Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Apparatus
2.3. Cell Lines and Cell Culture
2.4. Preparation of GO-Based Fluorescent Aptasensor
2.5. Fluorescence Labeling Based on FAM-W3-GO
2.6. Target Cells’ Detection by GO-Based Fluorescent Assay
2.7. Specificity Analysis
2.8. Whole Blood Sample Analysis
2.9. Statistical Analyses
3. Results and Discussion
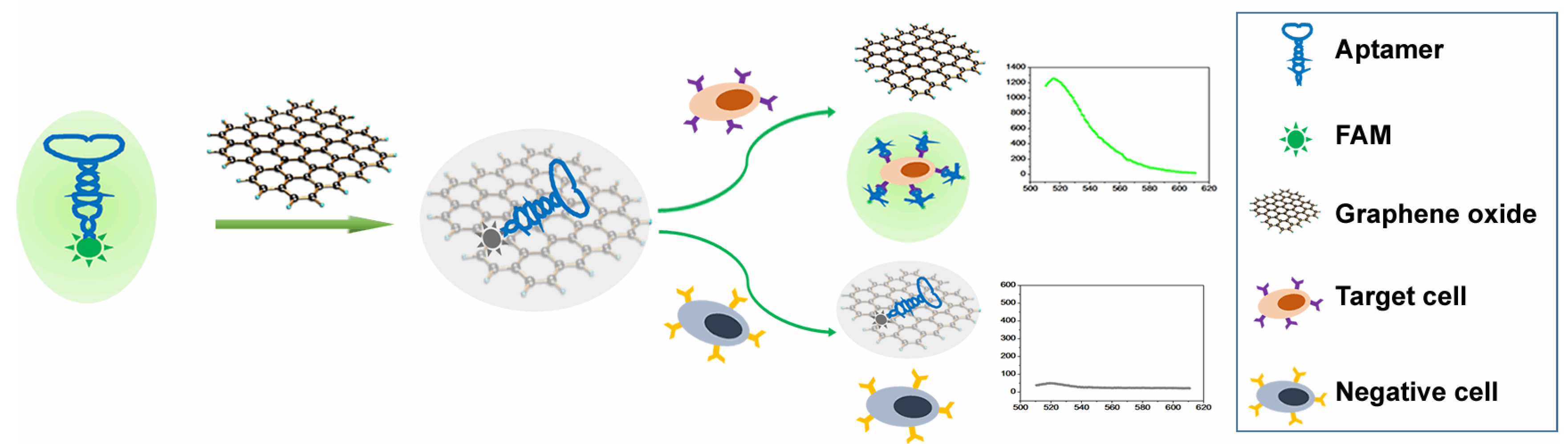
3.1. Principle of FAM-W3-GO for Cancer Cell Detection
3.2. The Feasibility of mCRC Cell Detection Using FAM-W3-GO
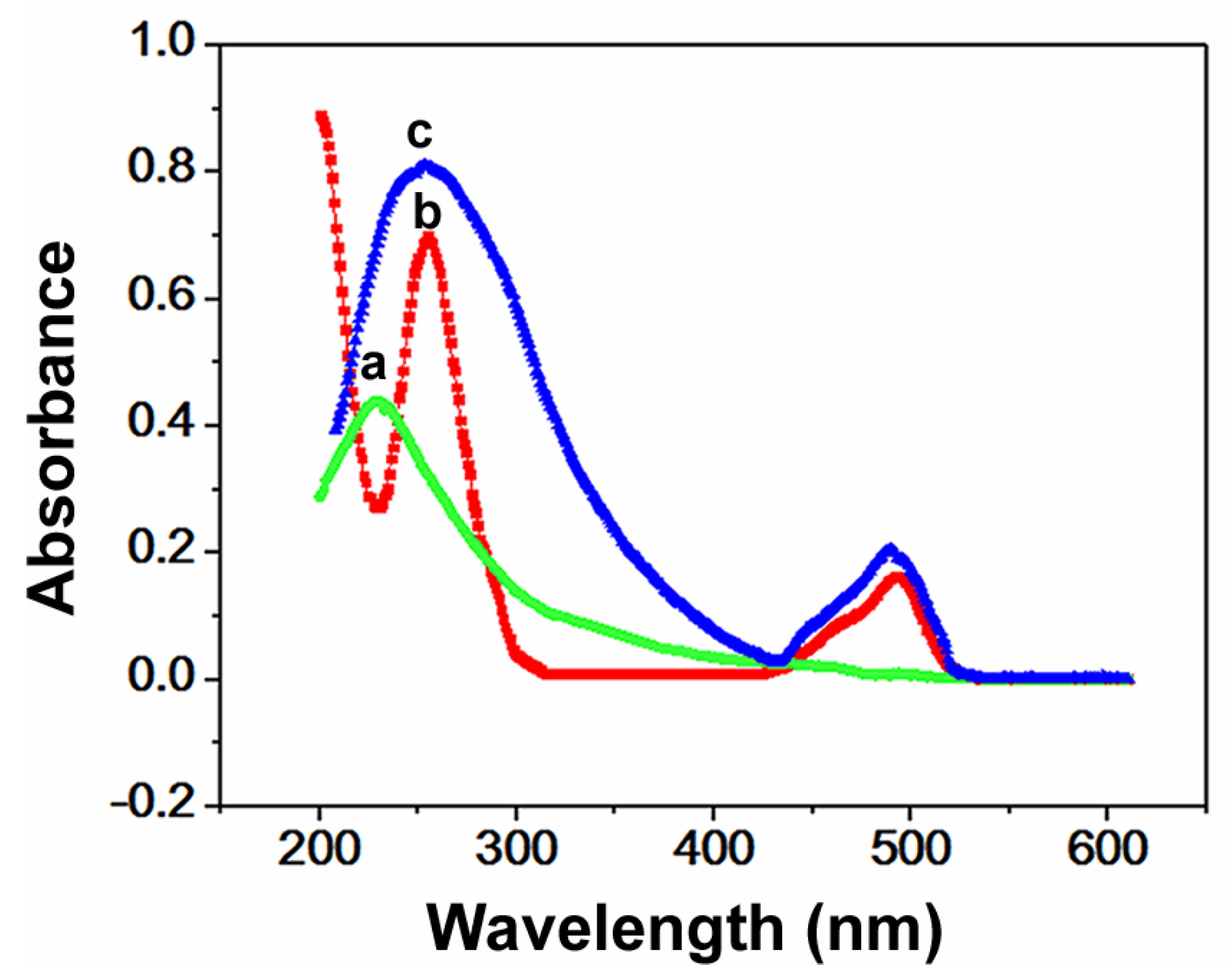
3.3. Optical Properties of FAM-W3-GO
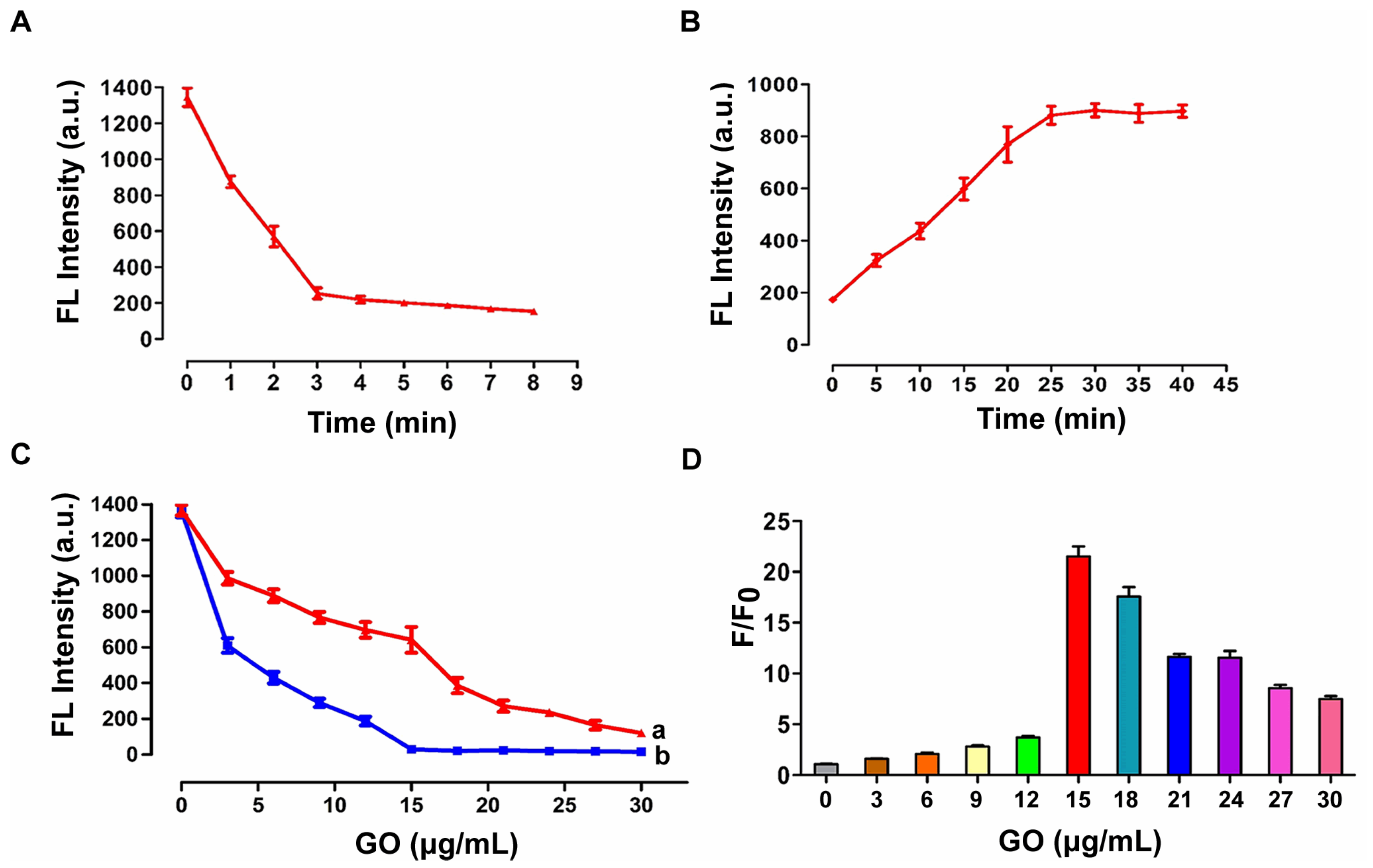
3.4. Optimization of Experimental Conditions
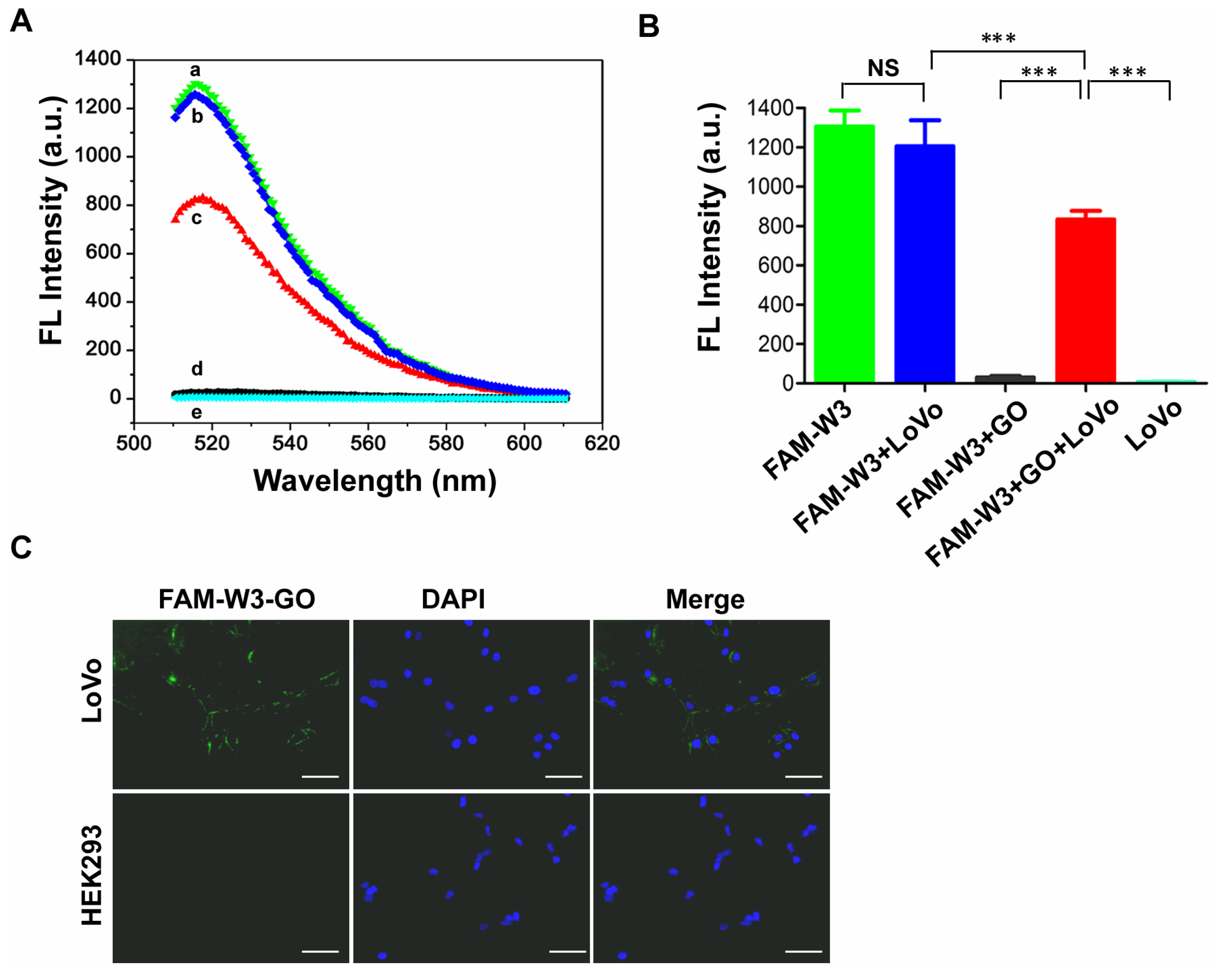
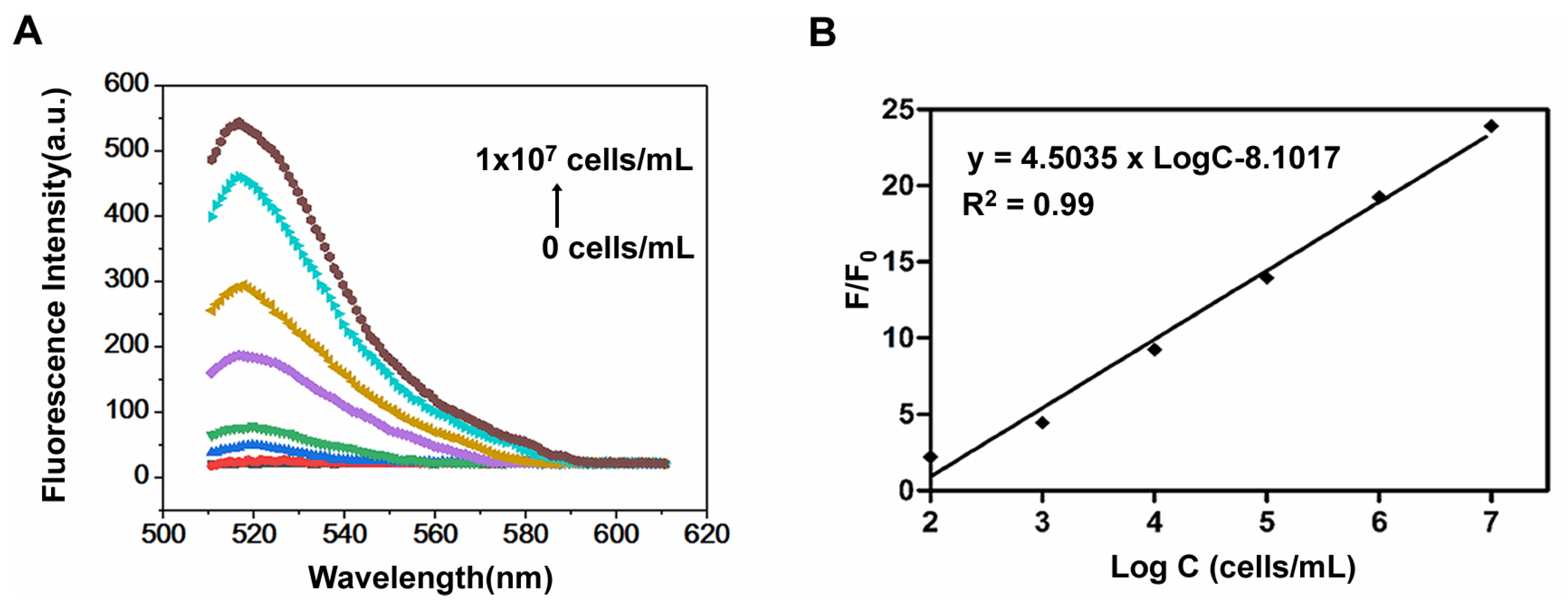
3.5. Detection of LoVo Cells with FAM-W3-GO
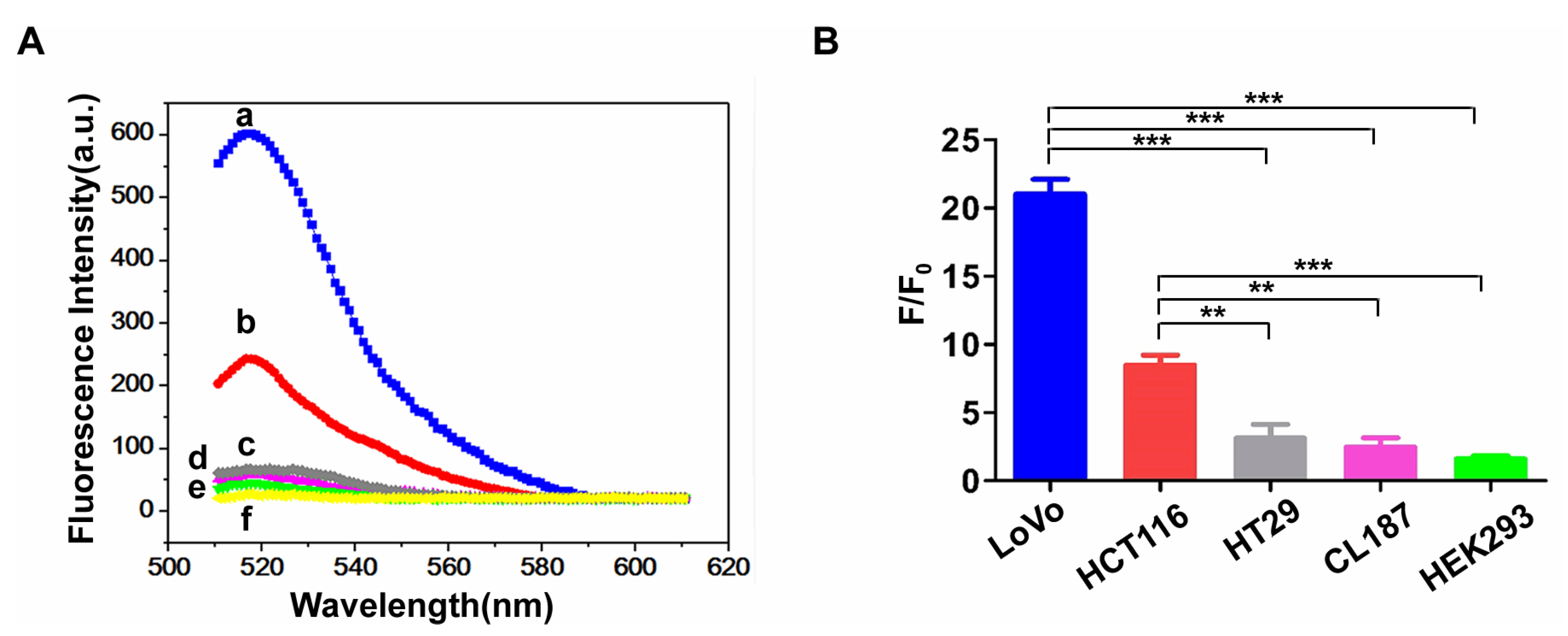
3.6. Specificity Analysis of FAM-W3-GO
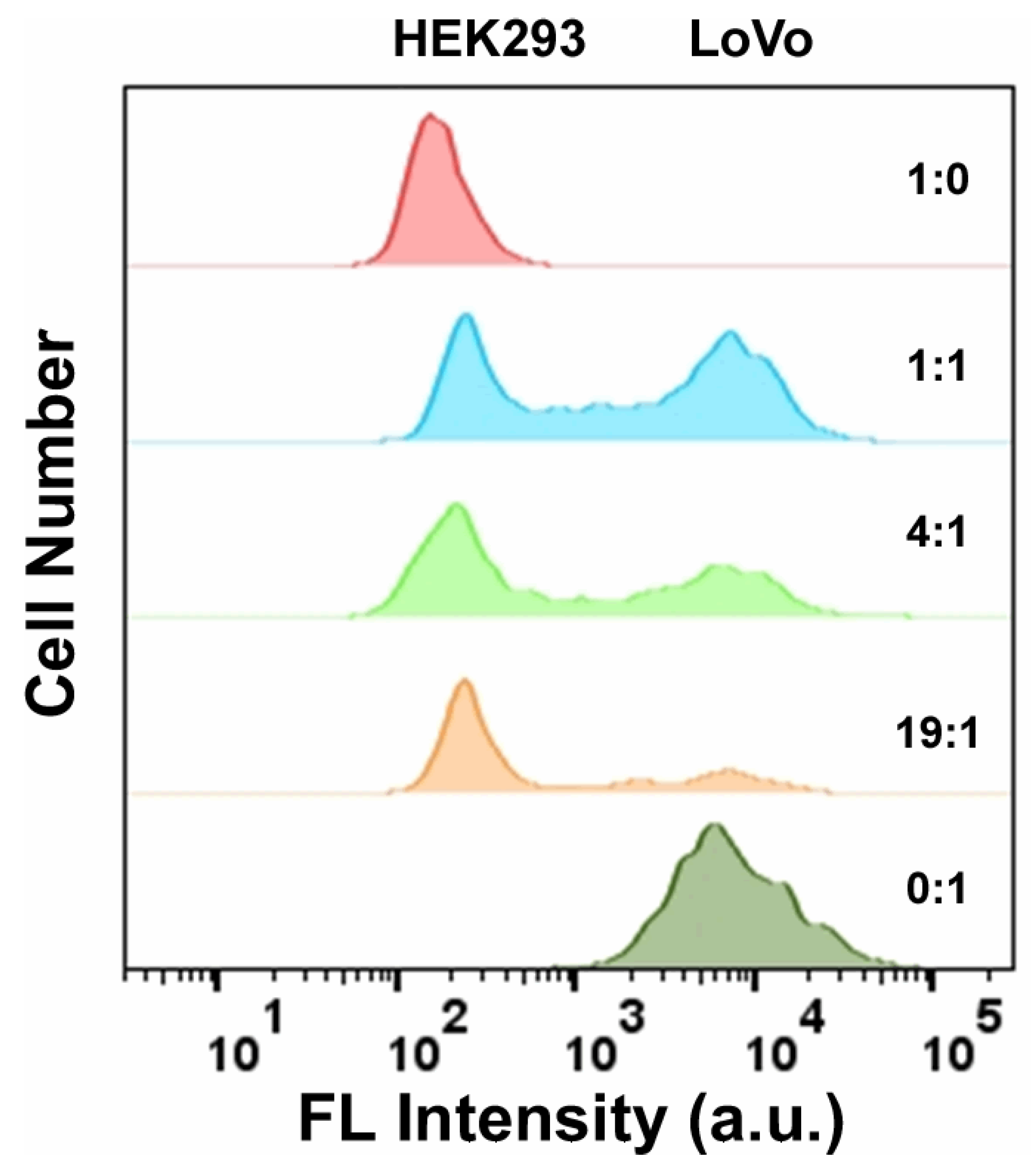
3.7. Detection of LoVo Cells in Whole Blood Samples Using FAM-W3-GO
