Electrochemical MIP Sensor for Butyrylcholinesterase
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
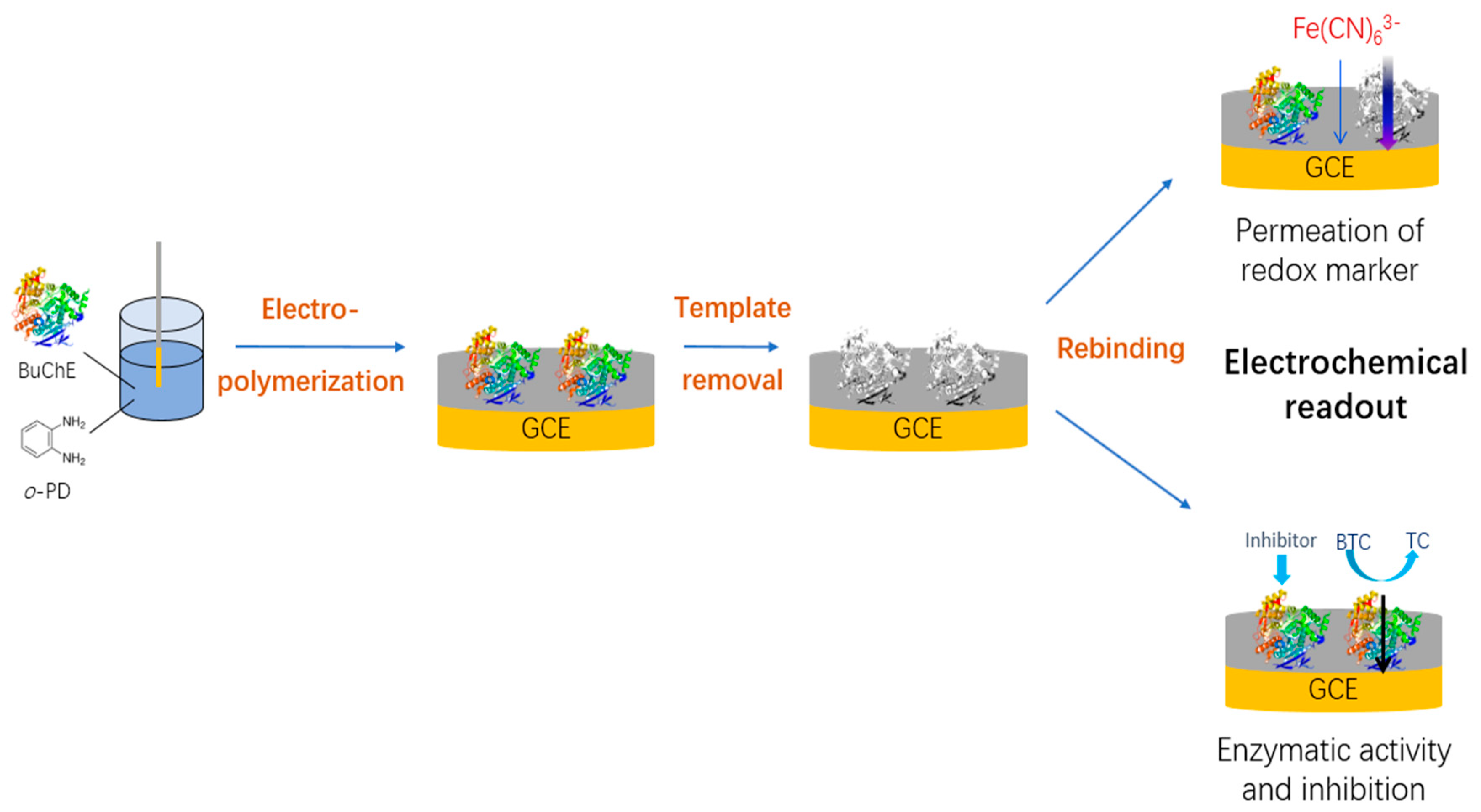
2.2. Preparation of BuChE Imprinted Electrodes
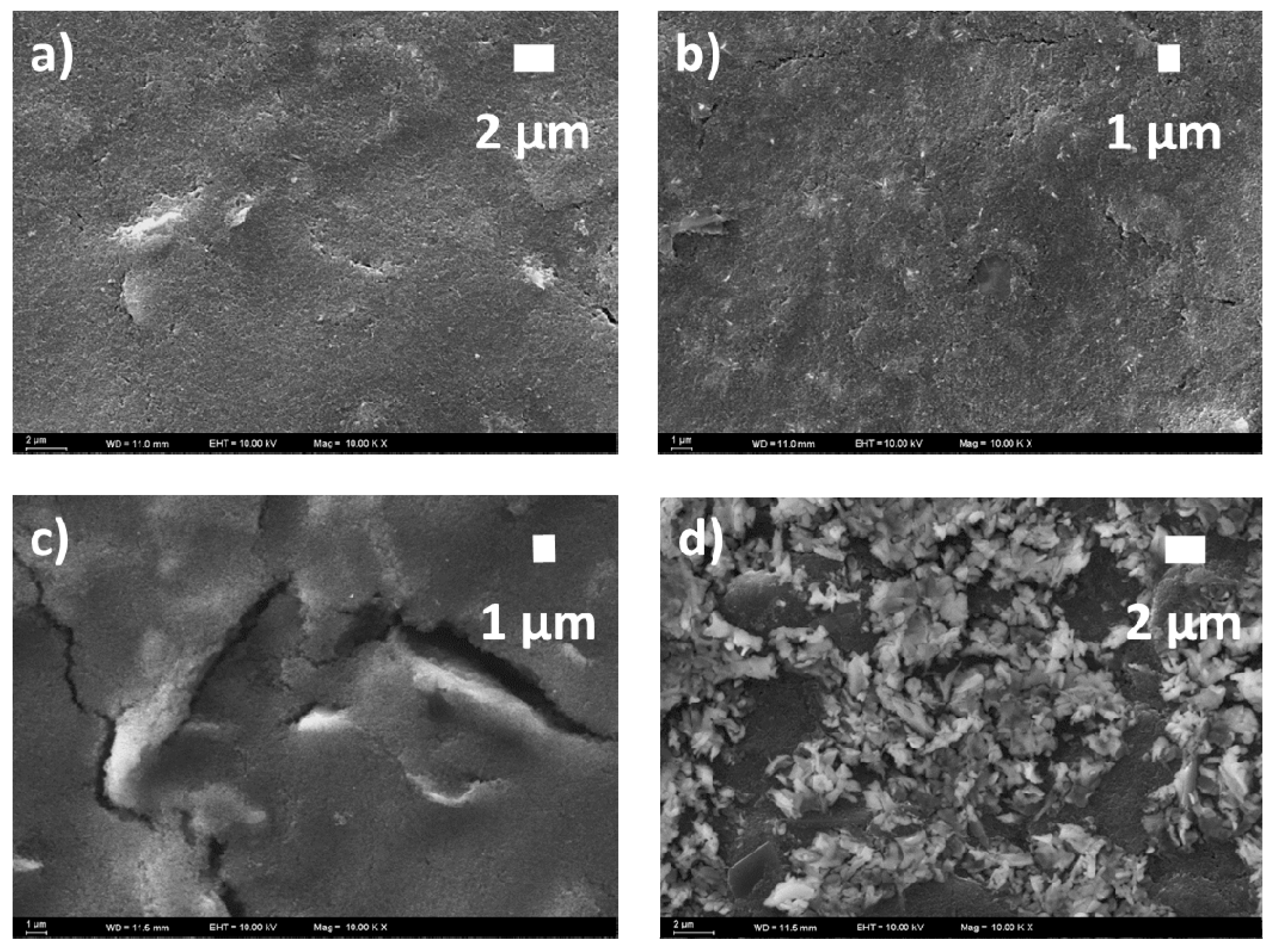
2.3. Electrochemical and Scanning Electron Microscope Measurements
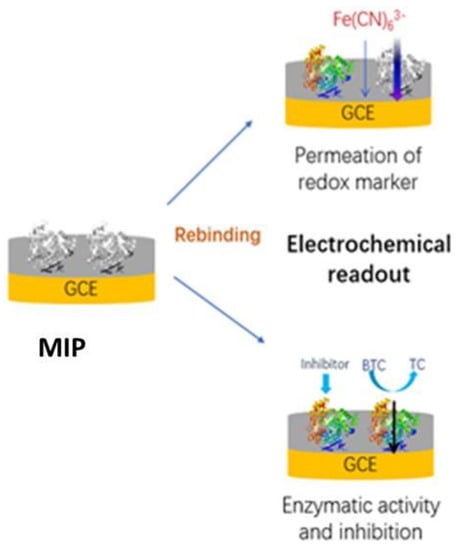
3. Results
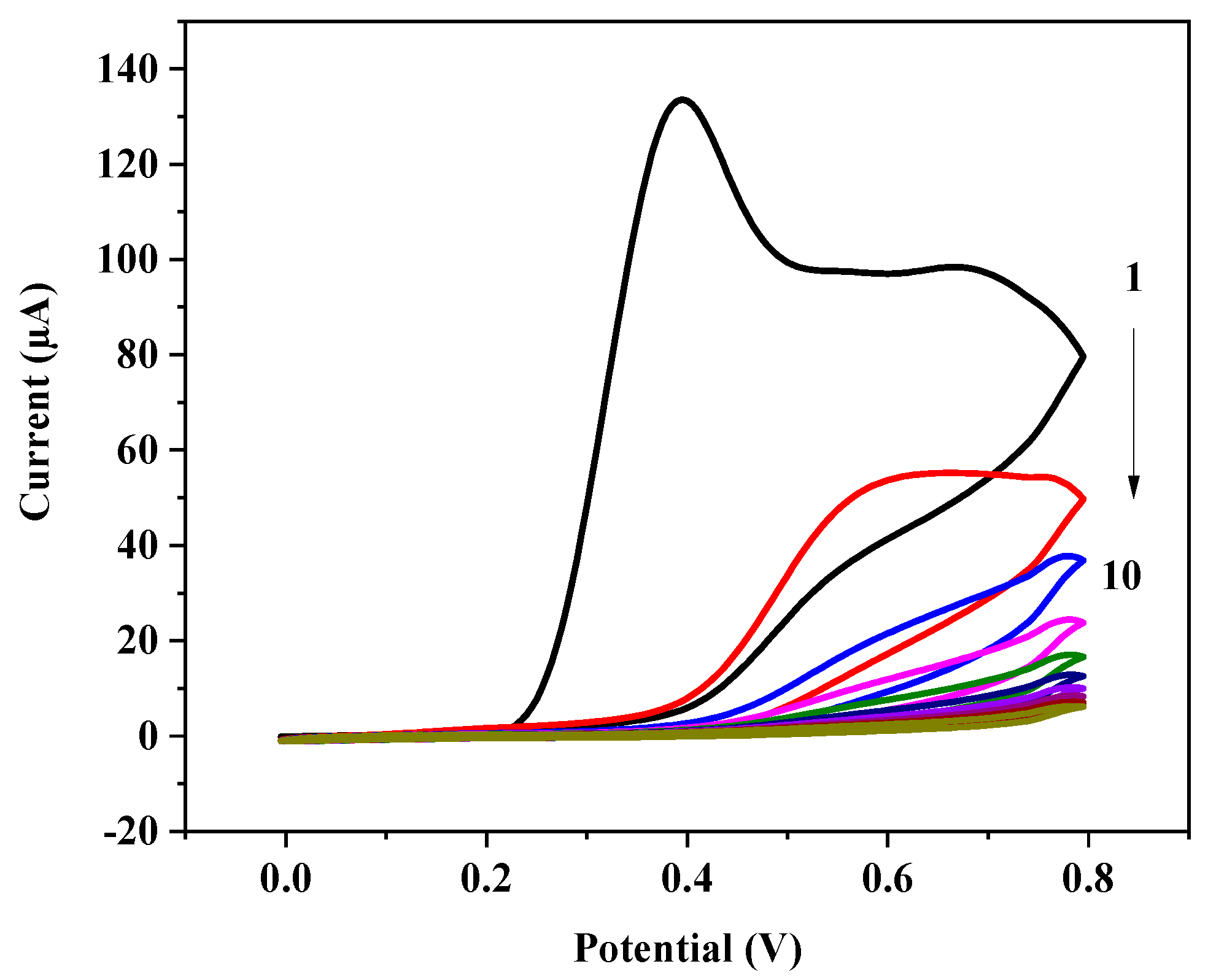
3.1. Electropolymerization and Template Removal
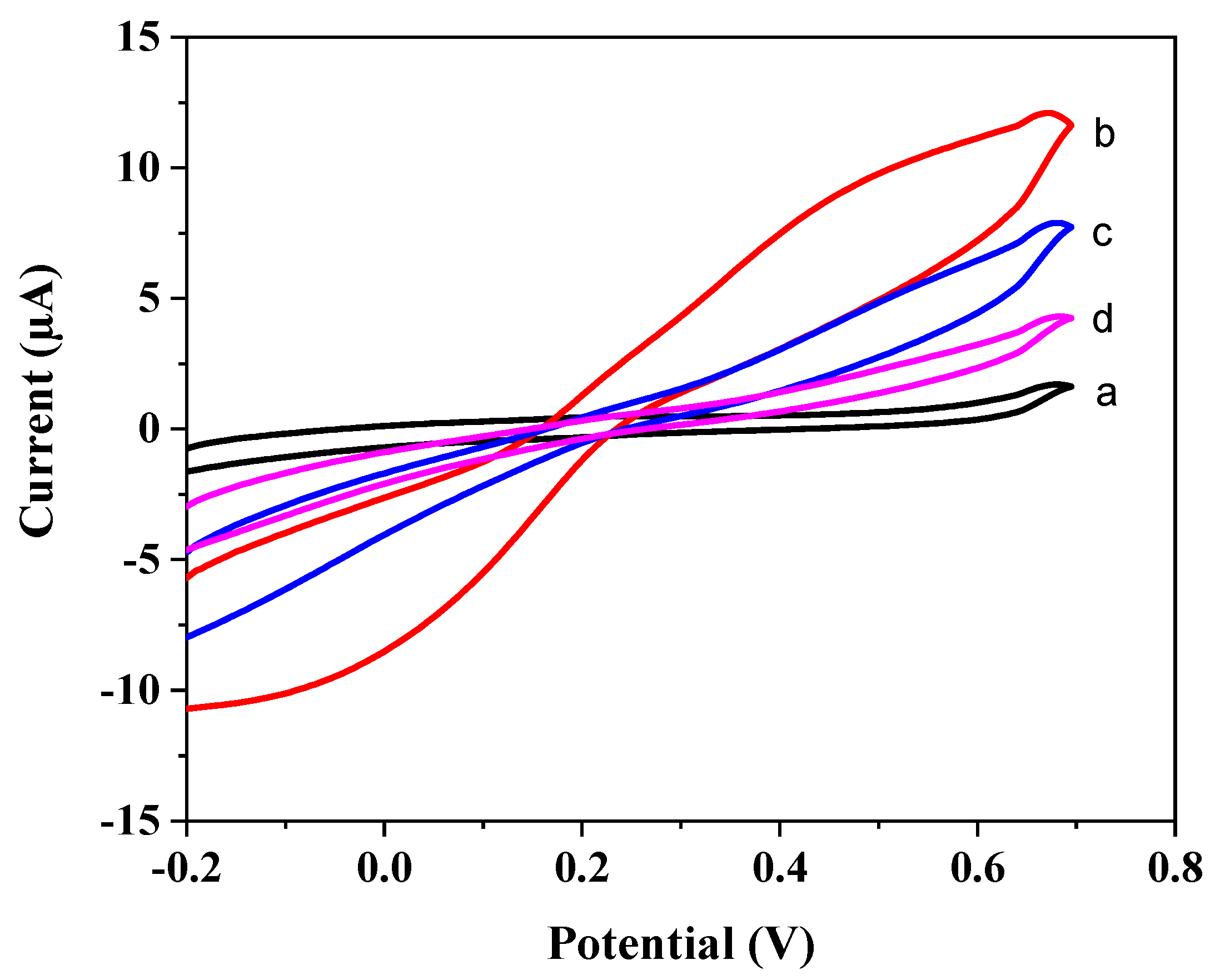
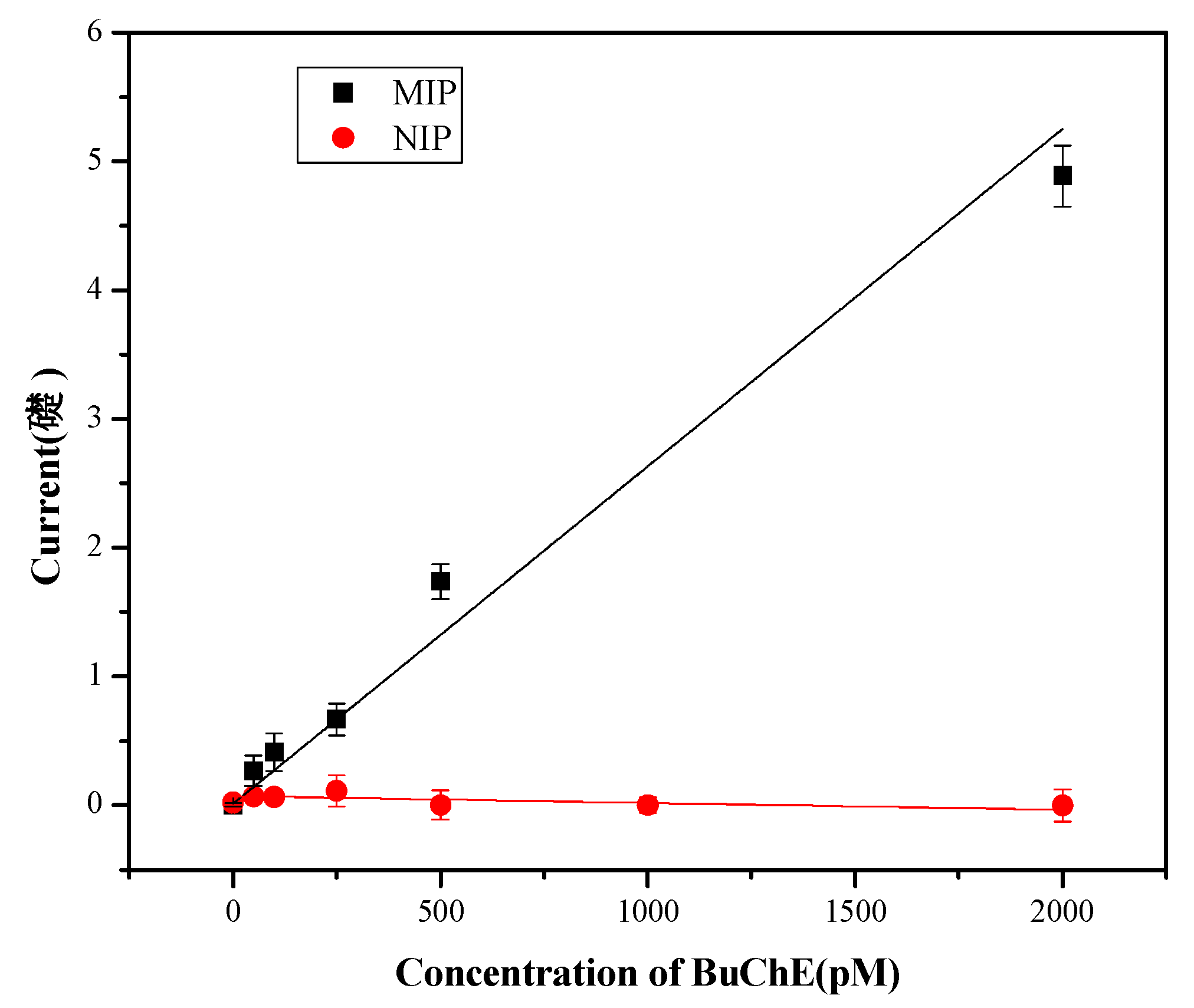
