.; Posner, M.R.; Goldmann, D.; Pier, G.B. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect. Immun. 2006, 74, 2742–2750. [Google Scholar] [CrossRef]
- Sanofi-Aventis. A Randomized, Double-blind, Placebo-controlled Trial to Assess the Pharmacokinetics, Pharmacodynamics and Safety of a Single Dose of SAR279356 in Patients Hospitalized in Intensive Care Unit and on Mechanical Ventilation. Available online: https://clinicaltrials.gov/ct2/show/NCT.T01389700 (accessed on 21 April 2022).
- Estelles, A.; Woischnig, A.K.; Liu, K.; Stephenson, R.; Lomongsod, E.; Nguyen, D.; Zhang, J.; Heidecker, M.; Yang, Y.; Simon, R.J.; et al. A High-Affinity Native Human Antibody Disrupts Biofilm from Staphylococcus aureus Bacteria and Potentiates Antibiotic Efficacy in a Mouse Implant Infection Model. Antimicrob. Agents Chemother. 2016, 60, 2292–2301. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Estelles, A.; Li, L.; Abdelhady, W.; Gonzales, R.; Bayer, A.S.; Tenorio, E.; Leighton, A.; Ryser, S.; Kauvar, L.M. A Human Biofilm-Disrupting Monoclonal Antibody Potentiates Antibiotic Efficacy in Rodent Models of both Staphylococcus aureus and Acinetobacter baumannii Infections. Antimicrob. Agents Chemother. 2017, 61, e00904-17. [Google Scholar] [CrossRef]
- LLC, T.B. Study to Evaluate Safety and Activity of TRL1068 in Prosthetic Joint Infections. Available online: https://clinicaltrials.gov/ct2/show/NCT04763759 (accessed on 21 April 2022).
- Wang, Y.; Cheng, L.I.; Helfer, D.R.; Ashbaugh, A.G.; Miller, R.J.; Tzomides, A.J.; Thompson, J.M.; Ortines, R.V.; Tsai, A.S.; Liu, H.; et al. Mouse model of hematogenous implant-related Staphylococcus aureus biofilm infection reveals therapeutic targets. Proc. Natl. Acad. Sci. USA 2017, 114, E5094–E5102. [Google Scholar] [CrossRef]
- Anderson, M.J.; Schaaf, E.; Breshears, L.M.; Wallis, H.W.; Johnson, J.R.; Tkaczyk, C.; Sellman, B.R.; Sun, J.; Peterson, M.L. Alpha-Toxin Contributes to Biofilm Formation among Staphylococcus aureus Wound Isolates. Toxins 2018, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, V.; Peng, L.; Damschroder, M.M.; Cheng, L.; Sadowska, A.; Tkaczyk, C.; Sellman, B.R.; Wu, H.; Dall’Acqua, W.F. Mechanisms of neutralization of a human anti-alpha-toxin antibody. J. Biol. Chem. 2014, 289, 29874–29880. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Robbie, G.J.; Wu, Y.; Esser, M.T.; Jensen, K.; Schwartz, H.I.; Bellamy, T.; Hernandez-Illas, M.; Jafri, H.S. Safety, Tolerability, and Pharmacokinetics of MEDI4893, an Investigational, Extended-Half-Life, Anti-Staphylococcus aureus Alpha-Toxin Human Monoclonal Antibody, in Healthy Adults. Antimicrob. Agents Chemother. 2017, 61, e01020-16. [Google Scholar] [CrossRef] [PubMed]
- Pharmaceuticals, A. AR-301 (Tosatoxumab). Available online: https://aridispharma.com/ar-301/ (accessed on 21 April 2022).