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Köhne, C.H.; Hitre, E.; Zaluski, J.; Chang Chien, C.R.; Makhson, A.; D’Haens, G.; Pintér, T.; Lim, R.; Bodoky, G.; et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 1408–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katelyn, N.S.; Katherine, H.R.T. Circulating Biomarkers in Breast Cancer. Clin. Breast Cancer 2022, 22, e319–e331. [Google Scholar]
- Barault, L.; Amatu, A.; Siravegna, G.; Ponzetti, A.; Moran, S.; Cassingena, A.; Mussolin, B.; Falcomata, C.; Binder, M.A.; Cristiano, A.; et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2018, 67, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Madanat-Harjuoja, L.M.; Klega, K.; Lu, Y.; Shulman, D.S.; Thorner, A.R.; Nag, A.; Tap, W.D.; Reinke, D.K.; Diller, L.; Ballman, K.V.; et al. Circulating Tumor DNA Is Associated with Response and Survival in Patients with Advanced Leiomyosarcoma. Clin. Cancer Res. 2022, 28, 2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, T.; Wu, Y.; Tan, Y.; Jiang, T.; Li, K.; Lou, B.; Chen, L.; Liu, Y.; Liu, Z. Aptamer based probes for living cell intracellular molecules detection. Biosens. Bioelectron. 2022, 208, 114231. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. Engl. 2021, 60, 16800–16823. [Google Scholar] [CrossRef]
