The Nanopore-Tweezing-Based, Targeted Detection of Nucleobases on Short Functionalized Peptide Nucleic Acid Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Buffer Solutions and Sample Preparation
2.3. Electrophysiology Experiments
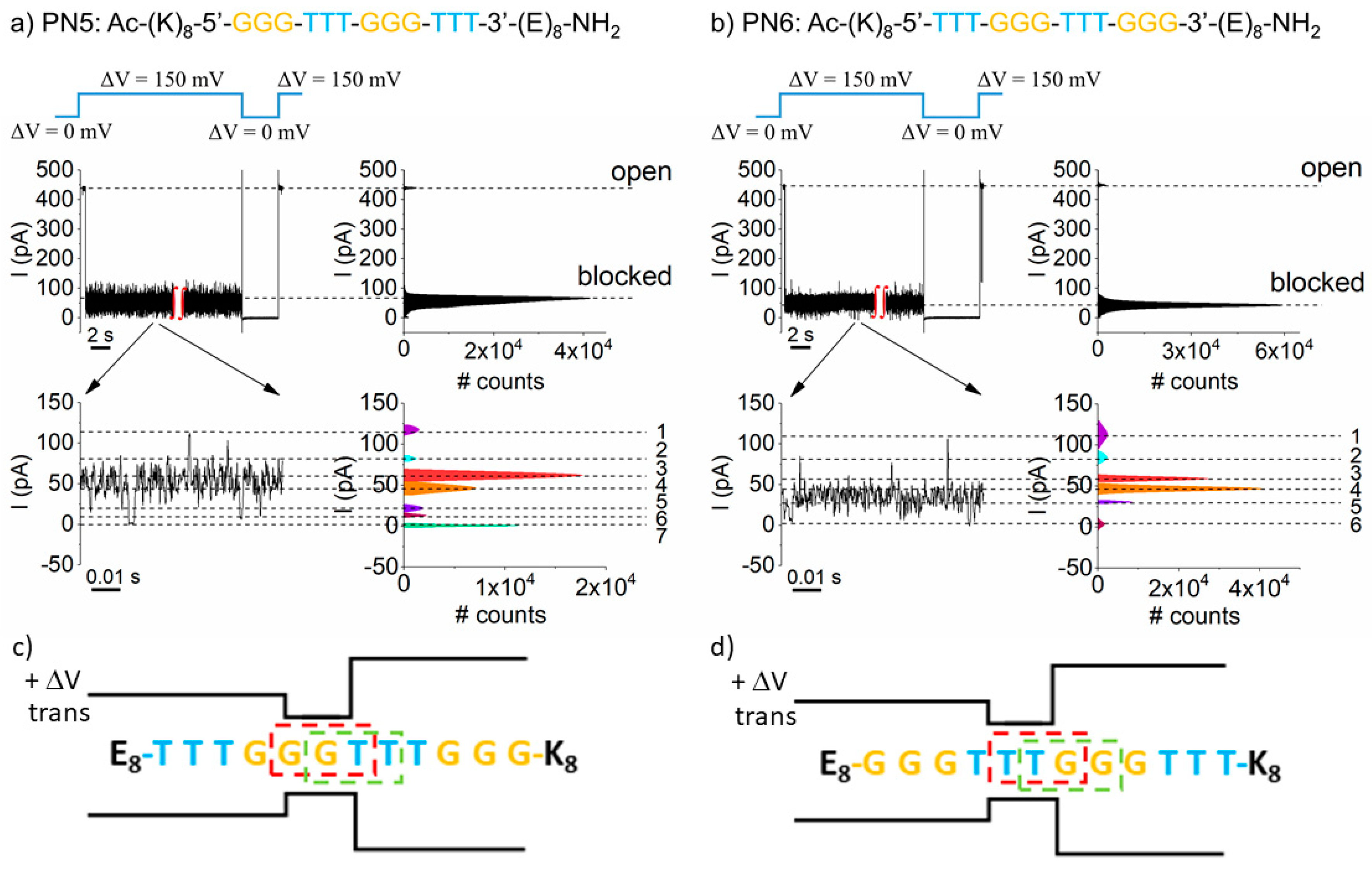
3. Results and Discussion
4. Use of Homopolymeric PNAs to Investigate Sequence Recognition with the Nanopore
5. Triplet Base Recognition in a Heteropolymeric PNA Background
6. The PN6 (K8–T3–G3–T3–G3–E8) PNA-Induced Conductance Fluctuations in a Single α-HL Nanopore are Voltage-Dependent
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shendure, J.A.; Porreca, G.J.; Church, G.M. Current Protocols in Molecular Biology; Ausubel, F.M., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2008. [Google Scholar]
- Bayley, H. Sequencing Single Molecules of DNA. Curr. Opin. Chem. Biol. 2006, 10, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Q.; Wang, Z. The Evolution of Nanopore Sequencing. Front. Genet. 2015, 5, 449. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Discrimination of Oligonucleotides of Different Lengths with a Wild-Type Aerolysin Nanopore. Nat. Nanotechnol. 2016, 11, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.Z.; Pavlenok, M.; Derrington, I.M.; Niederweis, M.; Gundlach, J.H. Single-Molecule DNA Detection with an Engineered MspA Protein Nanopore. Proc. Natl. Acad. Sci. USA 2008, 105, 20647–20652. [Google Scholar] [CrossRef]
- Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous Base Identification for Single-Molecule Nanopore DNA Sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef]
- Wanunu, M. Nanopores: A Journey towards DNA Sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Robertson, J.W.F.; Chan, E.R.; Reiner, J.E.; Stanford, V.M. Nanoscopic Porous Sensors. Annu. Rev. Anal. Chem. 2008, 1, 737–766. [Google Scholar] [CrossRef]
- Venkatesan, B.M.; Bashir, R. Nanopore Sensors for Nucleic Acid Analysis. Nat. Nano 2011, 6, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.-Q.; Shim, J.W. Single Molecule Sensing by Nanopores and Nanopore devices. Analyst 2010, 135, 441–451. [Google Scholar] [CrossRef]
