On the Use of the Discrete Constant pH Molecular Dynamics to Describe the Conformational Space of Peptides
Abstract
1. Introduction
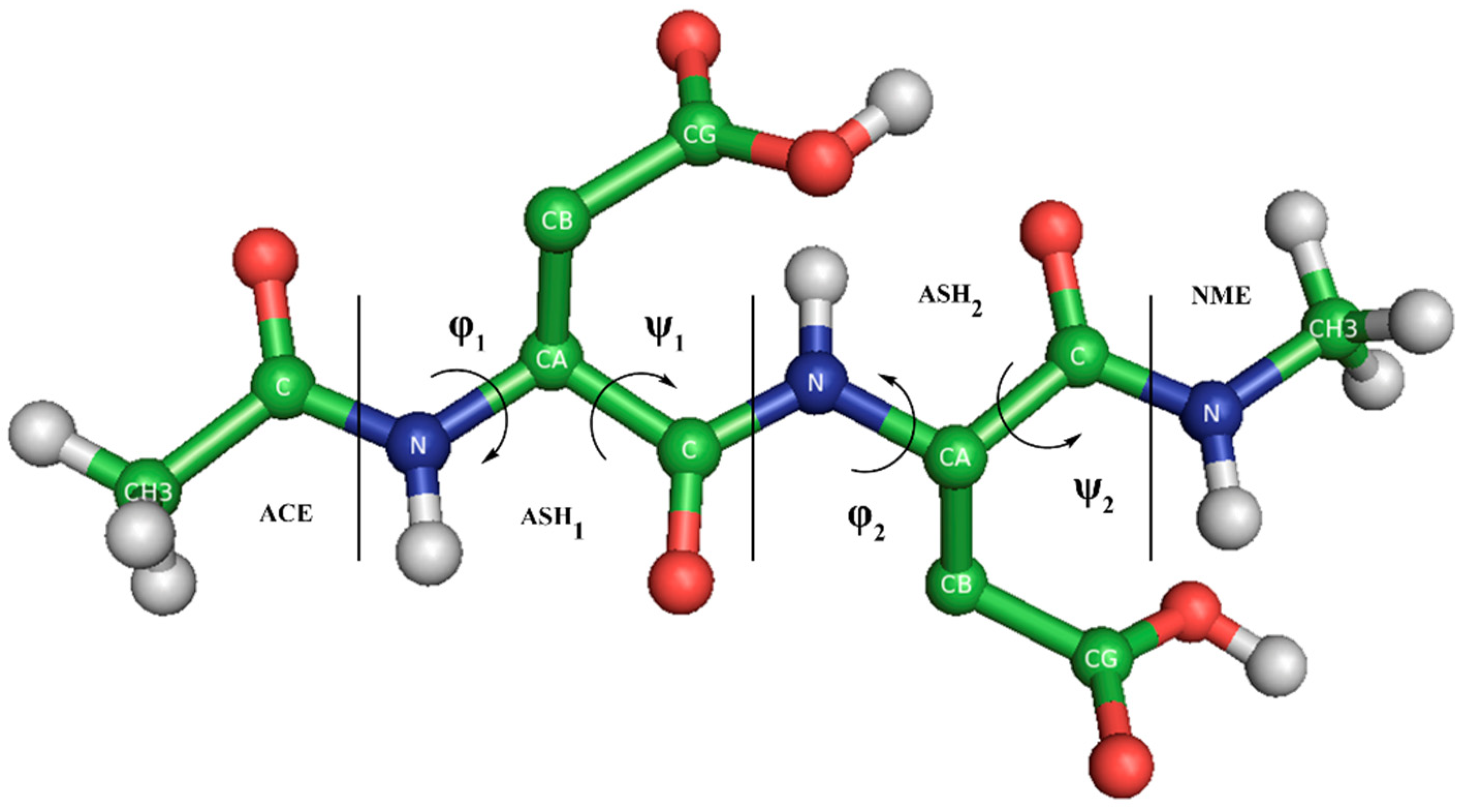
2. Materials and Methods
3. Results
3.1. Gibbs Free Energies in the Ramachandran Space
3.1.1. Basic pKa Amino Acids
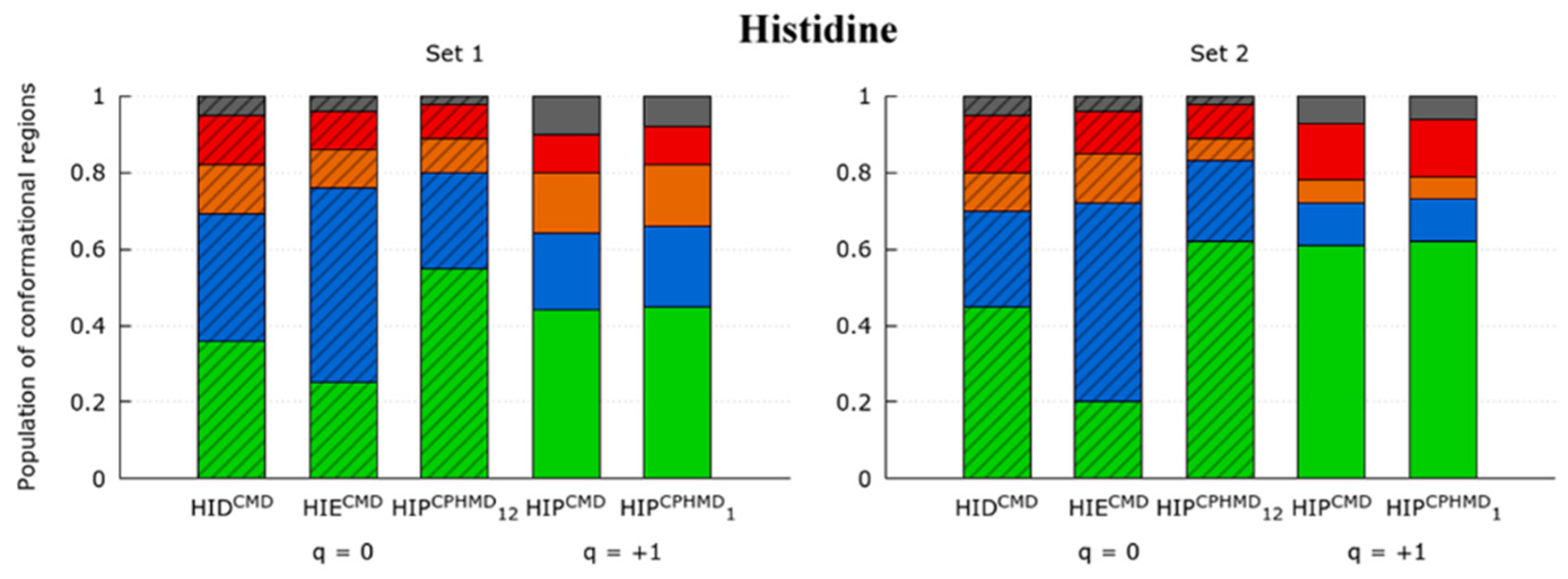
3.1.2. Histidine
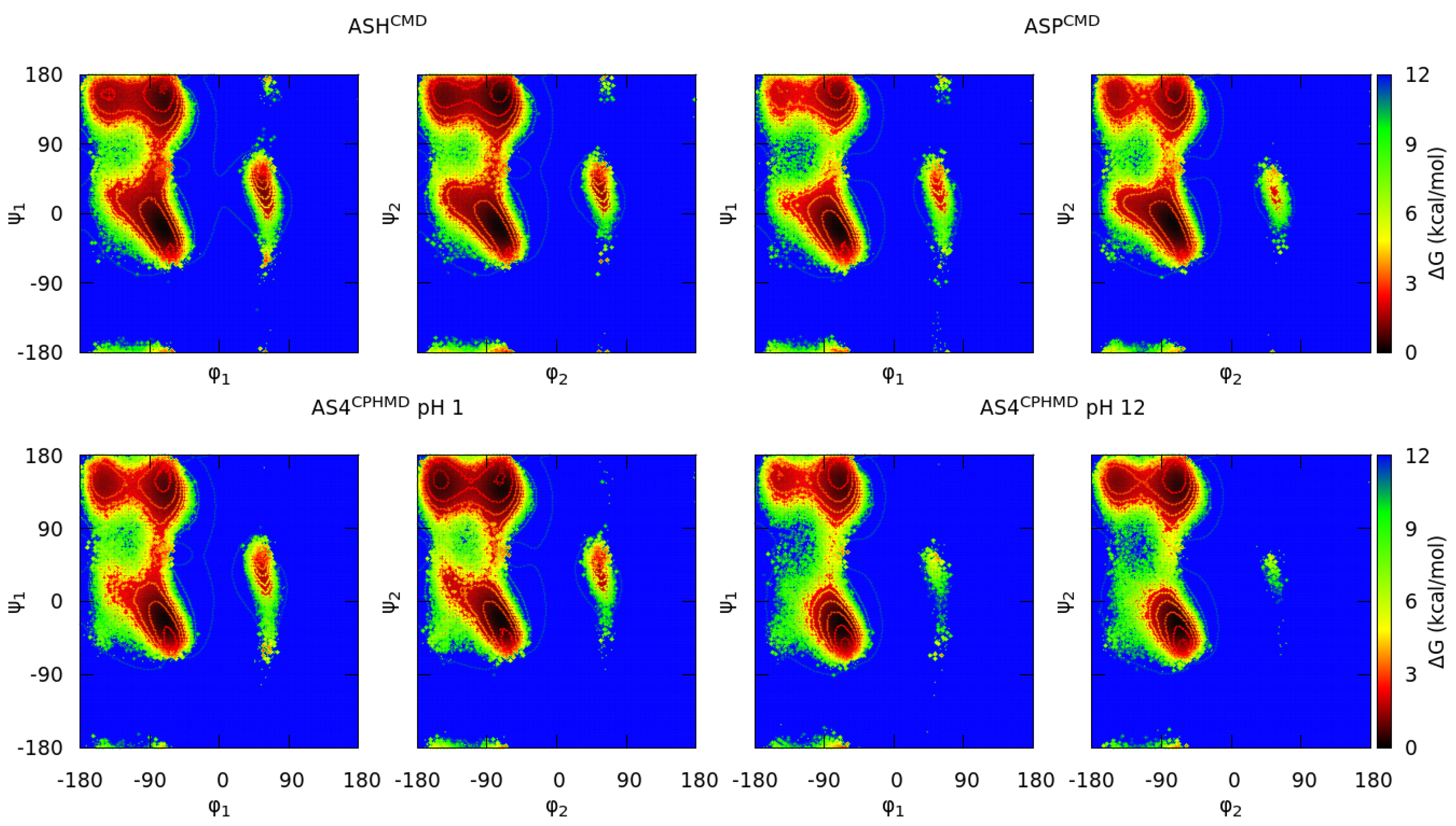
3.1.3. Acidic Amino Acids
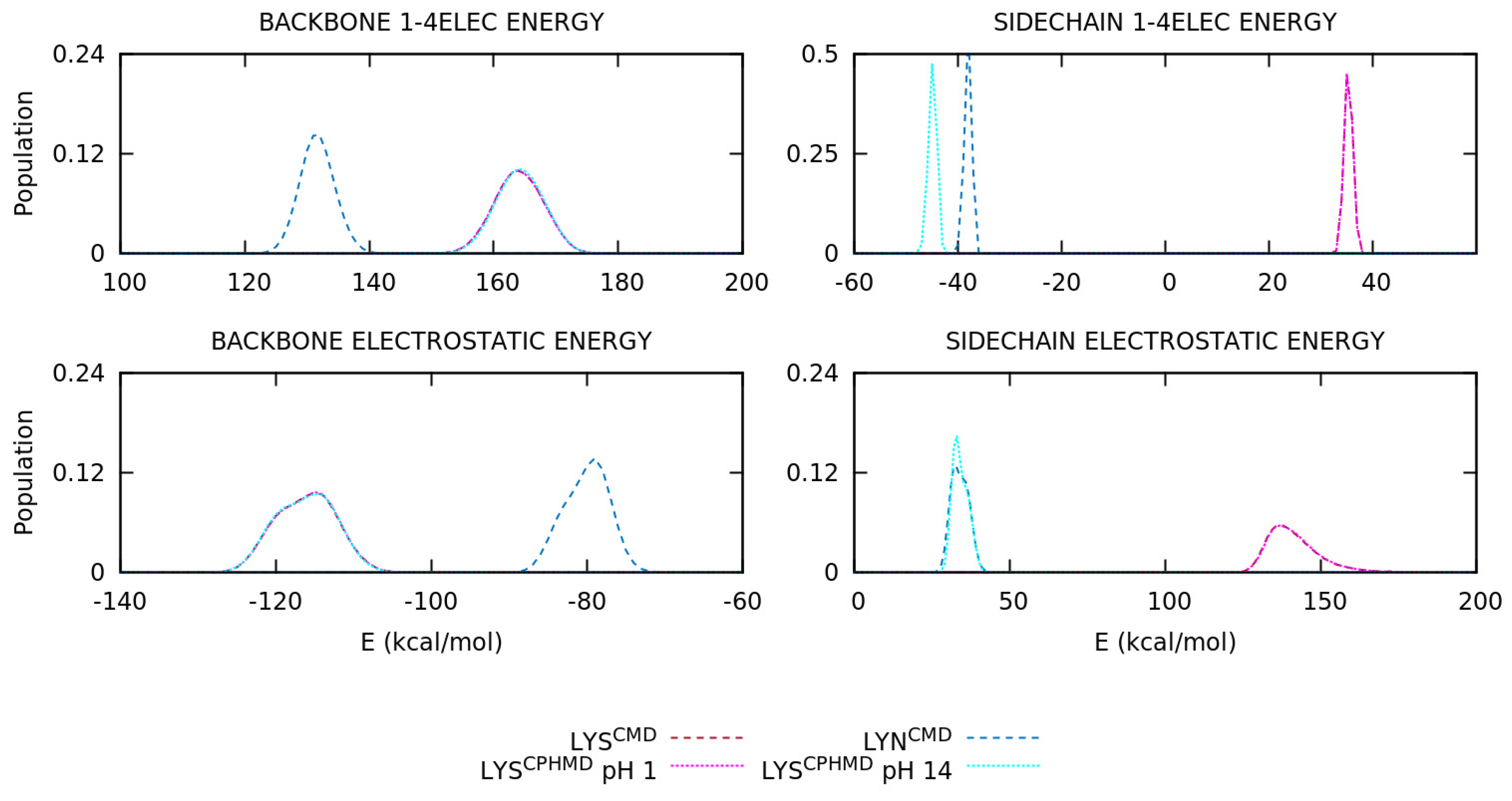
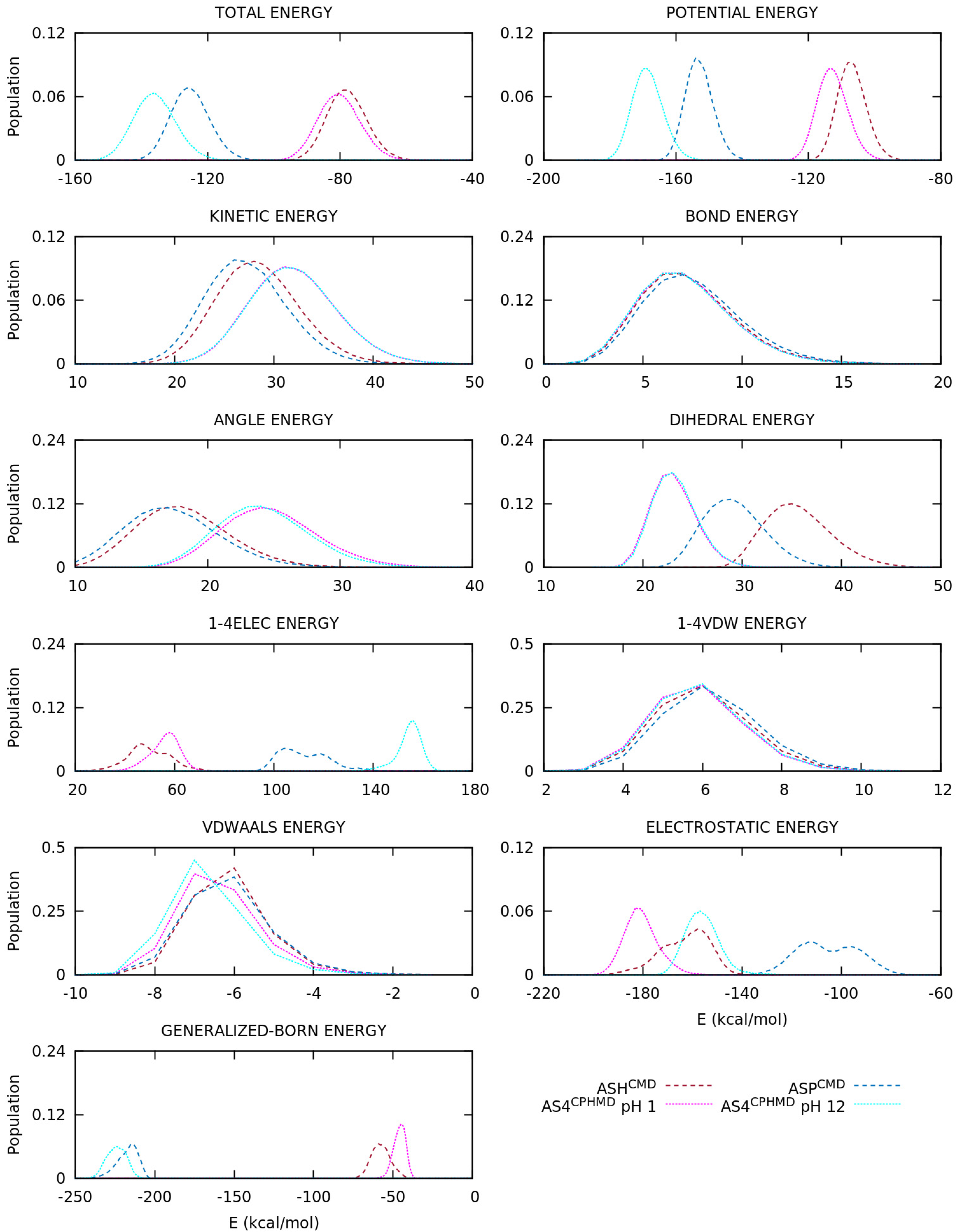
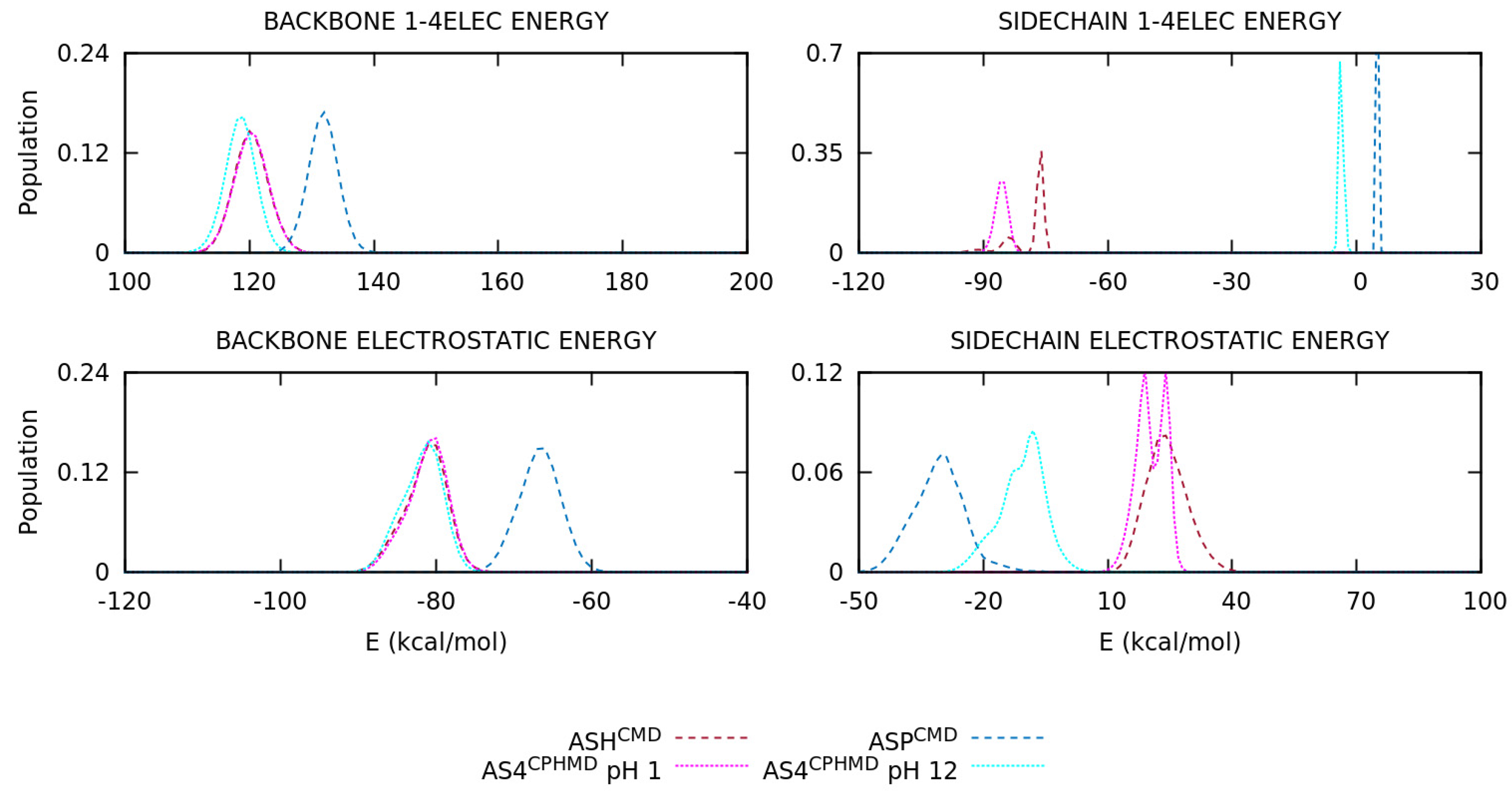
3.2. Energetic Contributions
3.3. Side Chain Orientation and Atom Distances
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bohacek, R.S.; McMartin, C.; Guida, W.C. The art and practice of structure-based drug design: A molecular modeling per-spective. Med. Res. Rev. 1996, 16, 3–50. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Aminpour, M.; Choi, H.-J.; Tuszyński, J.A. An Overview of Molecular Modeling for Drug Discovery with Specific Illustrative Examples of Applications. Molecules 2019, 24, 1693. [Google Scholar] [CrossRef]
- Klepeis, J.L.; Lindorff-Larsen, K.; Dror, R.O.; Shaw, D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009, 19, 120–127. [Google Scholar] [CrossRef]
- Hamelberg, D.; Mongan, J.; McCammon, J.A. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004, 120, 11919–11929. [Google Scholar] [CrossRef]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef]
- Barducci, A.; Bonomi, M.; Parrinello, M. Metadynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 826–843. [Google Scholar] [CrossRef]