- Centane, S.; Nyokong, T. Aptamer versus antibodies as probe for the impedimetric biosensors for human epidermal growth factor receptor. J. Inorg. Biochem. 2022, 230, 111764. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.H.; Li, Y.; Tang, Z.W.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, J.C.; Tan, W.H. Aptamer evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.M.; Bing, T.; Wang, R.; Jin, S.H.; Shangguan, D.H.; Chen, H. Cell-SELEX-based selection of ssDNA aptamers for specifically targeting BRAF V600E-mutated melanoma. Analyst 2021, 147, 187–195. [Google Scholar] [CrossRef]
- Subjakova, V.; Oravczova, V.; Hianik, T. Polymer Nanoparticles and Nanomoyors Modified by DNA/RNA Aptamers and Antibodies in Targeted Therapy of Cancer. Polymers 2021, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Y.; Xu, X.; Liu, Y.; Lin, B.; Zhang, M.; Zhang, J.; Wan, S.; Yang, C.; Tan, W. Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Zhou, L.L.; Zheng, M.; Fang, J. Selection of Metastatic Breast Cancer Cell-Specific Aptamers for the Capture of CTCs with a Metastatic Phenotype by Cell-SELEX. Mol. Ther. Nucleic Acids 2018, 12, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Chen, H.; Yu, M.; Fang, J. Targeted delivery of doxorubicin using a colorectal cancer-specific ssDNA aptamer. Anat. Rec. 2014, 297, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Biru, E.I.; Necolau, M.I.; Zainea, A.; Iovu, H. Graphene Oxide-Protein-Based Scaffolds for Tissue Engineering: Recent Advance and Applications. Polymers 2022, 14, 1032. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jia, L.; Zhang, Y.; Qu, Y.; Jia, B.; Moss, D.J. Graphene Oxide for Integrated Photonics and Flat Optics. Adv. Mater. 2021, 33, e2006415. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Zhang, W.; Huang, Z.; Huang, J.; Wang, J.; Li, W.; Gu, S. Graphene Oxide-Copper Nanocomposites Suppress Cariogenic Streptococcus mutants Biofilm Formation. Int. J. Nanomed. 2021, 16, 7727–7739. [Google Scholar] [CrossRef]
