Effects of the Temperature and Salt Concentration on the Structural Characteristics of the Protein (PDB Code 1BBL)
Abstract
:1. Introduction
2. Simulation Details and Methods
3. Results and Discussion
3.1. Effect of the Salt Solution
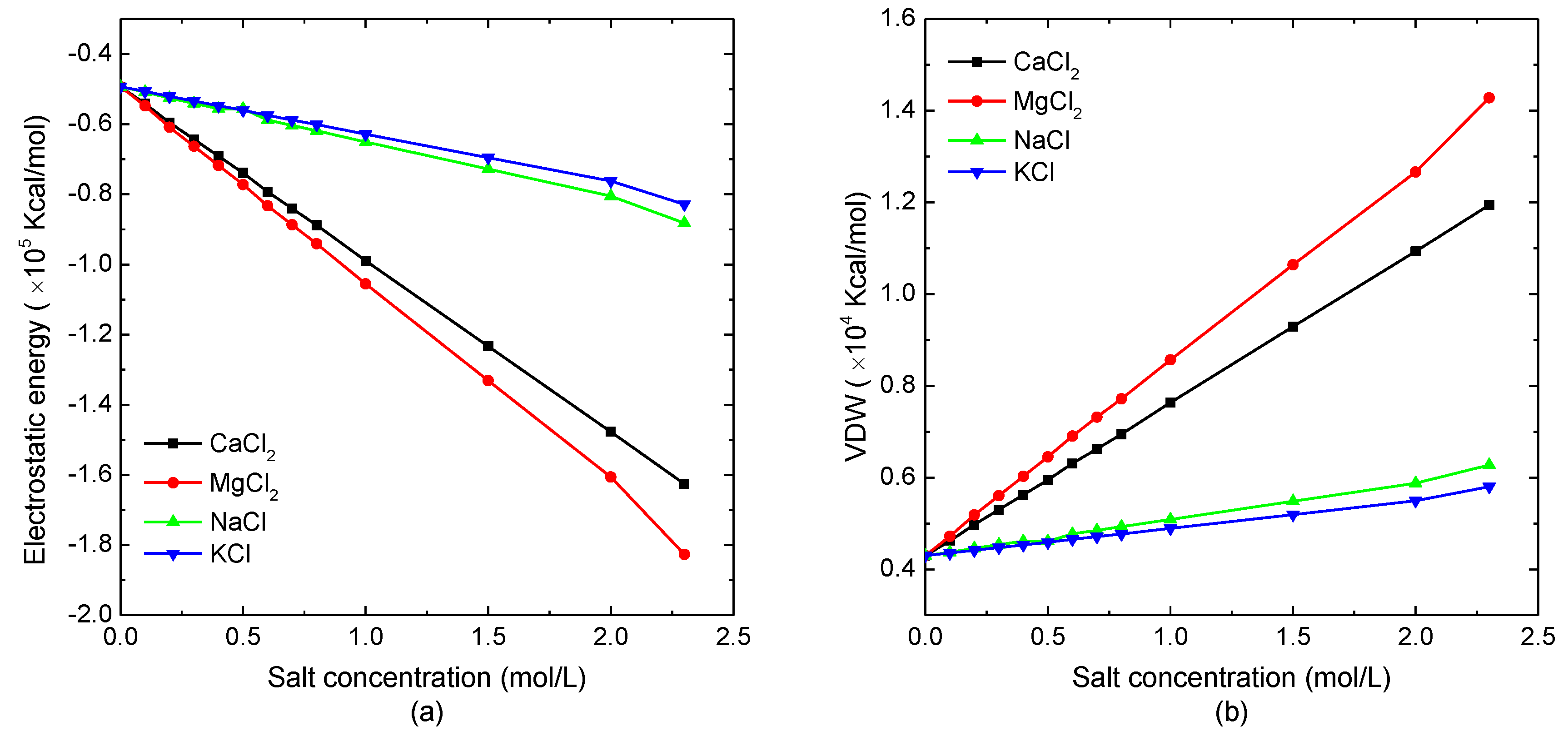
3.1.1. Energy Analysis
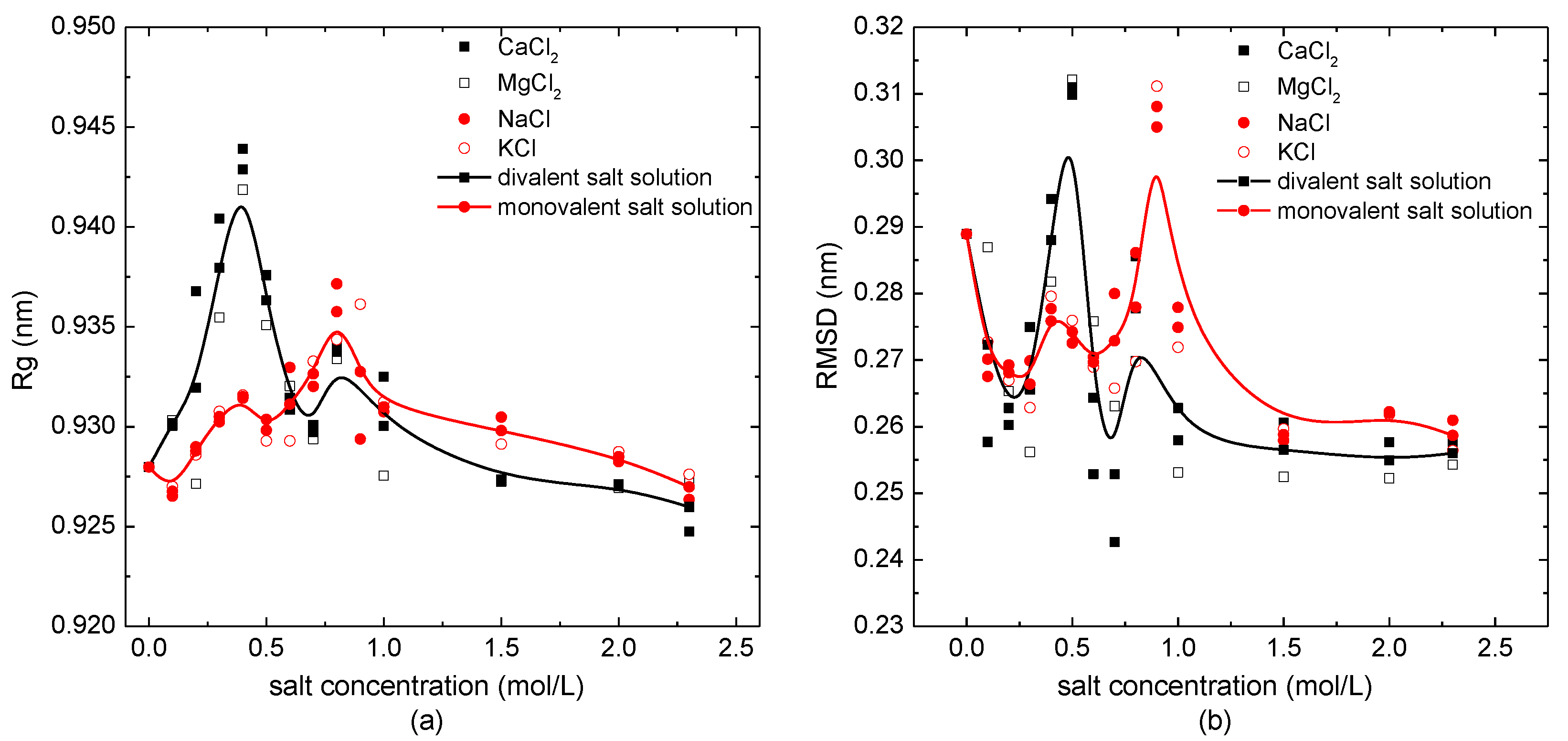
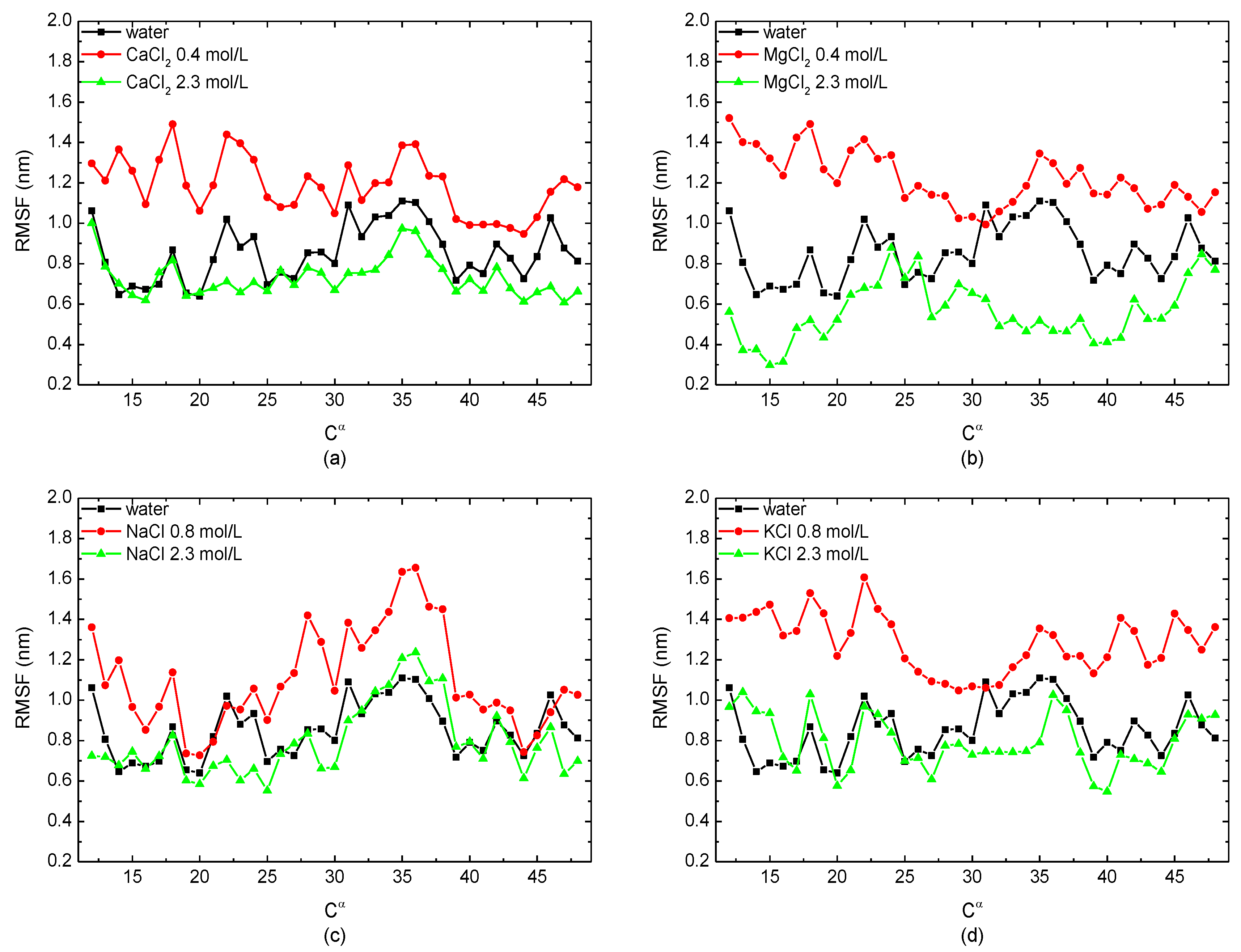
3.1.2. Structural Analysis
3.2. Effect of Temperature
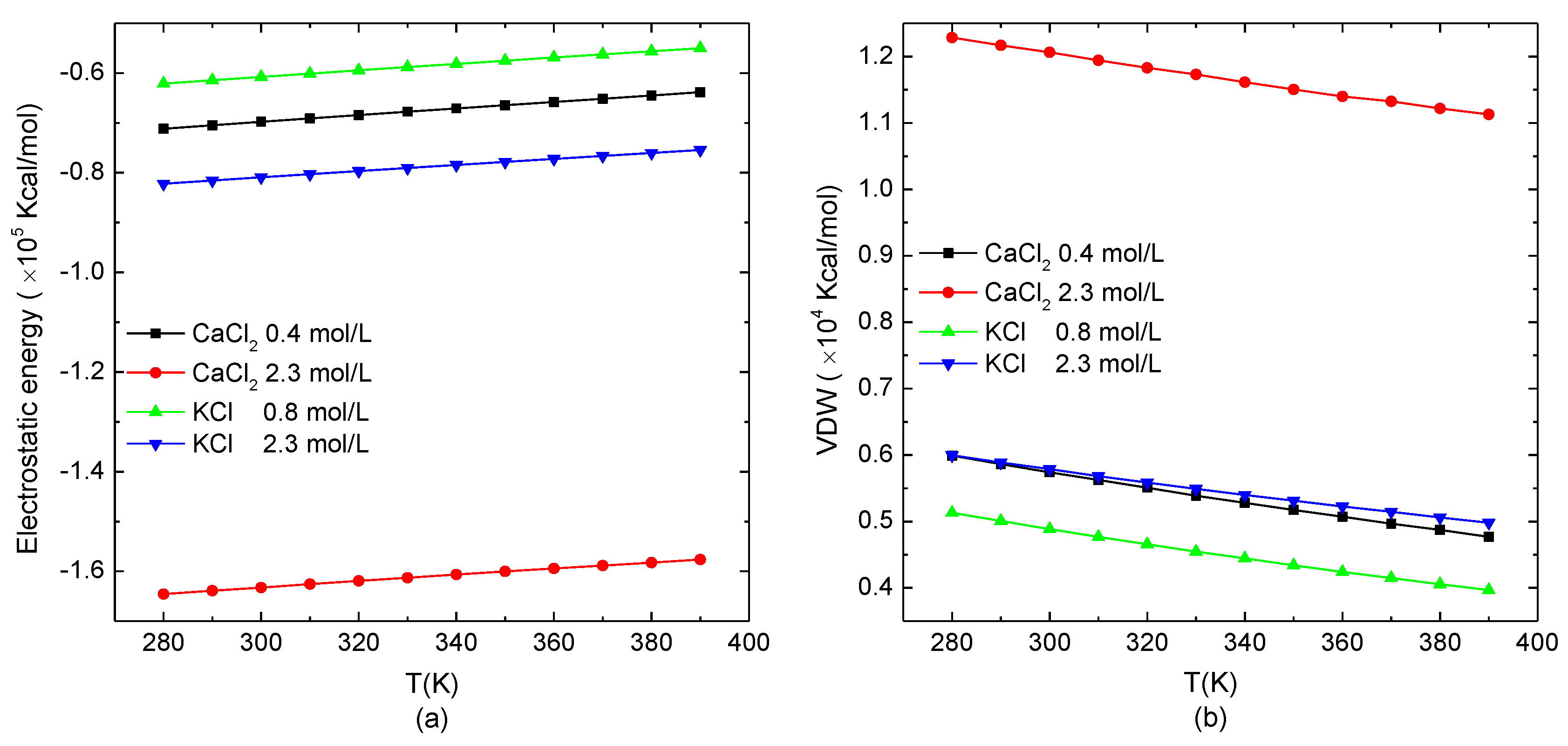
3.2.1. Energy Analysis
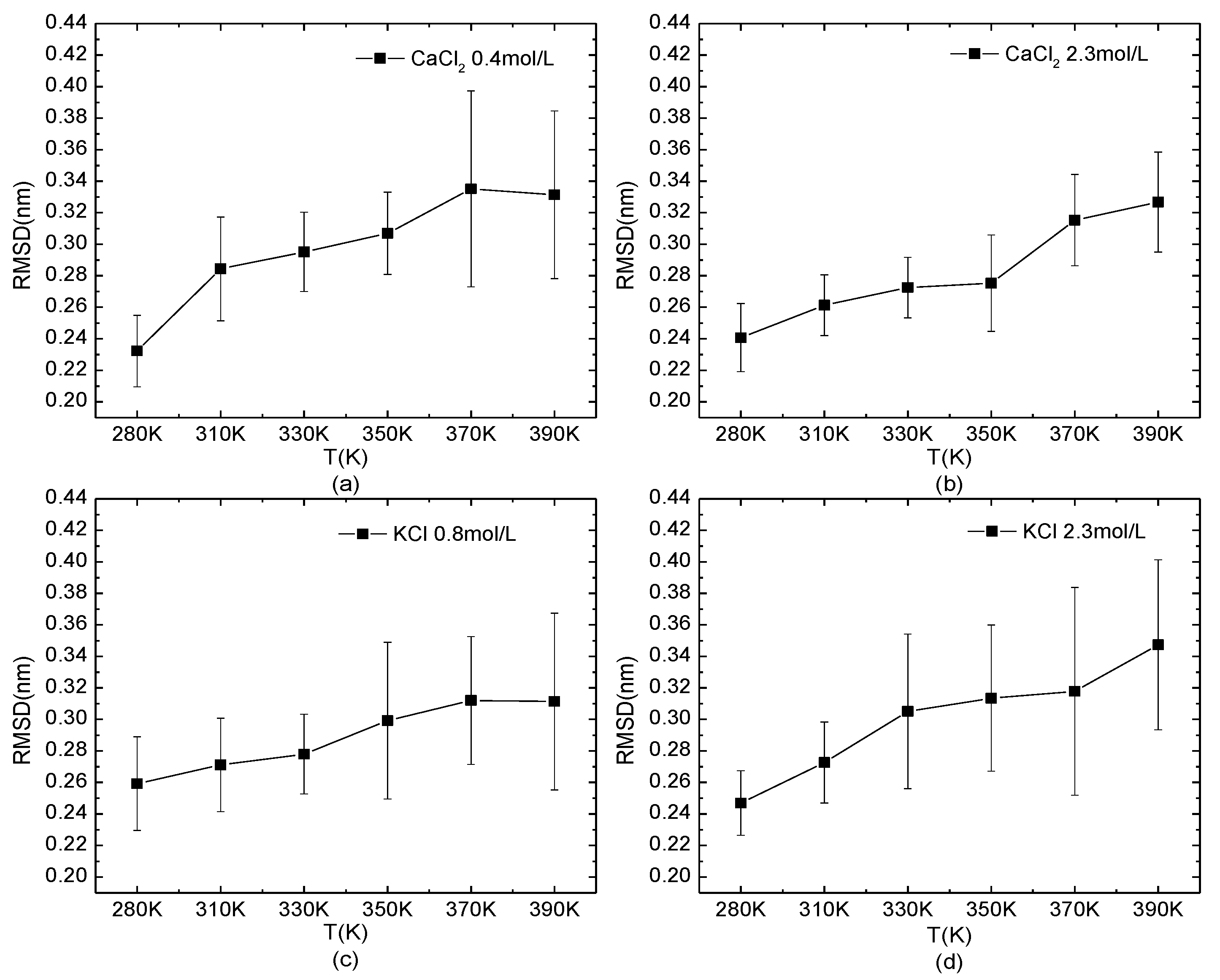
3.2.2. Structural Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anfinsen, C.B. Principles that govern folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counterions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Rozhkov, S.P.; Goryunov, A.S. Phase States of Water-Protein(Polypeptide)-Salt System and Reaction to External Environment Factors. Biophysics 2014, 59, 43–48. [Google Scholar] [CrossRef]
- Aleksandra, K.W.; Vasyl, M. Investigation of Biosensor Potential Component Stability Caused by Influence of External Condition. Ecol. Chem. Eng. 2019, 26, 665–674. [Google Scholar]