- Sugita, Y.; Okamotoab, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Asokan, A.; Cho, M.J. Exploitation of intracellular pH gradients in the cellular delivery of macromolecules. J. Pharm. Sci. 2002, 91, 903–913. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, 537–541. [Google Scholar] [CrossRef]
- Barazorda-Ccahuana, H.L.; Gómez, B.; Mas, F.; Madurga, S. Effect of pH on the Supramolecular Structure of Helicobacter pylori Urease by Molecular Dynamics Simulations. Polymer 2020, 12, 2713. [Google Scholar] [CrossRef] [PubMed]
- Beroza, P.; Fredkin, D.R.; Okamura, M.Y.; Feher, G. Protonation of interacting residues in a protein by a Monte Carlo method: Application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. USA 1991, 88, 5804–5808. [Google Scholar] [CrossRef] [PubMed]
- Mertz, J.E.; Pettitt, B.M. Molecular Dynamics At a Constant pH. Int. J. Supercomput. Appl. High Perform. Comput. 1994, 8, 47–53. [Google Scholar] [CrossRef]
- Baptista, A.M.; Martel, P.J.; Petersen, S.B. Simulation of protein conformational freedom as a function of pH: Constant-pH molecular dynamics using implicit titration. Proteins Struct. Funct. Bioinform. 1997, 27, 523–544. [Google Scholar] [CrossRef]
- Börjesson, U.; Hünenberger, P.H. Explicit-solvent molecular dynamics simulation at constant pH: Methodology and application to small amines. J. Chem. Phys. 2001, 114, 9706–9719. [Google Scholar] [CrossRef]