- Chen, X.W.; Hai, X.; Wang, J.H. Graphene/graphene oxide and their derivatives in the separation/isolation and preconcentration of protein species: A review. Anal. Chim. Acta 2016, 922, 1–10. [Google Scholar] [CrossRef]
- Li, G.Y.; Zeng, J.X.; Liu, H.L.; Ding, P.; Liang, J.T.; Nie, X.M.; Zhou, Z.D. A fluorometric aptamer nanoprobe for alpha-fetoprotein by exploiting the FRET between 5-carboxyfluorescein and palladium ananoparticels. Microchim. Acta 2019, 186, 314. [Google Scholar] [CrossRef]
- Tan, J.; Lai, Z.Q.; Zhong, L.P.; Zhang, Z.H.; Zheng, R.; Su, J.; Huang, Y.; Huang, P.P.; Song, H.; Yang, N.; et al. A Graphene Oxide-Based Fluorescent Aptasensor for the Turn-on Detection of CCRF-CEM. Nanoscale Res. Lett. 2018, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.X.; Lv, J.J.; Chen, L.; Li, S.B.; Mou, X.J.; Xu, Y. A fluorescent probe composed of quantum dot labeled aptamer and graphene oxide for the determination of the lipopolysaccharide endotoxin. Microchim. Acta 2019, 186, 122. [Google Scholar] [CrossRef] [PubMed]