- Thomsen, I.P.; Sapparapu, G.; James, D.B.A.; Cassat, J.E.; Nagarsheth, M.; Kose, N.; Putnam, N.; Boguslawski, K.M.; Jones, L.S.; Wood, J.B.; et al. Monoclonal Antibodies Against the Staphylococcus aureus Bicomponent Leukotoxin AB Isolated Following Invasive Human Infection Reveal Diverse Binding and Modes of Action. J. Infect. Dis. 2017, 215, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Burnie, J.P.; Matthews, R.C.; Carter, T.; Beaulieu, E.; Donohoe, M.; Chapman, C.; Williamson, P.; Hodgetts, S.J. Identification of an immunodominant ABC transporter in methicillin-resistant Staphylococcus aureus infections. Infect. Immun. 2000, 68, 3200–3209. [Google Scholar] [CrossRef]
- Lam, H.; Kesselly, A.; Stegalkina, S.; Kleanthous, H.; Yethon, J.A. Antibodies to PhnD inhibit staphylococcal biofilms. Infect. Immun. 2014, 82, 3764–3774. [Google Scholar] [CrossRef][Green Version]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penades, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Broker, B.M.; Holtfreter, S.; Bekeredjian-Ding, I. Immune control of Staphylococcus aureus-regulation and counter-regulation of the adaptive immune response. Int. J. Med. Microbiol. 2014, 304, 204–214. [Google Scholar] [CrossRef]
- Lindsay, J.A.; Moore, C.E.; Day, N.P.; Peacock, S.J.; Witney, A.A.; Stabler, R.A.; Husain, S.E.; Butcher, P.D.; Hinds, J. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 2006, 188, 669–676. [Google Scholar] [CrossRef]
- Murphy, E.; Lin, S.L.; Nunez, L.; Andrew, L.; Fink, P.S.; Dilts, D.A.; Hoiseth, S.K.; Jansen, K.U.; Anderson, A.S. Challenges for the evaluation of Staphylococcus aureus protein based vaccines: Monitoring antigenic diversity. Hum. Vaccin. 2011, 7 (Suppl. S1), 51–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Herman-Bausier, P.; Labate, C.; Towell, A.M.; Derclaye, S.; Geoghegan, J.A.; Dufrene, Y.F. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc. Natl. Acad. Sci. USA 2018, 115, 5564–5569. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, D.; Francois, P.; Vaudaux, P.; Foster, T.J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 1994, 11, 237–248. [Google Scholar] [CrossRef]
- Ni Eidhin, D.; Perkins, S.; Francois, P.; Vaudaux, P.; Hook, M.; Foster, T.J. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 1998, 30, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Wann, E.R.; Gurusiddappa, S.; Hook, M. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 2000, 275, 13863–13871. [Google Scholar] [CrossRef] [PubMed]
- Ponnuraj, K.; Bowden, M.G.; Davis, S.; Gurusiddappa, S.; Moore, D.; Choe, D.; Xu, Y.; Hook, M.; Narayana, S.V. A "dock, lock, and latch" structural model for a staphylococcal adhesin binding to fibrinogen. Cell 2003, 115, 217–228. [Google Scholar] [CrossRef]
- Brady, R.A.; Mocca, C.P.; Burns, D.L. Immunogenicity analysis of Staphylococcus aureus clumping factor A genetic variants. Clin. Vaccine Immunol. 2013, 20, 1338–1340. [Google Scholar] [CrossRef] [PubMed]
- Becsek, B.; Pietrasanta, L.; Obrist, D. Turbulent Systolic Flow Downstream of a Bioprosthetic Aortic Valve: Velocity Spectra, Wall Shear Stresses, and Turbulent Dissipation Rates. Front. Physiol. 2020, 11, 577188. [Google Scholar] [CrossRef]
- Zhang, P.; Yeo, J.H.; Qian, P.; Hwang, N.H. Shear stress investigation across mechanical heart valve. ASAIO J. 2007, 53, 530–536. [Google Scholar] [CrossRef]
- Hawkins, J.; Kodali, S.; Matsuka, Y.V.; McNeil, L.K.; Mininni, T.; Scully, I.L.; Vernachio, J.H.; Severina, E.; Girgenti, D.; Jansen, K.U.; et al. A recombinant clumping factor A-containing vaccine induces functional antibodies to Staphylococcus aureus that are not observed after natural exposure. Clin. Vaccine Immunol. 2012, 19, 1641–1650. [Google Scholar] [CrossRef]
- Ganesh, V.K.; Liang, X.; Geoghegan, J.A.; Cohen, A.L.V.; Venugopalan, N.; Foster, T.J.; Hook, M. Lessons from the Crystal Structure of the S. aureus Surface Protein Clumping Factor A in Complex With Tefibazumab, an Inhibiting Monoclonal Antibody. EBioMedicine 2016, 13, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Patti, J.M. A humanized monoclonal antibody targeting Staphylococcus aureus. Vaccine 2004, 22 (Suppl. S1), S39–S43. [Google Scholar] [CrossRef] [PubMed]