- Cao, C.; Li, M.Y.; Cirauqui, N.; Wang, Y.Q.; dal Peraro, M.; Tian, H.; Long, Y.-T. Mapping the Sensing Spots of Aerolysin for Single Oligonucleotides Analysis. Nat. Commun. 2018, 9, 2823. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef]
- Akeson, M.; Branton, D.; Kasianowicz, J.J.; Brandin, E.; Deamer, D.W. Microsecond Time-Scale Discrimination among Polycytidylic acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments within Single RNA Molecules. Biophys. J. 1999, 77, 3227–3233. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.; Branton, D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 1079–1084. [Google Scholar] [CrossRef]
- Fologea, D.; Uplinger, J.; Thomas, B.; McNabb, D.S.; Li, J. Slowing DNA Translocation in a Solid-State Nanopore. Nano Lett. 2005, 5, 1734–1737. [Google Scholar] [CrossRef]
- Deamer, D.W.; Branton, D. Characterization of Nucleic Acids by Nanopore Analysis. Acc. Chem. Res. 2002, 35, 817–825. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef]
- Howorka, S.; Cheley, S.; Bayley, H. Sequence-Specific Detection of Individual DNA Strands Using Engineered Nanopores. Nat. Biotechnol. 2001, 19, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Ashkenasy, N.; Sánchez-Quesada, J.; Bayley, H.; Ghadiri, M.R. Recognizing a Single Base in an Individual DNA Strand: A Step Toward Nanopore DNA Sequencing. Angew. Chem. Int. Ed. 2005, 44, 1401–1404. [Google Scholar] [CrossRef] [PubMed]
- Derrington, I.M.; Butler, T.Z.; Collins, M.D.; Manrao, E.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Nanopore DNA Sequencing with MspA. Proc. Natl. Acad. Sci. USA 2010, 107, 16060–16065. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C. Solid-State Nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef]
- NHGRI. “Revolutionary Genome Sequencing Technologies—The $1000 Genome,” in NIH. 2004. Available online: http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04-003.html (accessed on 3 March 2021).