- Safavipour, M.; Kharaziha, M.; Amjadi, E.; Karimzadeh, F.; Allafchian, A. TiO2 nanotubes/reduced GO nanoparticles for sensitive detection of breast cancer cells and photothermal performance. Talanta 2020, 208, 120369. [Google Scholar] [CrossRef]
- Huang, X.; He, Z.; Guo, D.; Liu, Y.; Song, J.; Yung, B.C.; Lin, L.; Yu, G.; Zhu, J.J.; Xiong, Y.; et al. Nanoquenchers to Enhance No-Wash Fluorescence Biosensors for Ratiometric Detection of Cancer Biomarkers. Theranostics 2018, 8, 3461–3473. [Google Scholar] [CrossRef]
- Li, W.M.; Bing, T.; Wei, J.Y.; Chen, Z.Z.; Shangguan, D.H.; Fang, J. Cell-SELEX-based selection of aptamers that recognized distinct targets on metastatic colorectal cancer cells. Biomaterials 2014, 35, 6998–7007. [Google Scholar] [CrossRef] [PubMed]
- De Larrea, C.F.; Staehr, M.; Lopez, A.V.; Ng, K.Y.; Chen, Y.; Godfrey, W.D.; Purdon, T.J.; Ponomarev, V.; Wendel, H.G.; Brentjens, R.J.; et al. Defining an Optimal Dual-Targeted CAR T-cell Therapy Approach Simultaneously Targeting BCMA and GPRC5D to Prevent BCMA Escape-Driven Relapse in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 146–154. [Google Scholar] [CrossRef]