- Rozalska, B.; Wadstrom, T. Protective opsonic activity of antibodies against fibronectin-binding proteins (FnBPs) of Staphylococcus aureus. Scand. J. Immunol. 1993, 37, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Tkaczyk, C.; Semenova, E.; Shi, Y.Y.; Rosenthal, K.; Oganesyan, V.; Warrener, P.; Stover, C.K.; Sellman, B.R. Alanine Scanning Mutagenesis of the MEDI4893 (Suvratoxumab) Epitope Reduces Alpha Toxin Lytic Activity In Vitro and Staphylococcus aureus Fitness in Infection Models. Antimicrob. Agents Chemother. 2018, 62, e01033-18. [Google Scholar] [CrossRef]
- Loughman, A.; Sweeney, T.; Keane, F.M.; Pietrocola, G.; Speziale, P.; Foster, T.J. Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 2008, 8, 74. [Google Scholar] [CrossRef]
- Burke, F.M.; McCormack, N.; Rindi, S.; Speziale, P.; Foster, T.J. Fibronectin-binding protein B variation in Staphylococcus aureus. BMC Microbiol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Herman-Bausier, P.; El-Kirat-Chatel, S.; Foster Timothy, J.; Geoghegan Joan, A.; Dufrêne Yves, F.; Torres, V.; Pier Gerald, B. Staphylococcus aureus Fibronectin-Binding Protein A Mediates Cell-Cell Adhesion through Low-Affinity Homophilic Bonds. mBio 2015, 6, e00413-15. [Google Scholar] [CrossRef]
- Devaraj, A.; Buzzo, J.; Rocco, C.J.; Bakaletz, L.O.; Goodman, S.D. The DNABII family of proteins is comprised of the only nucleoid associated proteins required for nontypeable Haemophilus influenzae biofilm structure. Microbiologyopen 2018, 7, e00563. [Google Scholar] [CrossRef]
- Provenza, G.; Provenzano, M.; Visai, L.; Burke, F.M.; Geoghegan, J.A.; Stravalaci, M.; Gobbi, M.; Mazzini, G.; Arciola, C.R.; Foster, T.J.; et al. Functional analysis of a murine monoclonal antibody against the repetitive region of the fibronectin-binding adhesins fibronectin-binding protein A and fibronectin-binding protein B from Staphylococcus aureus. FEBS J. 2010, 277, 4490–4505. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef]
- O’Gara, J.P. ica and beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007, 270, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Cywes-Bentley, C.; Skurnik, D.; Zaidi, T.; Roux, D.; Deoliveira, R.B.; Garrett, W.S.; Lu, X.; O’Malley, J.; Kinzel, K.; Zaidi, T.; et al. Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, E2209–E2218. [Google Scholar] [CrossRef] [PubMed]
- Burgui, S.; Gil, C.; Solano, C.; Lasa, I.; Valle, J. A Systematic Evaluation of the Two-Component Systems Network Reveals That ArlRS Is a Key Regulator of Catheter Colonization by Staphylococcus aureus. Front. Microbiol. 2018, 9, 342. [Google Scholar] [CrossRef]
- Atkin, K.E.; MacDonald, S.J.; Brentnall, A.S.; Potts, J.R.; Thomas, G.H. A different path: Revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014, 588, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Cywes-Bentley, C.; Zhang, Y.F.; Pons, S.; Konkol, M.; Kearns, D.B.; Little, D.J.; Howell, P.L.; Skurnik, D.; Pier, G.B. Identification of Poly-N-acetylglucosamine as a Major Polysaccharide Component of the Bacillus subtilis Biofilm Matrix. J. Biol. Chem. 2015, 290, 19261–19272. [Google Scholar] [CrossRef]
- Haji-Ghassemi, O.; Blackler, R.J.; Martin Young, N.; Evans, S.V. Antibody recognition of carbohydrate epitopesdagger. Glycobiology 2015, 25, 920–952. [Google Scholar] [CrossRef]
- Cerca, N.; Maira-Litran, T.; Jefferson, K.K.; Grout, M.; Goldmann, D.A.; Pier, G.B. Protection against Escherichia coli infection by antibody to the Staphylococcus aureus poly-N-acetylglucosamine surface polysaccharide. Proc. Natl. Acad. Sci. USA 2007, 104, 7528–7533. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Lau, G.W. DNABII targeting antibodies as vaccines against biofilm diseases. EBioMedicine 2020, 58, 102921. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Carcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef]