- Cockroft, S.L.; Chu, J.; Amorin, M.; Ghadiri, M.R. A Single-Molecule Nanopore Device Detects DNA Polymerase Activity with Single-Nucleotide Resolution. J. Am. Chem. Soc. 2008, 130, 818–820. [Google Scholar] [CrossRef]
- Astier, Y.; Braha, O.; Bayley, H. Toward Single Molecule DNA Sequencing: Direct Identification of Ribonucleoside and Deoxyribonucleoside 5′-Monophosphates by Using an Engineered Protein Nanopore Equipped with a Molecular Adapter. J. Am. Chem. Soc. 2006, 128, 1705–1710. [Google Scholar] [CrossRef] [PubMed]
- Mathé, J.; Aksimentiev, A.; Nelson, D.R.; Schulten, K.; Meller, A. Orientation Discrimination of Single-Stranded DNA Inside the alpha-Hemolysin Membrane Channel. Proc. Natl. Acad. Sci. USA 2005, 102, 12377–12382. [Google Scholar] [CrossRef] [PubMed]
- Henrickson, S.E.; Misakian, M.; Robertson, B.; Kasianowicz, J.J. Driven DNA Transport into an Asymmetric Nanometer-Scale Pore. Phys. Rev. Lett. 2000, 85, 3057–3060. [Google Scholar] [CrossRef] [PubMed]
- Nakane, J.; Wiggin, M.; Marziali, A. A Nanosensor for Transmembrane Capture and Identification of Single Nucleic Acid Molecules. Biophys. J. 2004, 87, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Quesada, J.; Saghatelian, A.; Cheley, S.; Bayley, H.; Ghadiri, M.R. Single DNA Rotaxanes of a Transmembrane Pore Protein. Angew. Chem. Int. Ed. 2004, 43, 3063–3067. [Google Scholar] [CrossRef] [PubMed]
- Purnell, R.F.; Mehta, K.K.; Schmidt, J.J. Nucleotide Identification and Orientation Discrimination of DNA Homopolymers Immobilized in a Protein Nanopore. Nano Lett. 2008, 8, 3029–3034. [Google Scholar] [CrossRef]
- Asandei, A.; Chinappi, M.; Lee, J.K.; Seo, C.H.; Mereuta, L.; Park, Y.; Luchian, T. Placement of Oppositely Charged Aminoacids at a Polypeptide Termini Determines the Voltage-Controlled Braking of Polymer Transport through Nanometer-Scale pores. Sci. Rep. 2015, 5, 10419. [Google Scholar] [CrossRef] [PubMed]
- Chinappi, M.; Luchian, T.; Cecconi, F. Nanopore Tweezers: Voltage-Controlled Trapping and Releasing of Analytes. Phys. Rev. E 2015, 92, 032714. [Google Scholar] [CrossRef]
- Asandei, A.; Di Muccio, G.; Schiopu, I.; Mereuta, L.; Dragomir, I.S.; Chinappi, M.; Luchian, T. Nanopore-Based Protein Sequencing Using Biopores: Current Achievements and Open Challenges. Small Methods 2020, 4, 1900595. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA Hybridizes to Complementary Oligonucleotides Obeying the Watson-Crick Hydrogen-Bonding Rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Haaima, G. Peptide Nucleic acid (PNA). A DNA Mimic with a Pseudopeptide Backbone. Chem. Soc. Rev. 1997, 26, 73–78. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide Nucleic Acids: Protocols and Applications, 2nd ed.; Horizon Bioscience: Cambridge, UK, 2004. [Google Scholar]
- Marin, V.L.; Roy, S.; Armitage, B.A. Recent Advances in the Development of Peptide Nucleic Acid as a Gene-Targeted Drug. Expert Opin. Biol. Ther. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Gupta, A.; Mishra, A.; Puri, N. Peptide Nucleic Acids: Advanced Tools for Biomedical Applications. J. Biotechnol. 2017, 259, 148–159. [Google Scholar] [CrossRef]
- Ciuca, A.; Asandei, A.; Schiopu, I.; Apetrei, A.; Mereuta, L.; Seo, C.H.; Park, Y.; Luchian, T. Single-Molecule, Real-Time Dissecting of Peptide Nucleic Acid-DNA Duplexes with a Protein Nanopore Tweezer. Anal. Chem. 2018, 90, 7682–7690. [Google Scholar] [CrossRef]