- Shen, Y.; Li, M.; Liu, T.; Liu, J.; Xie, Y.; Zhang, J.; Xu, S.; Liu, H. A dual-functional HER2 aptamer-conjugated, pH-activated mesoporous silica nanocarrier-based drug delivery system provides in vitro synergistic cytotoxicity in Her2-positive breast cancer cells. Int. J. Nanomed. 2019, 14, 4029–4044. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, C.; Martinelli, G.; Papayannidis, C.; Cortes, J.E. Hedgehog Pathway Inhibitors: A new Therapeutic Class for the Treatment of Acute Myeloid Leukemia. Blood Cancer Discov. 2020, 1, 134–145. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Ma, W.J.; Zou, S.Z.; Chen, B.; Cheng, H.; He, X.X.; Wang, K.M. Terminal deoxynucleotidyl transferase-initiated molecule beacons arrayed aptamer probe for sensitive detection of metastatic colorectal cancer cells. Talanta 2019, 202, 152–158. [Google Scholar] [CrossRef]
| Detection Method | Linear Range | Detection Limit (/mL) | Reference |
|---|---|---|---|
| Terminal deoxynucleotidyl | 23–1.5 × 104 | 115 | [31] |
| FAM | 1.2 × 102–2 × 104 | 600 | [31] |
