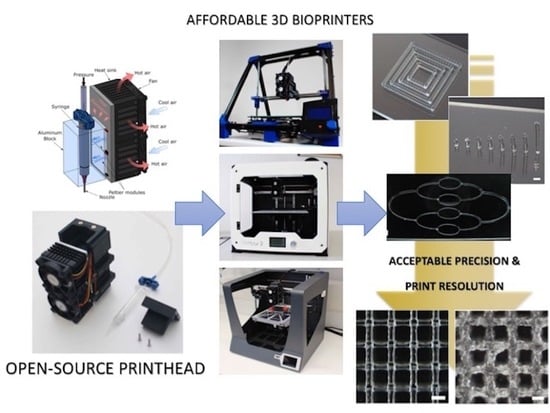
A Versatile Open-Source Printhead for Low-Cost 3D Microextrusion-Based Bioprinting
Abstract
1. Introduction
2. Materials and Methods
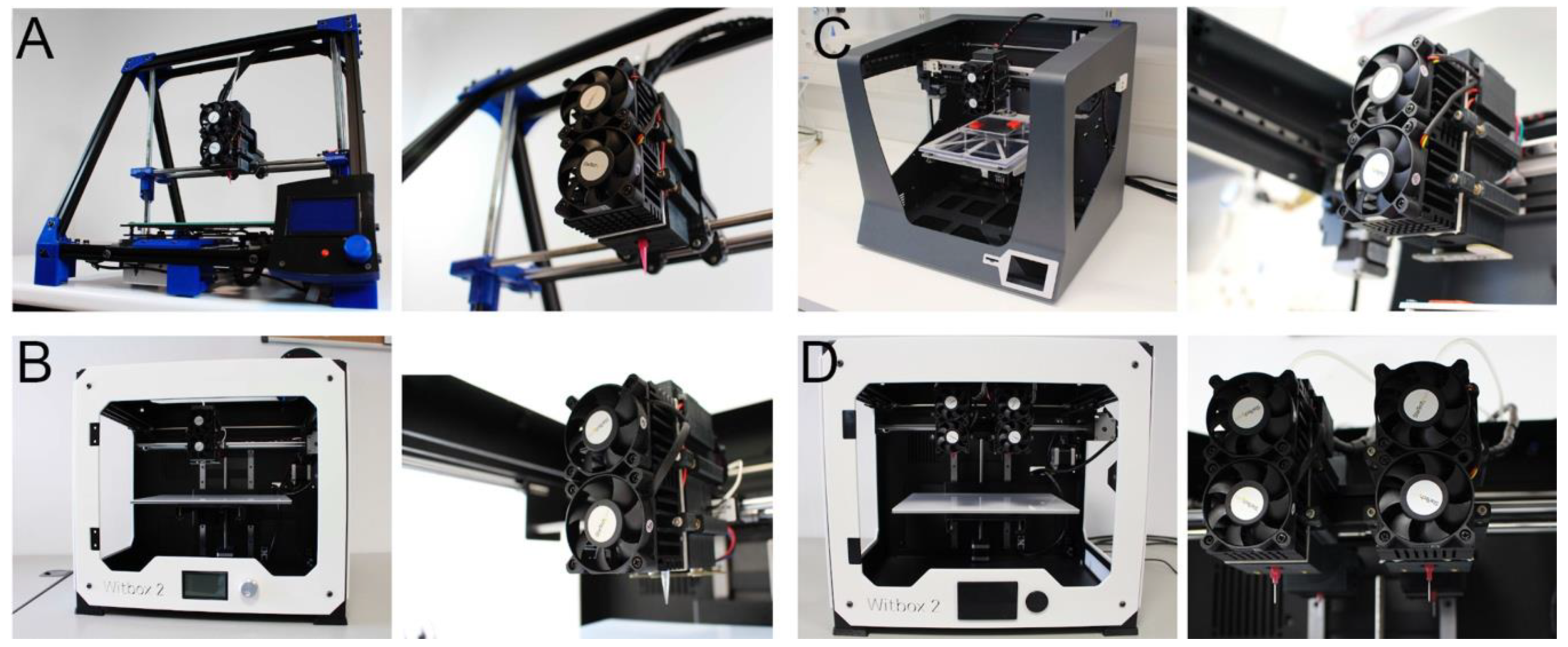
2.1. Printhead Design and Fabrication
2.2. Specifications and Modifications of the 3D Printers
2.3. Thermal Performance of the Printhead
2.4. Hydrogel Preparation
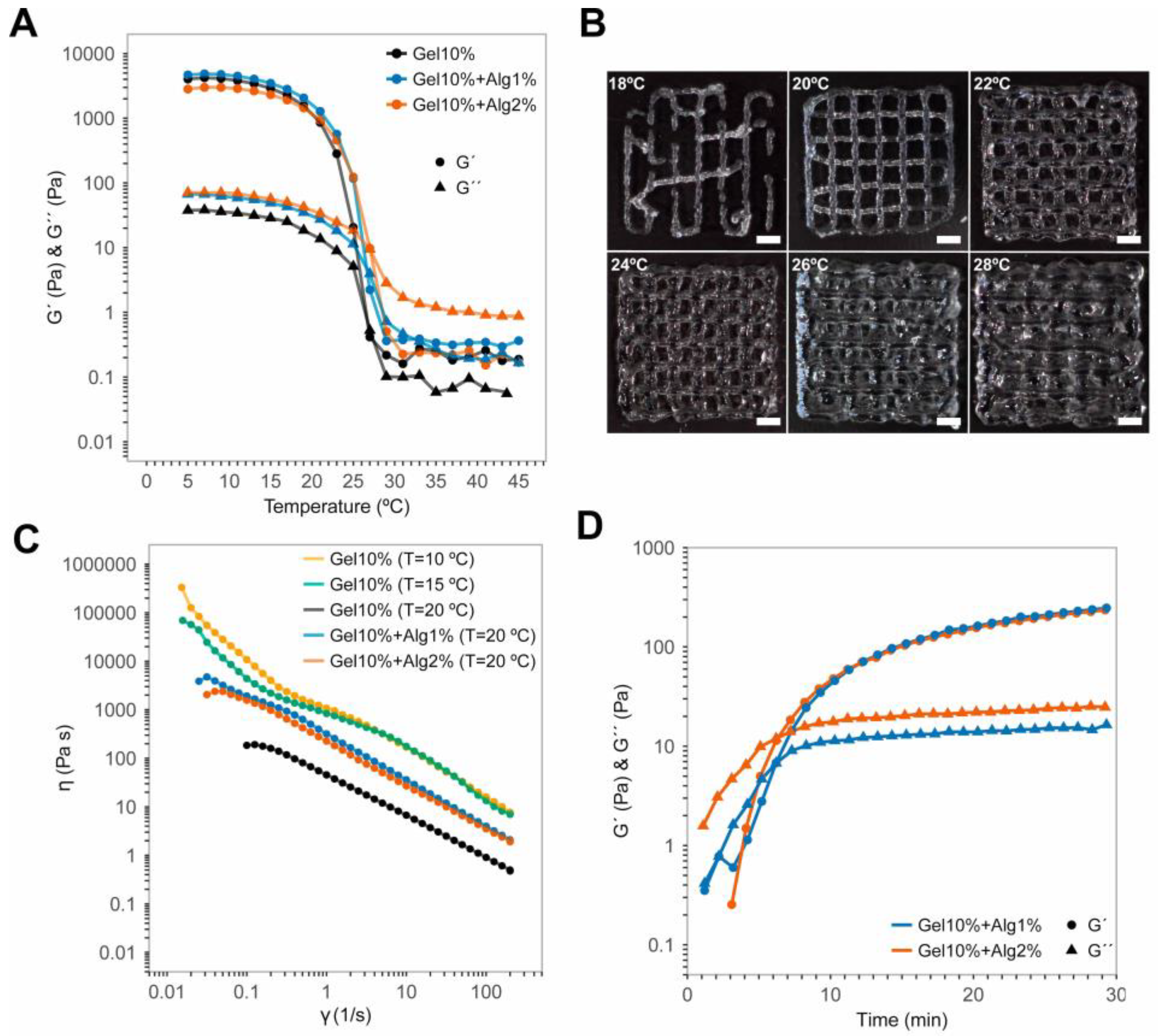
2.5. Rheological Characterization of the Hydrogels