- Mereuta, L.; Asandei, A.; Schiopu, I.; Park, Y.; Luchian, T. Nanopore-Assisted, Sequence-Specific Detection, and Single-Molecule Hybridization Analysis of Short, Single-Stranded DNAs. Anal. Chem. 2019, 91, 8630–8637. [Google Scholar] [CrossRef]
- Marchelli, R.; Corradini, R.; Manicardi, A.; Sforza, S.; Tedeschi, T.; Fabbri, E.; Borgatti, M.; Bianchi, N.; Gambari, R. Targets in Gene Therapy; You, Y., Ed.; BoD—Books on Demand: Rijeka, Croatia, 2011. [Google Scholar]
- Tian, K.; He, Z.; Wang, Y.; Chen, S.-J.; Gu, L.-Q. Designing a Polycationic Probe for Simultaneous Enrichment and Detection of MicroRNAs in a Nanopore. ACS Nano 2013, 7, 3962–3969. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.-Q. Nanopore-Based Detection of Circulating microRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Zhou, S.; Roozbahani, G.M.; Zhang, Y.; Wang, D.; Guan, X. Displacement Chemistry-Based Nanopore Analysis of Nucleic Acids in Complicated Matrices. Chem. Commun. 2018, 54, 13977–13980. [Google Scholar] [CrossRef]
- Wong, C.T.A.; Muthukumar, M. Polymer Translocation through α-Hemolysin Pore with Tunable Polymer-Pore Electrostatic Interaction. J. Chem. Phys. 2010, 133, 045101. [Google Scholar] [CrossRef] [PubMed]
- Asandei, A.; Chinappi, M.; Kang, H.-K.; Seo, C.H.; Mereuta, L.; Park, Y.; Luchian, T. Acidity-Mediated, Electrostatic Tuning of Asymmetrically Charged Peptides Interactions with Protein Nanopores. ACS Appl. Mater. Interfaces 2015, 7, 16706–16714. [Google Scholar] [CrossRef]
- Mereuta, L.; Luchian, T.; Park, Y.; Hahm, K.-S. Single-Molecule Investigation of the Interactions between Reconstituted Planar Lipid Membranes and an Analogue of the HP(2–20) Antimicrobial Peptide. Biochem. Biophys. Res. Commun. 2008, 373, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Mereuta, L.; Asandei, A.; Luchian, T. Meet Me on the Other Side: Trans-Bilayer Modulation of a Model Voltage-Gated Ion Channel Activity by Membrane Electrostatics Asymmetry. PLoS ONE 2011, 6, e25276. [Google Scholar] [CrossRef][Green Version]
- Asandei, A.; Rossini, A.E.; Chinappi, M.; Park, Y.; Luchian, T. Protein Nanopore-Based Discrimination between Selected Neutral Amino Acids from Polypeptides. Langmuir 2017, 33, 14451–14459. [Google Scholar] [CrossRef] [PubMed]
- Asandei, A.; Dragomir, I.S.; Di Muccio, G.; Chinappi, M.; Park, Y.; Luchian, T. Single-Molecule Dynamics and Discrimination between Hydrophilic and Hydrophobic Amino Acids in Peptides, through Controllable, Stepwise Translocation across Nanopores. Polymers 2018, 10, 885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Pérez, L.R.; Soskine, M.; Maglia, G.; Joo, C.; Dekker, C.; Aksimentiev, A. Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate. ACS Nano 2019, 13, 2398–2409. [Google Scholar] [CrossRef] [PubMed]
- Pérez, L.R.; Wong, C.H.; Maglia, G.; Dekker, C.; Joo, C. Label-Free Detection of Post-translational Modifications with a Nanopore. Nano Lett. 2019, 19, 7957–7964. [Google Scholar] [CrossRef]
- Ralph, R.K.; Connors, W.J.; Khorana, H.G. Secondary Structure and Aggregation in Deoxyguanosine Oligonucleotides. J. Am. Chem. Soc. 1962, 84, 2265–2266. [Google Scholar] [CrossRef]
- Stoddart, D.; Heron, A.J.; Mikhailova, E.; Maglia, G.; Bayley, H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. USA 2009, 106, 7702–7707. [Google Scholar] [CrossRef] [PubMed]
- Asandei, A.; Ciuca, A.; Apetrei, A.; Schiopu, I.; Mereuta, L.; Seo, C.H.; Park, Y.; Luchian, T. Nanoscale Investigation of Generation 1 PAMAM Dendrimers Interaction with a Protein Nanopore. Sci. Rep. 2017, 7, 6167. [Google Scholar] [CrossRef] [PubMed]