| Graphene Oxide | 1 × 102–1 × 107 | 70 | This work |
| Spiked (Number of Cells/500 μL) | Measured (Number of Cells/500 μL) | Recovery (%) | RSD (%) |
|---|---|---|---|
| 100 | 102 ± 2 | 102.0 ± 1.5 | 1.49 |
| 500 | 466 ± 9 | 93.2 ± 1.7 | 1.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhang, S.; Hsiao, Y.-C.; Wang, Q.; Yu, J.-S.; Li, W. Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells. Polymers 2022, 14, 3040. https://doi.org/10.3390/polym14153040
Chen H, Zhang S, Hsiao Y-C, Wang Q, Yu J-S, Li W. Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells. Polymers. 2022; 14(15):3040. https://doi.org/10.3390/polym14153040
Chicago/Turabian StyleChen, Hang, Shurui Zhang, Yung-Chin Hsiao, Qun Wang, Jau-Song Yu, and Wanming Li. 2022. "Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells" Polymers 14, no. 15: 3040. https://doi.org/10.3390/polym14153040
APA StyleChen, H., Zhang, S., Hsiao, Y.-C., Wang, Q., Yu, J.-S., & Li, W. (2022). Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells. Polymers, 14(15), 3040. https://doi.org/10.3390/polym14153040