- Maira-Litran, T.; Kropec, A.; Goldmann, D.A.; Pier, G.B. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal Poly-N-acetyl-beta-(1-6)-glucosamine. Infect. Immun. 2005, 73, 6752–6762. [Google Scholar] [CrossRef]
- Skurnik, D.; Cywes-Bentley, C.; Pier, G.B. The exceptionally broad-based potential of active and passive vaccination targeting the conserved microbial surface polysaccharide PNAG. Expert Rev. Vaccines 2016, 15, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Gening, M.L.; Maira-Litran, T.; Kropec, A.; Skurnik, D.; Grout, M.; Tsvetkov, Y.E.; Nifantiev, N.E.; Pier, G.B. Synthetic {beta}-(1->6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect. Immun. 2010, 78, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Soliman, C.; Walduck, A.K.; Yuriev, E.; Richards, J.S.; Cywes-Bentley, C.; Pier, G.B.; Ramsland, P.A. Structural basis for antibody targeting of the broadly expressed microbial polysaccharide poly-N-acetylglucosamine. J. Biol. Chem. 2018, 293, 5079–5089. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef]
- Rocco, C.J.; Davey, M.E.; Bakaletz, L.O.; Goodman, S.D. Natural antigenic differences in the functionally equivalent extracellular DNABII proteins of bacterial biofilms provide a means for targeted biofilm therapeutics. Mol. Oral Microbiol. 2017, 32, 118–130. [Google Scholar] [CrossRef]
- Swinger, K.K.; Rice, P.A. IHF and HU: Flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004, 14, 28–35. [Google Scholar] [CrossRef]
- Devaraj, A.; Justice, S.S.; Bakaletz, L.O.; Goodman, S.D. DNABII proteins play a central role in UPEC biofilm structure. Mol. Microbiol. 2015, 96, 1119–1135. [Google Scholar] [CrossRef]
- Goodman, S.D.; Obergfell, K.P.; Jurcisek, J.A.; Novotny, L.A.; Downey, J.S.; Ayala, E.A.; Tjokro, N.; Li, B.; Justice, S.S.; Bakaletz, L.O. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011, 4, 625–637. [Google Scholar] [CrossRef]
- Kamashev, D.; Balandina, A.; Rouviere-Yaniv, J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999, 18, 5434–5444. [Google Scholar] [CrossRef]
- Azam, T.A.; Ishihama, A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999, 274, 33105–33113. [Google Scholar] [CrossRef]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Nagaraja, V.; Ramakumar, S. Structural and evolutionary analyses reveal determinants of DNA binding specificities of nucleoid-associated proteins HU and IHF. Mol. Phylogenet Evol. 2017, 107, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Im, H.; Jee, J.G.; Jang, S.B.; Yoon, H.J.; Kwon, A.R.; Kang, S.M.; Lee, B.J. beta-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 3273–3289. [Google Scholar] [CrossRef] [PubMed]
- Novotny, L.A.; Jurcisek, J.A.; Goodman, S.D.; Bakaletz, L.O. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine 2016, 10, 33–44. [Google Scholar] [CrossRef]
- Oscherwitz, J.; Cease, K.B. Identification and validation of a linear protective neutralizing epitope in the beta-pore domain of alpha toxin. PLoS ONE 2015, 10, e0116882. [Google Scholar] [CrossRef]
- Walker, B.; Bayley, H. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J. Biol. Chem. 1995, 270, 23065–23071. [Google Scholar] [CrossRef]
- Biswas, R.; Voggu, L.; Simon, U.K.; Hentschel, P.; Thumm, G.; Gotz, F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006, 259, 260–268. [Google Scholar] [CrossRef]
- Koedijk, D.; Pastrana, F.R.; Hoekstra, H.; Berg, S.V.D.; Back, J.W.; Kerstholt, C.; Prins, R.C.; Bakker-Woudenberg, I.; van Dijl, J.M.; Buist, G. Differential epitope recognition in the immunodominant staphylococcal antigen A of Staphylococcus aureus by mouse versus human IgG antibodies. Sci. Rep. 2017, 7, 8141. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Percy, M.G.; Grundling, A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 2014, 68, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Leonard, G.D.; Fojo, T.; Bates, S.E. The role of ABC transporters in clinical practice. Oncologist 2003, 8, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Tabor, D.E.; Yu, L.; Mok, H.; Tkaczyk, C.; Sellman, B.R.; Wu, Y.; Oganesyan, V.; Slidel, T.; Jafri, H.; McCarthy, M.; et al. Staphylococcus aureus Alpha-Toxin Is Conserved among Diverse Hospital Respiratory Isolates Collected from a Global Surveillance Study and Is Neutralized by Monoclonal Antibody MEDI4893. Antimicrob. Agents Chemother. 