- Lee, M.S.; Salsbury, F.R.; Brooks, C.L. Constant-pH molecular dynamics using continuous titration coordinates. Proteins Struct. Funct. Bioinform. 2004, 56, 738–752. [Google Scholar] [CrossRef]
- Khandogin, J.; Brooks, C.L. Constant pH Molecular Dynamics with Proton Tautomerism. Biophys. J. 2005, 89, 141–157. [Google Scholar] [CrossRef]
- Donnini, S.; Tegeler, F.; Groenhof, G.; Grubmüller, H. Constant pH Molecular Dynamics in Explicit Solvent with λ-Dynamics. J. Chem. Theory Comput. 2011, 7, 1962–1978. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.A.; Shen, J.K. Continuous Constant pH Molecular Dynamics in Explicit Solvent with pH-Based Replica Exchange. J. Chem. Theory Comput. 2011, 7, 2617–2629. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.B.; Hulbert, B.S.; Zhou, H.; Brooks, C.L. Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism. Proteins Struct. Funct. Bioinform. 2014, 82, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Brooks, C.L. λ-dynamics: A new approach to free energy calculations. J. Chem. Phys. 1996, 105, 2414–2423. [Google Scholar] [CrossRef]
- Baptista, A.M.; Teixeira, V.H.; Soares, C.M. Constant-pH molecular dynamics using stochastic titration. J. Chem. Phys. 2002, 117, 4184–4200. [Google Scholar] [CrossRef]
- Bürgi, R.; Kollman, P.A.; Van Gunsteren, W.F. Simulating proteins at constant pH: An approach combining molecular dynamics and Monte Carlo simulation. Proteins Struct. Funct. Bioinform. 2002, 47, 469–480. [Google Scholar] [CrossRef]
- Mongan, J.; Case, D.A.; McCammon, J.A. Constant pH molecular dynamics in generalized Born implicit solvent. J. Comput. Chem. 2004, 25, 2038–2048. [Google Scholar] [CrossRef]