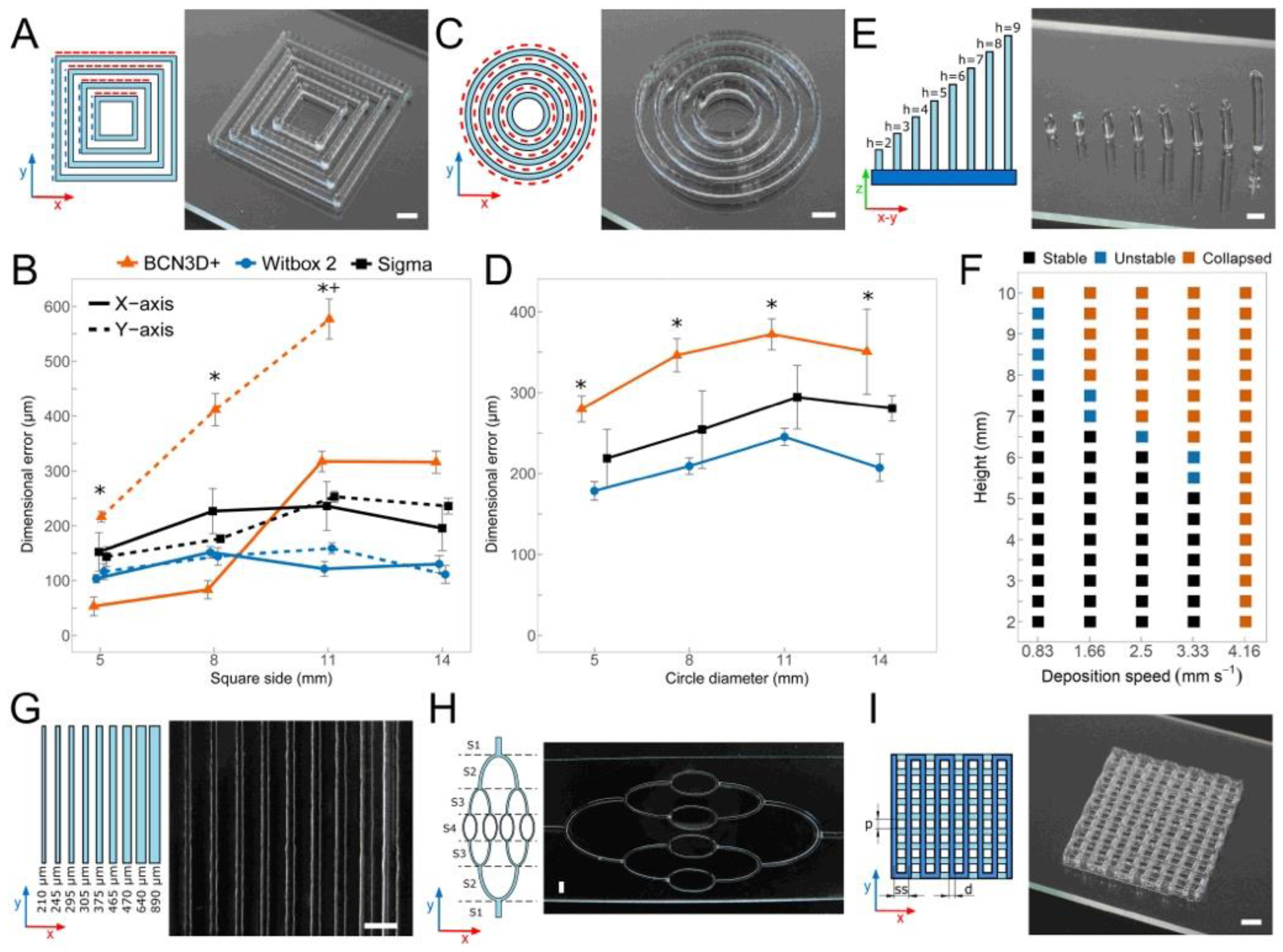
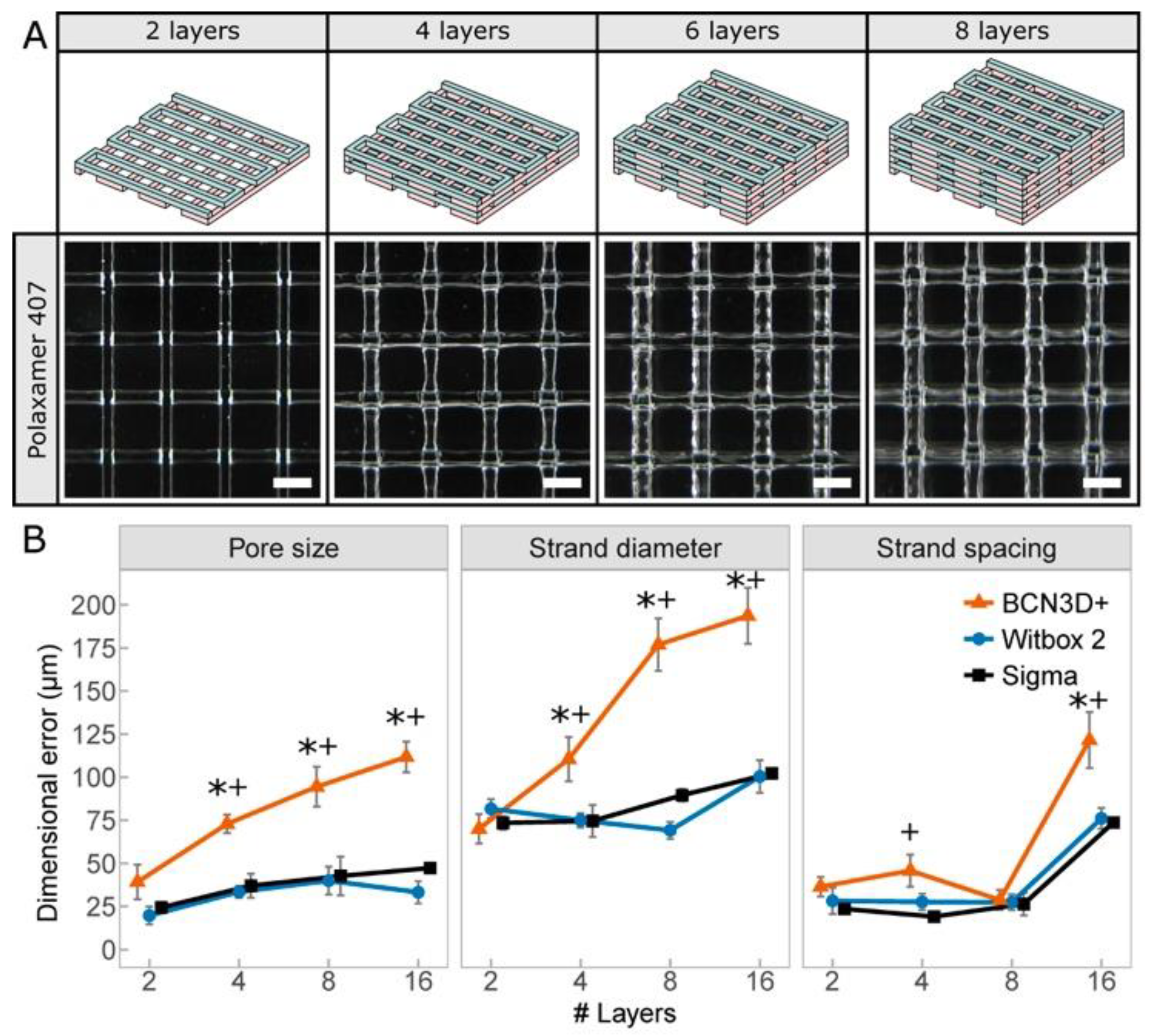
2.6. Benchmark Printing Six Calibration Models with P407 Hydrogels
- Concentric squares [43]: four concentric empty squares of sides 5, 8, 11, and 14 mm were printed varying the number of layers stacked (1, 2, 4, 8, and 16 layers). Squares were aligned with the XY-axes. Square sides in X-Y directions were measured separately. In the case of concentric squares, and the following two models (circles and multilayer lattice structures), the dimensional errors were calculated as the difference between the dimensions of the extruded model and the values of the CAD model.
- Concentric circles [44]: concentric empty circles of diameters 5, 8, 11, and 14 mm were printed varying the number of layers stacked (1, 2, 4, 8, and 16 layers). Circles involved XY-axes movements at the same time. The diameter of all circles was measured and compared to the model diameter to find the accuracy of the combined XY-axes.