- Yuan, G.C.; Kienzle, P.A.; Satija, S.K. Salting Up and Salting Down of Bovine Serum Albumin Layers at the Air-Water Interface. Langmuir 2020, 36, 15240–15246. [Google Scholar] [CrossRef]
- Phillips, R.K.R.; Omanovic, S.; Roscoe, S.G. Electrochemical Studies of the Effect of Temperature on the Adsorption of Yeast Alcohol Dehydrogenase at Pt. Langmuir 2001, 17, 2471–2477. [Google Scholar] [CrossRef]
- Mehan, S.; Chinchalikar, A.J.; Kumar, S.; Aswal, V.K.; Schweins, R. Small-angle neutron scattering study of structure and interaction during salt-induced liquid-liquid phase transition in protein solutions. Phys. Rev. E 2013, 87, 062708. [Google Scholar]
- Feng, H.; Zhou, Z.; Bai, Y. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc. Natl. Acad. Sci. USA 2005, 102, 5026–5031. [Google Scholar] [CrossRef] [Green Version]
- Simone, A.D.; Zagari, A.; Derreumaux, P. Structural and Hydration Properties of the Partially Unfolded States of the Prion Protein. Biophys. J. 2007, 93, 1284–1292. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.X.; Yuan, R.B.; Cui, R.F.; Qiao, L.Y. The dynamics and self-assembly of chemically self-propelled sphere dimmers. Nanoscale 2021, 13, 1055–1060. [Google Scholar] [CrossRef]