- Meng, Y.; Roitberg, A.E. Constant pH Replica Exchange Molecular Dynamics in Biomolecules Using a Discrete Protonation Model. J. Chem. Theory Comput. 2010, 6, 1401–1412. [Google Scholar] [CrossRef]
- Itoh, S.G.; Damjanović, A.; Brooks, B.R. pH replica-exchange method based on discrete protonation states. Proteins Struct. Funct. Bioinform. 2011, 79, 3420–3436. [Google Scholar] [CrossRef]
- Swails, J.M.; York, D.M.; Roitberg, A.E. Constant pH Replica Exchange Molecular Dynamics in Explicit Solvent Using Discrete Protonation States: Implementation, Testing, and Validation. J. Chem. Theory Comput. 2014, 10, 1341–1352. [Google Scholar] [CrossRef]
- Dlugosz, M.; Antosiewicz, J.M.; Robertson, A.D. Constant- p H molecular dynamics study of protonation-structure relationship in a heptapeptide derived from ovomucoid third domain. Phys. Rev. E 2004, 69, 021915. [Google Scholar] [CrossRef]
- Machuqueiro, M.; Baptista, A.M. Acidic range titration of HEWL using a constant-pH molecular dynamics method. Proteins Struct. Funct. Bioinform. 2008, 72, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Swails, J.M.; Roitberg, A.E. Enhancing Conformation and Protonation State Sampling of Hen Egg White Lysozyme Using pH Replica Exchange Molecular Dynamics. J. Chem. Theory Comput. 2012, 8, 4393–4404. [Google Scholar] [CrossRef] [PubMed]