- Multilayer lattice structures [45]: pore size (p), strand diameter (d) and strand spacing (ss) were measured varying the number of layers stacked (2, 4, 8, and 16 layers). Predefined values of p = 1.3 mm, d = 0.2 mm, and ss = 1.5 mm were used. When creating lattice models, pore size, strand diameter, and strand spacing were the main quantitative parameters to define the print resolution.
- Straight filaments [46]: 30 mm long straight filaments with different strand widths were printed aligned with the Y-axis using the same tapered nozzle, but varying the deposition speed from 5 to 16.6 m/s.
- Vertical pillars [47]: Pillars were printed without stacking layers by moving along the Z-axis at the same x-y coordinates until the desired pillar height from 2 to 10 mm was reached. Printing parameters, such as pressure and deposition speed (0.83 to 4.16 mm/s), were adjusted to withstand their vertical shape and avoid the collapse. Stability was evaluated based on the final straightness of the pillars for different heights and the outcome categorized into three categories: (i) stable, if no bending was observed, (ii) unstable, if the pillar bent to one side, and (iii) collapsed, if the post bent utterly touching the glass slide.
- Hierarchical networks of filaments with varying diameter [7]: the printed model simulates the potential creation of a hierarchical vascular network. The connected network of curved filaments was printed in four different sections with the same nozzle size at different speeds to change the printed diameter.
2.7. Stem Cells Isolation and Expansion
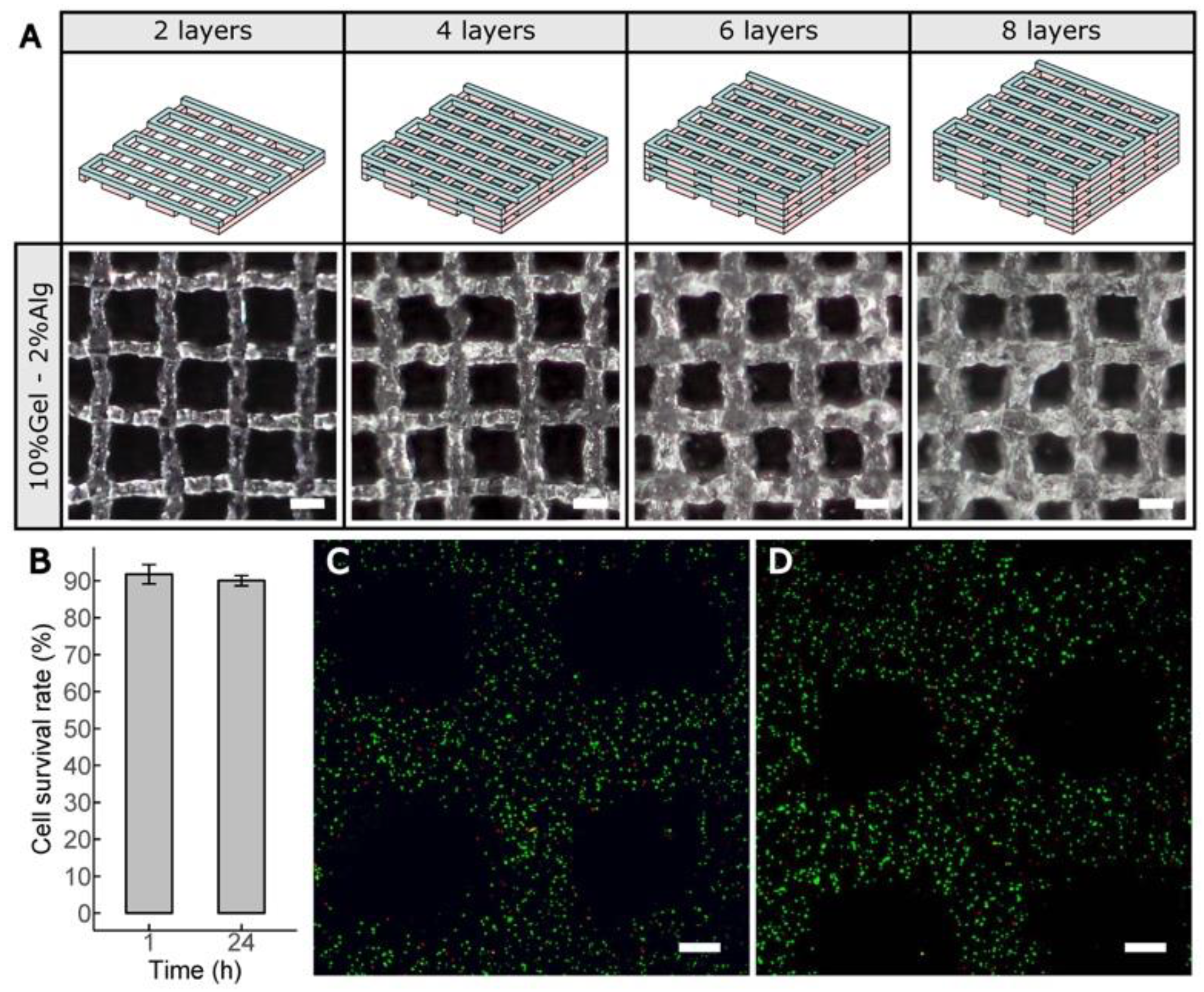
2.8. Cell-Laden Constructs Bioprinted Using Gel–Alg Blends
2.9. Cell Viability Assay
2.10. Statistical Analysis
3. Results
3.1. Temperature Limits, Control, and Performance of the Printhead
3.2. Rheology of the Gel–Alg Bioinks
3.3. Print Resolutions Using Three Different 3D Printing Platforms
3.4. Bioprinting Cell-Laden Lattice-Shaped Constructs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | three-dimensional |
| TE | tissue engineering |
| MEBB | microextrusion-based bioprinting |
| DIY | do-it-yourself |
| Al | Aluminum |
| STL | stereolithography |
| ABS | acrylonitrile butadiene styrene |
| P407 | Poloxamer 407 |
| PBS | phosphate buffered saline |
| CAD | computer-aided design |
| ID | inner diameter |
| hASCs | human adipose-derived mesenchymal stem cells |
| HIV | human immunodeficiency virus |
| DMEM | Dulbecco’s modified Eagle’s medium |
| ANOVA | one-way analysis of variance |
References
- Watson, C.J.E.; Dark, J.H. Organ transplantation: Historical perspective and current practice. Br. J. Anaesth. 2012, 108, i29–i42. [Google Scholar] [CrossRef]
- Port, F.K.; Merion, R.M.; Roys, E.C.; Wolfe, R.A. Trends in Organ Donation and Transplantation in the United States, 1997–2006. Am. J. Transplant. 2008, 8, 911–921. [Google Scholar] [CrossRef]
- Wiegers, T.; Bouwman, R.; Friele, R.; Schoten, S.v.; Coppen, R. Study on the Uptake and Impact of the EU Action Plan on Organ Donation and Transplantation (2009–2015) in the EU Member States; EB-02-17-747-EN-N; European Commission: Brussels, Belgium, 2017; p. 458. [Google Scholar]
- Bajaj, P.; Schweller, R.M.; Khademhosseini, A.; West, J.L.; Bashir, R. 3D Biofabrication Strategies for Tissue Engineering and Regenerative Medicine. Annu. Rev. Biomed. Eng. 2014, 16, 247–276. [Google Scholar] [CrossRef]
- Jakab, K.; Norotte, C.; Marga, F.; Murphy, K.; Vunjak-Novakovic, G.; Forgacs, G. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2010, 2, 022001. [Google Scholar] [CrossRef]
- Mironov, V.; Trusk, T.; Kasyanov, V.; Little, S.; Swaja, R.; Markwald, R. Biofabrication: A 21st century manufacturing paradigm. Biofabrication 2009, 1, 022001. [Google Scholar] [CrossRef]