- Guy, A.T.; Piggot, T.J.; Khalid, S. Single-Stranded DNA within Nanopores: Conformational Dynamics and Implications for Sequencing; A Molecular Dynamics Simulation Study. Biophys. J. 2012, 103, 1028–1036. [Google Scholar] [CrossRef]
- Buhot, A.; Halperin, A. Effects of Stacking on the Configurations and Elasticity of Single-Stranded Nucleic Acids. Phys. Rev. E 2004, 70, 020902. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Gerstein, M. Calculation of Standard Atomic Volumes for RNA and Comparison with Proteins: RNA is packed more tightly. J. Mol. Biol. 2005, 346, 477–492. [Google Scholar] [CrossRef]
- Aalberts, D.P.; Parman, J.M.; Goddard, N.L. Single-Strand Stacking Free Energy from DNA Beacon kinetics. Biophys. J. 2003, 84, 3212–3217. [Google Scholar] [CrossRef]
- Smith, S.B.; Cui, Y.J.; Bustamante, C. Overstretching B-DNA: The Elastic Response of Individual Double-Stranded and Single-Stranded DNA Molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef] [PubMed]



| PN1 | Ac-(K)11-5′-GGG-GGG-3′-(E)11-NH2 |
| PN2 | Ac-(K)8-5′-TTT-TTT-TTT-TTT-3′-(E)8-NH2 |
| PN3 | Ac-(K)8-5′-AAA-AAA-AAA-AAA-3′-(E)8-NH2 |
| PN4 | Ac-(K)8-5′-CCC-CCC-CCC-CCC-3′-(E)8-NH2 |
| PN5 | Ac-(K)8-5′-GGG-TTT-GGG-TTT-3′-(E)8-NH2 |
| PN6 | Ac-(K)8-5′-TTT-GGG-TTT-GGG-3′-(E)8-NH2 |
| PN1: K11 − G6 − E11 | PN4: K8 − C12 − E8 | |
| Total relative blockade | −0.746 ± 0.007 | −0.849 ± 0.012 |
| 1 | −0.735 ± 0.002 | −0.779 ± 0.007 |
| 2 | −0.756 ± 0.007 | −0.813 ± 0.003 |
| 3 | −0.779 ± 0.003 | −0.839 ± 0.003 |
| 4 | −0.800 ± 0.002 | −0.857 ± 0.001 |
| 5 | −0.817 ± 0.003 | −0.873 ± 0.002 |
| 6 | −0.862 ±0.011 | −0.901 ± 0.005 |
| 7 | - | −0.941 ± 0.005 |
| PN3: K8 − A12 − E8 | PN2: K8 − T12 − E8 | |
| Total relative blockade | −0.830 ± 0.023 | −0.844 ± 0.034 |
| 1 | −0.736 ± 0.005 | −0.701 ± 0.009 |
| 2 | −0.791 ± 0.005 | −0.760 ± 0.012 |
| 3 | −0.82 ±0.002 | −0.846 ± 0.003 |
| 4 | −0.872 ± 0.004 | −0.940 ± 0.006 |
| 5 | −0.943 ± 0.002 | −0.97 ± 0.014 |
| 6 | −0.973 ± 0.001 | - |
| PN5: K8 − (G3 − T3)2 − E8 | PN6: K8 − (T3 − G3)2 − E8 | |
| Total relative blockade | −0.839 ± 0.021 | −0.871 ± 0.007 |
| 1 | −0.742 ± 0.004 | −0.748 ± 0.005 |
| 2 | −0.809 ± 0.003 | −0.822 ± 0.001 |
| 3 | −0.858 ± 0.003 | −0.875 ± 0.003 |
| 4 | −0.898 ± 0.004 | −0.901 ± 0.002 |
| 5 | −0.943 ± 0.002 | −0.95 ± 0.001 |
| 6 | −0.978 ± 0.001 | −0.988 ± 0.001 |
| 7 | −0.996 ± 0.0005 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dragomir, I.S.; Asandei, A.; Schiopu, I.; Bucataru, I.C.; Mereuta, L.; Luchian, T. The Nanopore-Tweezing-Based, Targeted Detection of Nucleobases on Short Functionalized Peptide Nucleic Acid Sequences. Polymers 2021, 13, 1210. https://doi.org/10.3390/polym13081210
Dragomir IS, Asandei A, Schiopu I, Bucataru IC, Mereuta L, Luchian T. The Nanopore-Tweezing-Based, Targeted Detection of Nucleobases on Short Functionalized Peptide Nucleic Acid Sequences. Polymers. 2021; 13(8):1210. https://doi.org/10.3390/polym13081210
Chicago/Turabian StyleDragomir, Isabela S., Alina Asandei, Irina Schiopu, Ioana C. Bucataru, Loredana Mereuta, and Tudor Luchian. 2021. "The Nanopore-Tweezing-Based, Targeted Detection of Nucleobases on Short Functionalized Peptide Nucleic Acid Sequences" Polymers 13, no. 8: 1210. https://doi.org/10.3390/polym13081210
APA StyleDragomir, I. S., Asandei, A., Schiopu, I., Bucataru, I. C., Mereuta, L., & Luchian, T. (2021). The Nanopore-Tweezing-Based, Targeted Detection of Nucleobases on Short Functionalized Peptide Nucleic Acid Sequences. Polymers, 13(8), 1210. https://doi.org/10.3390/polym13081210