- Di Russo, N.V.; Estrin, D.A.; Marti, M.A.; Roitberg, A.E. pH-Dependent Conformational Changes in Proteins and Their Effect on Experimental pKas: The Case of Nitrophorin. PLoS Comput. Biol. 2012, 8, e1002761. [Google Scholar] [CrossRef] [PubMed]
- McDougal, O.M.; Granum, D.M.; Swartz, M.; Rohleder, C.; Maupin, C.M. pKa Determination of Histidine Residues in α-Conotoxin MII Peptides by 1H NMR and Constant pH Molecular Dynamics Simulation. J. Phys. Chem. B 2013, 117, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Gupta, P.L.; Roitberg, A.E. pH-Dependent Conformational Changes Due to Ionizable Residues in a Hydrophobic Protein Interior: The Study of L25K and L125K Variants of SNase. J. Phys. Chem. B 2019, 123, 5742–5754. [Google Scholar] [CrossRef] [PubMed]
- Khandogin, J.; Chen, J.; Brooks, C.L., III. Exploring atomistic details of pH-dependent peptide folding. Proc. Natl. Acad. Sci. USA 2006, 103, 18546–18550. [Google Scholar] [CrossRef]
- Williams, S.L.; Blachly, P.G.; McCammon, J.A. Measuring the successes and deficiencies of constant pH molecular dynamics: A blind prediction study. Proteins Struct. Funct. Bioinform. 2011, 79, 3381–3388. [Google Scholar] [CrossRef]
- Machuqueiro, M.; Baptista, A.M. Is the prediction of pK a values by constant-pH molecular dynamics being hindered by inherited problems? Proteins Struct. Funct. Bioinform. 2011, 79, 3437–3447. [Google Scholar] [CrossRef]
- Wallace, J.A.; Wang, Y.; Shi, C.; Pastoor, K.J.; Nguyen, B.-L.; Xia, K.; Shen, J.K. Toward accurate prediction of pKa values for internal protein residues: The importance of conformational relaxation and desolvation energy. Proteins Struct. Funct. Bioinform. 