- Chen, X.; Duan, D.H.; Zhu, S.Y.; Zhang, J.L. Molecular dynamics simulation of the temperature-induced unfolding of the animal prion protein. J. Mol. Model. 2013, 19, 4433–4441. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, Y.; Guo, F.; He, L.; Zhang, L. Sliding dynamics of multi-rings on a semiflexible polymer in poly[n] catenanes. Soft Matter 2021, 17, 2557–2567. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Limon, A.; Aguilar-Toala, J.E.; Liceaga, A.M. Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for Studying Food Proteins and Bioactive Peptides. J. Agric. Food. Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Yi, Z.; Glass, D.C.; Hong, L.; Tyagi, M.; Baudry, J.; Jain, N.; Smith, J.C. Temperature-dependent dynamical transitions of different classes of amino acid residue in a globular protein. J. Am. Chem. Soc. 2012, 134, 19576–19579. [Google Scholar] [CrossRef]
- Xu, Z.; Lazim, R.; Sun, T.; Mei, Y.; Zhang, D. Solvent effect on the folding dynamics and structure of E6-associated protein characterized from ab initio protein folding simulations. J. Chem. Phys. 2012, 136, 135102. [Google Scholar] [CrossRef]
- Zhang, Q.; Shao, D.Q.; Xu, P.; Jiang, Z.T. Effects of an Electric Field on the Conformational Transition of the Protein: Pulsed and Oscillating Electric Fields with Different Frequencies. Polymers 2022, 14, 123. [Google Scholar] [CrossRef]
- Panuszko, A.; Wojciechowski, M.; Bruzdziak, P.; Rakowska, P.; Stangret, J. Characteristics of hydration water around hen egg lysozyme as the protein model in aqueous solution. FTIR spectroscopy and molecular dynamics simulation. Phys. Chem. Chem. Phys. 2012, 14, 15765–15773. [Google Scholar] [CrossRef]
- Sang, P.; Liu, S.Q.; Yang, L.Q. New Insight into Mechanisms of Protein Adaptation to High Temperatures: A Comparative Molecular Dynamics Simulation Study of Thermophilic and Mesophilic Subtilisin-Like Serine Proteases. Int. J. Mol. Sci. 2020, 21, 3128. [Google Scholar] [CrossRef]
- Dong, Y.W.; Liao, M.L.; Meng, X.L.; Somero, G.N. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine mollusks. Proc. Natl. Acad. Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef] [Green Version]