- Kolesky, D.B.; Truby, R.L.; Gladman, A.S.; Busbee, T.A.; Homan, K.A.; Lewis, J.A. 3D Bioprinting of Vascularized, Heterogeneous Cell-Laden Tissue Constructs. Adv. Mater. 2014, 26, 3124–3130. [Google Scholar] [CrossRef]
- Kuss, M.; Duan, B. 3D Bioprinting for Cardiovascular Tissue Engineering. In Rapid Prototyping in Cardiac Disease: 3D Printing the Heart; Farooqi, K.M., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 167–182. [Google Scholar] [CrossRef]
- Albanna, M.; Binder, K.W.; Murphy, S.V.; Kim, J.; Qasem, S.A.; Zhao, W.; Tan, J.; El-Amin, I.B.; Dice, D.D.; Marco, J.; et al. In Situ Bioprinting of Autologous Skin Cells Accelerates Wound Healing of Extensive Excisional Full-Thickness Wounds. Sci. Rep. 2019, 9, 1856. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Breitenkamp, K.; Finn, M.G.; Lotz, M.; D’Lima, D.D. Direct Human Cartilage Repair Using Three-Dimensional Bioprinting Technology. Tissue Eng. Part A 2012, 18, 1304–1312. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Hasan, A.; Kaarela, O.; Byambaa, B.; Sheikhi, A.; Gaharwar, A.K.; Khademhosseini, A. Advancing Frontiers in Bone Bioprinting. Adv. Healthc. Mater. 2019, 8, 1801048. [Google Scholar] [CrossRef]
- Blaeser, A.; Duarte Campos, D.F.; Puster, U.; Richtering, W.; Stevens, M.M.; Fischer, H. Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv. Healthc. Mater. 2016, 5, 326–333. [Google Scholar] [CrossRef]
- Liu, W.; Heinrich, M.A.; Zhou, Y.; Akpek, A.; Hu, N.; Liu, X.; Guan, X.; Zhong, Z.; Jin, X.; Khademhosseini, A.; et al. Extrusion Bioprinting of Shear-Thinning Gelatin Methacryloyl Bioinks. Adv. Healthc. Mater. 2017, 6, 1601451. [Google Scholar] [CrossRef] [PubMed]
- Tse, C.; Whiteley, R.; Yu, T.; Stringer, J.; MacNeil, S.; Haycock, J.W.; Smith, P.J. Inkjet printing Schwann cells and neuronal analogue NG108-15 cells. Biofabrication 2016, 8, 015017. [Google Scholar] [CrossRef]
- Kang, H.-W.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016, 34, 312. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Stone, A.L.; Parkhill, R.L.; Stewart, R.L.; Simpkins, M.W.; Kachurin, A.M.; Warren, W.L.; Williams, S.K. Three-Dimensional BioAssembly Tool for Generating Viable Tissue-Engineered Constructs. Tissue Eng. 2004, 10, 1566–1576. [Google Scholar] [CrossRef]
- Moroni, L.; De Wijn, J.R.; Van Blitterswijk, C.A. Three-dimensional fiber-deposited PEOT/PBT copolymer scaffolds for tissue engineering: Influence of porosity, molecular network mesh size, and swelling in aqueous media on dynamic mechanical properties. J. Biomed. Mater. Res. Part A 2005, 75A, 957–965. [Google Scholar] [CrossRef]
- Pereira, F.D.A.S.; Parfenov, V.; Khesuani, Y.D.; Ovsianikov, A.; Mironov, V. Commercial 3D Bioprinters. In 3D Printing and Biofabrication; Ovsianikov, A., Yoo, J., Mironov, V., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 535–549. [Google Scholar] [CrossRef]
- Soltan, N.; Ning, L.; Mohabatpour, F.; Papagerakis, P.; Chen, X. Printability and Cell Viability in Bioprinting Alginate Dialdehyde-Gelatin Scaffolds. ACS Biomater. Sci. Eng. 2019, 5, 2976–2987. [Google Scholar] [CrossRef]
- Neufurth, M.; Wang, X.; Schröder, H.C.; Feng, Q.; Diehl-Seifert, B.; Ziebart, T.; Steffen, R.; Wang, S.; Müller, W.E.G. Engineering a morphogenetically active hydrogel for bioprinting of bioartificial tissue derived from human osteoblast-like SaOS-2 cells. Biomaterials 2014, 35, 8810–8819. [Google Scholar] [CrossRef]
- Melchels, F.P.W.; Blokzijl, M.M.; Levato, R.; Peiffer, Q.C.; Ruijter, M.d.; Hennink, W.E.; Vermonden, T.; Malda, J. Hydrogel-based reinforcement of 3D bioprinted constructs. Biofabrication 2016, 8, 035004. [Google Scholar] [CrossRef]