2016, 60, 5312–5321. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Valour, F.; Nguyen, N.T.Q.; Doan, T.M.N.; Koelkebeck, H.; Richardson, C.; Cheng, L.I.; Sellman, B.R.; Tkaczyk, C.; Diep, B.A. Multimechanistic Monoclonal Antibody Combination Targeting Key Staphylococcus aureus Virulence Determinants in a Rabbit Model of Prosthetic Joint Infection. Antimicrob. Agents Chemother. 2021, 65, e0183220. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Turner, A.P.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef]
- Canfarotta, F.; Poma, A.; Guerreiro, A.; Piletsky, S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 2016, 11, 443–455. [Google Scholar] [CrossRef]
- Piletsky, S.; Canfarotta, F.; Poma, A.; Bossi, A.M.; Piletsky, S. Molecularly Imprinted Polymers for Cell Recognition. Trends Biotechnol. 2020, 38, 368–387. [Google Scholar] [CrossRef]
- Liu, R.; Poma, A. Advances in Molecularly Imprinted Polymers as Drug Delivery Systems. Molecules 2021, 26, 3589. [Google Scholar] [CrossRef]
- Poma, A.; Whitcombe, M.J.; Piletsky, S. Plastic Antibodies. In Designing Receptors for the Next Generation of Biosensors; Piletsky, S.A., Whitcombe, M.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 105–129. [Google Scholar]
- Poma, A.; Guerreiro, A.; Whitcombe, M.J.; Piletska, E.V.; Turner, A.P.; Piletsky, S.A. Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template-“Plastic Antibodies”. Adv. Funct. Mater. 2013, 23, 2821–2827. [Google Scholar] [CrossRef]
- Moczko, E.; Poma, A.; Guerreiro, A.; Perez de Vargas Sansalvador, I.; Caygill, S.; Canfarotta, F.; Whitcombe, M.J.; Piletsky, S. Surface-modified multifunctional MIP nanoparticles. N. Nanoscale 2013, 5, 3733–3741. [Google Scholar] [CrossRef]
- Subrahmanyam, S.; Guerreiro, A.; Poma, A.; Moczko, E.; Piletska, E.; Piletsky, S. Optimisation of experimental conditions for synthesis of high affinity MIP nanoparticles. Eur. Polym. J. 2013, 49, 100–105. [Google Scholar] [CrossRef]
- Poma, A.; Guerreiro, A.; Caygill, S.; Moczko, E.; Piletsky, S. Automatic reactor for solid-phase synthesis of molecularly imprinted polymeric nanoparticles (MIP NPs) in water. RSC Adv. 2014, 4, 4203–4206. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, A.; Poma, A.; Karim, K.; Moczko, E.; Takarada, J.; de Vargas-Sansalvador, I.P.; Turner, N.; Piletska, E.; de Magalhaes, C.S.; Glazova, N.; et al. Influence of surface-imprinted nanoparticles on trypsin activity. Adv. Healthc. Mater. 2014, 3, 1426–1429. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Brahmbhatt, H.; Watts, J.K.; Turner, N.W. Nucleoside-Tailored Molecularly Imprinted Polymeric Nanoparticles (MIP NPs). Macromolecules 2014, 47, 6322–6330. [Google Scholar] [CrossRef]
- Poma, A.; Brahmbhatt, H.; Pendergraff, H.M.; Watts, J.K.; Turner, N.W. Generation of novel hybrid aptamer-molecularly imprinted polymeric nanoparticles. Adv. Mater. 2015, 27, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Brahmbhatt, H.; Poma, A.; Pendergraff, H.M.; Watts, J.K.; Turner, N.W. Improvement of DNA recognition through molecular imprinting: Hybrid oligomer imprinted polymeric nanoparticles (oligoMIP NPs). Biomater. Sci. 2016, 4, 281–287. [Google Scholar] [CrossRef]
- Canfarotta, F.; Lezina, L.; Guerreiro, A.; Czulak, J.; Petukhov, A.; Daks, A.; Smolinska-Kempisty, K.; Poma, A.; Piletsky, S.; Barlev, N.A. Specific Drug Delivery to Cancer Cells with Double-Imprinted Nanoparticles against Epidermal Growth Factor Receptor. Nano Lett. 2018, 18, 4641–4646. [Google Scholar] [CrossRef]
- Bedwell, T.S.; Anjum, N.; Ma, Y.; Czulak, J.; Poma, A.; Piletska, E.; Whitcombe, M.J.; Piletsky, S.A. New protocol for optimisation of polymer composition for imprinting of peptides and proteins. RSC Adv. 2019, 9, 27849–27855. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).