- Julió Plana, L.; Nadra, A.D.; Estrin, D.A.; Luque, F.J.; Capece, L. Thermal stability of globins: Implications of flexibility and heme coordination studied by molecular dynamics simulations. J. Chem. Inf. Model. 2019, 59, 441–452. [Google Scholar] [CrossRef]
- Jephthah, S.; Staby, L.; Kragelund, B.B.; Skepo, M. Temperature dependence of intrinsically disordered proteins in simulations: What are we missing? J. Chem. Theory Comput. 2019, 15, 2672–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuttke, R.; Hofmann, H.; Nettels, D.; Borgia, M.B.; Mittal, J.; Best, R.B.; Schuler, B. Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 5213–5218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pucci, F.; Dhanani, M.; Dehouck, Y.; Rooman, M. Protein thermostability prediction within homologous families using temperature-dependent statistical potentials. PLoS ONE 2014, 9, e91659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoque, M.A.; Ahmed, F.; Halim, M.A.; Molla, M.R.; Rana, S. Influence of salt and temperature on the interaction of bovine serum albumin with cetylpyridinium chloride: Insights from experimental and molecular dynamics simulation. J. Mol. Liq. 2018, 260, 121–130. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S.N. Mechanism of protein salting in and salting out by divalent cation salts: Balance between hydration and salt binding. Biochemistry 1984, 23, 5912–5923. [Google Scholar] [CrossRef]
- Okur, H.I.; Hladilková, J.; Rembert, K.B.; Cho, Y.; Heyda, J.; Dzubiella, J.; Cremer, P.S.; Jungwirth, P. Beyond the Hofmeister series: Ion-specific effects on proteins and their biological functions. J. Phys. Chem. B 2017, 121, 1997–2014. [Google Scholar] [CrossRef]
- Duong-Ly, K.C.; Gabelli, S.B. Salting out of Proteins Using Ammonium Sulfate Precipitation. Meth. Enzymol. 2014, 541, 85–94. [Google Scholar]
- Jiang, Y.; Zhu, Y.; Zheng, Y.; Liu, Z.; Zhong, Y.; Deng, Y.; Zhao, Y. Effects of salting-in/out-assisted extractions on structural, physicochemical and functional properties of Tenebrio Molitor larvae protein isolates. Food Chem. 2021, 338, 128158. [Google Scholar] [CrossRef]