2011, 79, 3364–3373. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Song, J. Extensive Assessment of Various Computational Methods for Aspartate’s pKa Shift. J. Chem. Inf. Model. 2017, 57, 1621–1639. [Google Scholar] [CrossRef] [PubMed]
- Mongan, J.; Case, D.A. Biomolecular simulations at constant pH. Curr. Opin. Struct. Biol. 2005, 15, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Morrow, B.H.; Shi, C.; Shen, J. Recent development and application of constant pH molecular dynamics. Mol. Simul. 2014, 40, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Machuqueiro, M.; Baptista, A.M. Constant-pH Molecular Dynamics with Ionic Strength Effects: Protonation-Conformation Coupling in Decalysine. J. Phys. Chem. B 2006, 110, 2927–2933. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.L.; de Oliveira, C.A.F.; McCammon, J.A. Coupling Constant pH Molecular Dynamics with Accelerated Molecular Dynamics. J. Chem. Theory Comput. 2010, 6, 560–568. [Google Scholar] [CrossRef]
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188. [Google Scholar] [CrossRef]
- Chen, W.; Wallace, J.A.; Yue, Z.; Shen, J. Introducing Titratable Water to All-Atom Molecular Dynamics at Constant pH. Biophys. J. 2013, 105, L15–L17. [Google Scholar] [CrossRef]
- Yeager, A.V.; Swails, J.M.; Miller, B.R. Improved Accuracy for Constant pH-REMD Simulations through Modification of Carboxylate Effective Radii. J. Chem. Theory Comput. 2017, 13, 4624–4635. [Google Scholar] [CrossRef]