- Müller, M.; Becher, J.; Schnabelrauch, M.; Zenobi-Wong, M. Nanostructured Pluronic hydrogels as bioinks for 3D bioprinting. Biofabrication 2015, 7, 035006. [Google Scholar] [CrossRef]
- Amir, M.; Ebrahim, M.; Danial, K.; Shu-Kai, H.; Matthew, M.; Ali, K. Bioprinters for organs-on-chips. Biofabrication 2019, 11, 4. [Google Scholar] [CrossRef]
- Faramarzi, N.; Yazdi, I.K.; Nabavinia, M.; Gemma, A.; Fanelli, A.; Caizzone, A.; Ptaszek, L.M.; Sinha, I.; Khademhosseini, A.; Ruskin, J.N.; et al. Patient-Specific Bioinks for 3D Bioprinting of Tissue Engineering Scaffolds. Adv. Healthc. Mater. 2018, 7, 1701347. [Google Scholar] [CrossRef]
- López-Marcial, G.R.; Zeng, A.Y.; Osuna, C.; Dennis, J.; García, J.M.; O’Connell, G.D. Agarose-Based Hydrogels as Suitable Bioprinting Materials for Tissue Engineering. ACS Biomater. Sci. Eng. 2018, 4, 3610–3616. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, D.Y.; Roh, H.H.; Kim, H.S.; Lee, J.W.; Lee, K.Y. Three-Dimensional Bioprinting of Cell-Laden Constructs Using Polysaccharide-Based Self-Healing Hydrogels. Biomacromolecules 2019, 20, 1860–1866. [Google Scholar] [CrossRef]
- Malone, E.; Lipson, H. Fab@Home: The personal desktop fabricator kit. Rapid Prototyp. J. 2007, 13, 245–255. [Google Scholar] [CrossRef]
- Hinton, T.J.; Jallerat, Q.; Palchesko, R.N.; Park, J.H.; Grodzicki, M.S.; Shue, H.-J.; Ramadan, M.H.; Hudson, A.R.; Feinberg, A.W. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 2015, 1, e1500758. [Google Scholar] [CrossRef]
- Goldstein, T.A.; Epstein, C.J.; Schwartz, J.; Krush, A.; Lagalante, D.J.; Mercadante, K.P.; Zeltsman, D.; Smith, L.P.; Grande, D.A. Feasibility of Bioprinting with a Modified Desktop 3D Printer. Tissue Eng. Part C Methods 2016, 22, 1071–1076. [Google Scholar] [CrossRef]
- Kim, B.S.; Lee, J.-S.; Gao, G.; Cho, D.-W. Direct 3D cell-printing of human skin with functional transwell system. Biofabrication 2017, 9, 025034. [Google Scholar] [CrossRef]
- Jones, R.; Haufe, P.; Sells, E.; Iravani, P.; Olliver, V.; Palmer, C.; Bowyer, A. RepRap—The replicating rapid prototyper. Robotica 2011, 29, 177–191. [Google Scholar] [CrossRef]
- Barrs, R.W.; Jia, J.; Silver, S.E.; Yost, M.; Mei, Y. Biomaterials for Bioprinting Microvasculature. Chem. Rev. 2020. [Google Scholar] [CrossRef]
- Diloksumpan, P.; De Ruijter, M.; Castilho, M.; Gbureck, U.; Vermonden, T.; Van Weeren, P.R.; Malda, J.; Levato, R. Combining multi-scale 3D printing technologies to engineer reinforced hydrogel-ceramic interfaces. Biofabrication 2020, 12, 025014. [Google Scholar] [CrossRef]
- Ying, G.; Jiang, N.; Parra-Cantu, C.; Tang, G.; Zhang, J.; Wang, H.; Chen, S.; Huang, N.-P.; Xie, J.; Zhang, Y.S. Bioprinted Injectable Hierarchically Porous Gelatin Methacryloyl Hydrogel Constructs with Shape-Memory Properties. Adv. Funct. Mater. 2020, 2003740. [Google Scholar] [CrossRef]
- Kaklamani, G.; Cheneler, D.; Grover, L.M.; Adams, M.J.; Bowen, J. Mechanical properties of alginate hydrogels manufactured using external gelation. J. Mech. Behav. Biomed. Mater. 2014, 36, 135–142. [Google Scholar] [CrossRef]
- Piras, C.C.; Smith, D.K. Multicomponent polysaccharide alginate-based bioinks. J. Mater. Chem. B 2020, 8, 8171–8188. [Google Scholar] [CrossRef]
- Wu, Z.; Li, Q.; Xie, S.; Shan, X.; Cai, Z. In vitro and in vivo biocompatibility evaluation of a 3D bioprinted gelatin-sodium alginate/rat Schwann-cell scaffold. Mater. Sci. Eng. C 2020, 109, 110530. [Google Scholar] [CrossRef]
- Luo, Y.; Li, Y.; Qin, X.; Wa, Q. 3D printing of concentrated alginate/gelatin scaffolds with homogeneous nano apatite coating for bone tissue engineering. Mater. Des. 2018, 146, 12–19. [Google Scholar] [CrossRef]
- Marlin; 1.1. Available online: http://marlinfw.org (accessed on 4 September 2017).