- Hubbard, T.J.P.; Ailey, B.; Brenner, S.E.; Murzin, A.G.; Chothia, C. SCOP: A Structural Classification of Proteins database. Nucleic Acids Res. 1999, 27, 254–256. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, D.; Zhang, Q.; Xu, P.; Jiang, Z. Effects of the Temperature and Salt Concentration on the Structural Characteristics of the Protein (PDB Code 1BBL). Polymers 2022, 14, 2134. https://doi.org/10.3390/polym14112134
Shao D, Zhang Q, Xu P, Jiang Z. Effects of the Temperature and Salt Concentration on the Structural Characteristics of the Protein (PDB Code 1BBL). Polymers. 2022; 14(11):2134. https://doi.org/10.3390/polym14112134
Chicago/Turabian StyleShao, Dongqing, Qun Zhang, Peng Xu, and Zhouting Jiang. 2022. "Effects of the Temperature and Salt Concentration on the Structural Characteristics of the Protein (PDB Code 1BBL)" Polymers 14, no. 11: 2134. https://doi.org/10.3390/polym14112134
APA StyleShao, D., Zhang, Q., Xu, P., & Jiang, Z. (2022). Effects of the Temperature and Salt Concentration on the Structural Characteristics of the Protein (PDB Code 1BBL). Polymers, 14(11), 2134. https://doi.org/10.3390/polym14112134