- Harris, R.C.; Shen, J. GPU-Accelerated Implementation of Continuous Constant pH Molecular Dynamics in Amber: pKa Predictions with Single-pH Simulations. J. Chem. Inf. Model. 2019, 59, 4821–4832. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Iii, T.E.C.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K. Amber 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C.L. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins Struct. Funct. Bioinform. 2004, 55, 383–394. [Google Scholar] [CrossRef]
- Perez, J.J.; Tomaš, M.; Rubio-Martinez, J. Assessment of the Sampling Performance of Multiple-Copy Dynamics versus a Unique Trajectory. J. Chem. Inf. Model. 2016, 56, 1950–1962. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, I.T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Rubio-Martinez, J.; Tomaš, M.; Perez, J.J. Effect of the solvent on the conformational behavior of the alanine dipeptide deduced from MD simulations. J. Mol. Graph. Model. 2017, 78, 118–128. [Google Scholar] [CrossRef]
- Williams, T.; Kelley, C.; Bröker, H.-B.; Campbell, J.; Cunningham, R.; Denholm, D.; Elber, G.; Fearick, R.; Grammes, C.; Hart, L.; et al. Gnuplot 4.6, 2012. Gnuplotting. Available online: http://www.gnuplotting.org/manpage-gnuplot-4-6/#x1-378000III (accessed on 29 December 2020).
- Pietropaolo, A.; Muccioli, L.; Zannoni, C.; La Mendola, D.; Maccarrone, G.; Pappalardo, A.G.; Rizzarelli, E. Unveiling the Role of Histidine and Tyrosine Residues on the Conformation of the Avian Prion Hexarepeat Domain. J. Phys. Chem. B 2008, 112, 5182–5188. [Google Scholar] [CrossRef]