- Repetier-Host-Mac; 0.56. Available online: https://www.repetier.com (accessed on 4 September 2017).
- Riegel, J.; Mayer, W.; Van Havre, Y. (2001–2017). FreeCAD (Version 0.16.6712). Available online: http://www.freecadweb.org (accessed on 3 March 2019).
- Slic3r: Open Source Toolpath Generator for 3D Printers. Available online: https://github.com/slic3r/Slic3r (accessed on 2 March 2019).
- Cruz Sanchez, F.A.; Boudaoud, H.; Muller, L.; Camargo, M. Towards a standard experimental protocol for open source additive manufacturing. Virtual Phys. Prototyp. 2014, 9, 151–167. [Google Scholar] [CrossRef]
- Seitz, H.; Polzin, C.; Spath, S. Characterization and evaluation of a PMMA-based 3D printing process. Rapid Prototyp. J. 2013, 19, 37–43. [Google Scholar] [CrossRef]
- Chung, J.H.Y.; Naficy, S.; Yue, Z.; Kapsa, R.; Quigley, A.; Moulton, S.E.; Wallace, G.G. Bio-ink properties and printability for extrusion printing living cells. Biomater. Sci. 2013, 1, 763–773. [Google Scholar] [CrossRef]
- Kang, K.H.; Hockaday, L.A.; Butcher, J.T. Quantitative optimization of solid freeform deposition of aqueous hydrogels. Biofabrication 2013, 5, 035001. [Google Scholar] [CrossRef]
- Hansen, C.J.; Wu, W.; Toohey, K.S.; Sottos, N.R.; White, S.R.; Lewis, J.A. Self-Healing Materials with Interpenetrating Microvascular Networks. Adv. Mater. 2009, 21, 4143–4147. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676. [Google Scholar] [CrossRef]
- Escobedo-Lucea, C.; Bellver, C.; Gandia, C.; Sanz-Garcia, A.; Esteban, F.J.; Mirabet, V.; Forte, G.; Moreno, I.; Lezameta, M.; Ayuso-Sacido, A.; et al. A Xenogeneic-Free Protocol for Isolation and Expansion of Human Adipose Stem Cells for Clinical Uses. PLoS ONE 2013, 8, e67870. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Sodupe Ortega, E.; Sanz-Garcia, A.; Pernia-Espinoza, A.; Escobedo-Lucea, C. Efficient Fabrication of Polycaprolactone Scaffolds for Printing Hybrid Tissue-Engineered Constructs. Materials 2019, 12, 613. [Google Scholar] [CrossRef]
- Ning, L.; Betancourt, N.; Schreyer, D.J.; Chen, X. Characterization of Cell Damage and Proliferative Ability during and after Bioprinting. ACS Biomater. Sci. Eng. 2018, 4, 3906–3918. [Google Scholar] [CrossRef]
- Stein, D.B. Handbook of Hydrogels: Properties, Preparation & Applications; Nova Science Publishers: Hauppauge, NY, USA, 2009. [Google Scholar]
- Wu, Q.; Therriault, D.; Heuzey, M.-C. Processing and Properties of Chitosan Inks for 3D Printing of Hydrogel Microstructures. ACS Biomater. Sci. Eng. 2018, 4, 2643–2652. [Google Scholar] [CrossRef]
- Gao, T.; Gillispie, G.J.; Copus, J.S.; Pr, A.K.; Seol, Y.-J.; Atala, A.; Yoo, J.J.; Lee, S.J. Optimization of gelatin–alginate composite bioink printability using rheological parameters: A systematic approach. Biofabrication 2018, 10, 034106. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Mao, S.; Sun, W.; Yao, R. The influence of printing parameters on cell survival rate and printability in microextrusion-based 3D cell printing technology. Biofabrication 2015, 7, 045002. [Google Scholar] [CrossRef]
- Narayanan, L.K.; Huebner, P.; Fisher, M.B.; Spang, J.T.; Starly, B.; Shirwaiker, R.A. 3D-Bioprinting of Polylactic Acid (PLA) Nanofiber–Alginate Hydrogel Bioink Containing Human Adipose-Derived Stem Cells. ACS Biomater. Sci. Eng. 2016, 2, 1732–1742. [Google Scholar] [CrossRef]
- Mahesh, M.; Wong, Y.S.; Fuh, J.Y.H.; Loh, H.T. A Six-sigma approach for benchmarking of RP&M processes. Int. J. Adv. Manuf. Technol. 2006, 31, 374–387. [Google Scholar] [CrossRef]
- Roberson, D.A.; Espalin, D.; Wicker, R.B. 3D printer selection: A decision-making evaluation and ranking model. Virtual Phys. Prototyp. 2013, 8, 201–212. [Google Scholar] [CrossRef]
- Scaravetti, D.; Dubois, P.; Duchamp, R. Qualification of rapid prototyping tools: Proposition of a procedure and a test part. Int. J. Adv. Manuf. Technol. 2008, 38, 683–690. [Google Scholar] [CrossRef]
- Zehnder, T.; Sarker, B.; Boccaccini, A.R.; Detsch, R. Evaluation of an alginate–gelatine crosslinked hydrogel for bioplotting. Biofabrication 2015, 7, 025001. [Google Scholar] [CrossRef] [PubMed]
- Stichler, S.; Böck, T.; Paxton, N.; Bertlein, S.; Levato, R.; Schill, V.; Smolan, W.; Malda, J.; Teßmar, J.; Blunk, T.; et al. Double printing of hyaluronic acid/poly(glycidol) hybrid hydrogels with poly(ε-caprolactone) for MSC chondrogenesis. Biofabrication 2017, 9, 044108. [Google Scholar] [CrossRef]
- Lee, J.; Kim, K.E.; Bang, S.; Noh, I.; Lee, C. A desktop multi-material 3D bio-printing system with open-source hardware and software. Int. J. Precis. Eng. Manuf. 2017, 18, 605–612. [Google Scholar] [CrossRef]
- Kolesky, D.B.; Homan, K.A.; Skylar-Scott, M.A.; Lewis, J.A. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [Google Scholar] [CrossRef]
- Richards, D.; Jia, J.; Yost, M.; Markwald, R.; Mei, Y. 3D Bioprinting for Vascularized Tissue Fabrication. Ann. Biomed. Eng. 2017, 45, 132–147. [Google Scholar] [CrossRef]
- Reid, J.A.; Mollica, P.A.; Johnson, G.D.; Ogle, R.C.; Bruno, R.D.; Sachs, P.C. Accessible bioprinting: Adaptation of a low-cost 3D-printer for precise cell placement and stem cell differentiation. Biofabrication 2016, 8, 025017. [Google Scholar] [CrossRef]
- Roehm, K.D.; Madihally, S.V. Bioprinted chitosan-gelatin thermosensitive hydrogels using an inexpensive 3D printer. Biofabrication 2017, 10, 015002. [Google Scholar] [CrossRef]
- Ouyang, L.; Yao, R.; Mao, S.; Chen, X.; Na, J.; Sun, W. Three-dimensional bioprinting of embryonic stem cells directs highly uniform embryoid body formation. Biofabrication 2015, 7, 044101. [Google Scholar] [CrossRef]
- Xu, W.; Molino, B.Z.; Cheng, F.; Molino, P.J.; Yue, Z.; Su, D.; Wang, X.; Willför, S.; Xu, C.; Wallace, G.G. On Low-Concentration Inks Formulated by Nanocellulose Assisted with Gelatin Methacrylate (GelMA) for 3D Printing toward Wound Healing Application. ACS Appl. Mater. Interfaces 2019, 11, 8838–8848. [Google Scholar] [CrossRef]
- Yeo, M.G.; Kim, G.H. A cell-printing approach for obtaining hASC-laden scaffolds by using a collagen/polyphenol bioink. Biofabrication 2017, 9, 025004. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.-H.; Lee, J.-S.; Kim, J.Y.; Cho, D.-W. Bioprinting of a mechanically enhanced three-dimensional dual cell-laden construct for osteochondral tissue engineering using a multi-head tissue/organ building system. J. Micromech. Microeng. 2012, 22, 085014. [Google Scholar] [CrossRef]
- Rocca, M.; Fragasso, A.; Liu, W.; Heinrich, M.A.; Zhang, Y.S. Embedded Multimaterial Extrusion Bioprinting. SLAS Technol. 2018, 23, 154–163. [Google Scholar] [CrossRef] [PubMed]

| Type | Temperature (°C) | Pore Size (mm) | Strand Width (mm) | Strand Spacing (mm) |
|---|---|---|---|---|
| CAD model | - | 1.75 | 0.25 | 2 |
| Printed model | 20 | 1.5 ± 0.02 | 0.4 ± 0.01 | 1.99 ± 0.02 |
| 22 | 1.05 ± 0.02 | 0.82 ± 0.01 | 1.99 ± 0.01 | |
| 24 | 0.99 ± 0.02 | 1 ± 0.01 | 1.99 ± 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanz-Garcia, A.; Sodupe-Ortega, E.; Pernía-Espinoza, A.; Shimizu, T.; Escobedo-Lucea, C. A Versatile Open-Source Printhead for Low-Cost 3D Microextrusion-Based Bioprinting. Polymers 2020, 12, 2346. https://doi.org/10.3390/polym12102346
Sanz-Garcia A, Sodupe-Ortega E, Pernía-Espinoza A, Shimizu T, Escobedo-Lucea C. A Versatile Open-Source Printhead for Low-Cost 3D Microextrusion-Based Bioprinting. Polymers. 2020; 12(10):2346. https://doi.org/10.3390/polym12102346
Chicago/Turabian StyleSanz-Garcia, Andres, Enrique Sodupe-Ortega, Alpha Pernía-Espinoza, Tatsuya Shimizu, and Carmen Escobedo-Lucea. 2020. "A Versatile Open-Source Printhead for Low-Cost 3D Microextrusion-Based Bioprinting" Polymers 12, no. 10: 2346. https://doi.org/10.3390/polym12102346
APA StyleSanz-Garcia, A., Sodupe-Ortega, E., Pernía-Espinoza, A., Shimizu, T., & Escobedo-Lucea, C. (2020). A Versatile Open-Source Printhead for Low-Cost 3D Microextrusion-Based Bioprinting. Polymers, 12(10), 2346. https://doi.org/10.3390/polym12102346