| Residue | CMD | CpHMD | Prot. State. | Intrinsic pKa | ||
|---|---|---|---|---|---|---|
| pH 1 | pH 12 | pH 14 | ||||
| ASP | ✓ | D | 4.0 | |||
| ASH | ✓ | P | ||||
| AS4 | ✓ | ✓ | T | |||
| GLU | ✓ | D | 4.4 | |||
| GLH | ✓ | P | ||||
| GL4 | ✓ | ✓ | T | |||
| HIE | ✓ | D | 7.1 (ε) 6.5 (δ) | |||
| HID | ✓ | D | ||||
| HIP | ✓ | ✓ | ✓ | P/T | ||
| CYM | ✓ | D | 8.5 | |||
| CYS | ✓ | ✓ | ✓ | P/T | ||
| TYR | ✓ | ✓ | P/T | 9.6 | ||
| LYN | ✓ | D | 10.4 | |||
| LYS | ✓ | ✓ | ✓ | P/T | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Privat, C.; Madurga, S.; Mas, F.; Rubio-Martínez, J. On the Use of the Discrete Constant pH Molecular Dynamics to Describe the Conformational Space of Peptides. Polymers 2021, 13, 99. https://doi.org/10.3390/polym13010099
Privat C, Madurga S, Mas F, Rubio-Martínez J. On the Use of the Discrete Constant pH Molecular Dynamics to Describe the Conformational Space of Peptides. Polymers. 2021; 13(1):99. https://doi.org/10.3390/polym13010099
Chicago/Turabian StylePrivat, Cristian, Sergio Madurga, Francesc Mas, and Jaime Rubio-Martínez. 2021. "On the Use of the Discrete Constant pH Molecular Dynamics to Describe the Conformational Space of Peptides" Polymers 13, no. 1: 99. https://doi.org/10.3390/polym13010099
APA StylePrivat, C., Madurga, S., Mas, F., & Rubio-Martínez, J. (2021). On the Use of the Discrete Constant pH Molecular Dynamics to Describe the Conformational Space of Peptides. Polymers, 13(1), 99. https://doi.org/10.3390/polym13010099