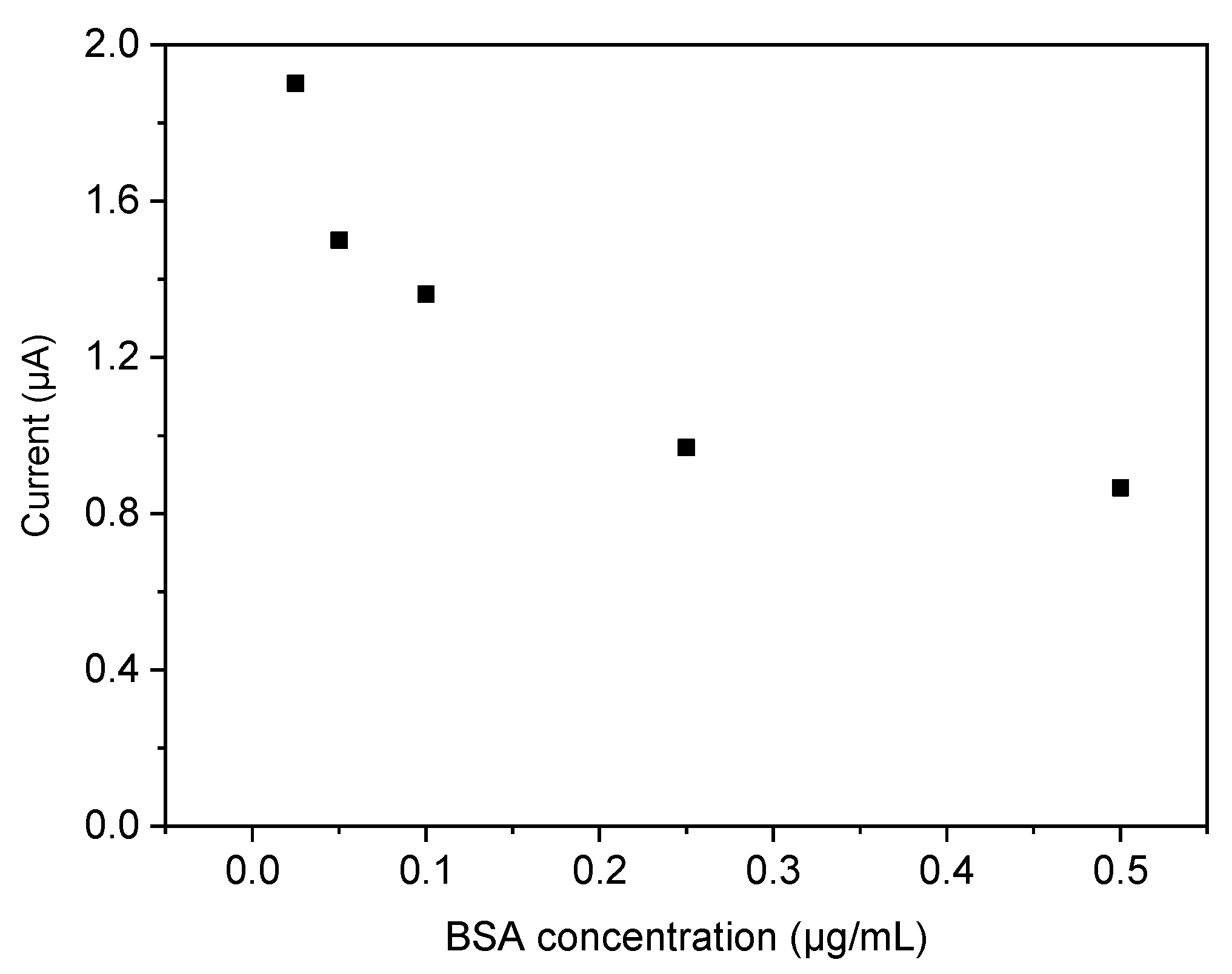
3.2. Rebinding
3.3. Inhibition of Butyrylcholinesterase by Anti-Alzheimer Drug Rivastigmine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wulff, G.; Sarhan, A. Use of Polymers with Enzyme-Analogous Structures for the Resolution of Racemates. Angew. Chem. Int. Ed. Engl. 1972, 11, 341–344. [Google Scholar]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Die Makromol. Chem. 1981, 182, 687–692. [Google Scholar] [CrossRef]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A Review of the Molecular Imprinting of Proteins. Biotechnol. Prog. 2006, 22, 1474–1489. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cao, S.; Whitcombe, M.J.; Piletsky, S.A. Size matters: Challenges in imprinting macromolecules. Prog. Polym. Sci. 2014, 39, 145–163. [Google Scholar] [CrossRef]
- Slinchenko, O.; Rachkov, A.; Miyachi, H.; Ogiso, M.; Minoura, N. Imprinted polymer layer for recognizing double-stranded DNA. Biosens. Bioelectron. 2004, 20, 1091–1097. [Google Scholar] [CrossRef]
- Hayden, O.; Lieberzeit, P.A.; Blaas, D.; Dickert, F.L. Artificial Antibodies for Bioanalyte Detection—Sensing Viruses and Proteins. Adv. Funct. Mater. 2006, 16, 1269–1278. [Google Scholar] [CrossRef]
- Kryscio, D.R.; Fleming, M.Q.; Peppas, N.A. Protein Conformational Studies for Macromolecularly Imprinted Polymers. Macromol. Biosci. 2012, 12, 1137–1144. [Google Scholar] [CrossRef]
- Menaker, A.; Syritski, V.; Reut, J.; Öpik, A.; Horváth, V.; Gyurcsányi, R.E. Electrosynthesized Surface-Imprinted Conducting Polymer Microrods for Selective Protein Recognition. Adv. Mater. 2009, 21, 2271–2275. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Kirsch, N.; Nicholls, I.A. Molecular imprinting science and technology: A survey of the literature for the years 2004–2011. J. Mol. Recognit. 2014, 27, 297–401. [Google Scholar]
- Erdőssy, J.; Horváth, V.; Yarman, A.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC Trends Anal. Chem. 2016, 79, 179–190. [Google Scholar] [CrossRef]
- Liao, J.-L.; Wang, Y.; Hjertén, S. A novel support with artificially created recognition for the selective removal of proteins and for affinity chromatography. Chromatographia 1996, 42, 259–262. [Google Scholar] [CrossRef]
- Takátsy, A.; Végvári, Á.; Hjertén, S.; Kilár, F. Universal method for synthesis of artificial gel antibodies by the imprinting approach combined with a unique electrophoresis technique for detection of minute structural differences of proteins, viruses and cells (bacteria). Ib. Gel antibodies against proteins (hemoglobins). Electrophoresis 2007, 28, 2345–2350. [Google Scholar] [PubMed]
- Schmidt, R.H.; Mosbach, K.; Haupt, K. A Simple Method for Spin-Coating Molecularly Imprinted Polymer Films of Controlled Thickness and Porosity. Adv. Mater. 2004, 16, 719–722. [Google Scholar] [CrossRef]
- Haupt, K.; Noworyta, K.; Kutner, W. Imprinted polymer-based enantioselective acoustic sensor using a quartz crystal microbalance. Anal. Commun. 1999, 36, 391–393. [Google Scholar] [CrossRef]
- Shi, H.; Tsal, W.B.; Garrison, M.D.; Ferrari, S.; Ratner, B.D. Template-imprinted nanostructured surfaces for protein recognition. Nature 1999, 398, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Hayden, O.; Dickert, F.L. Selective Microorganism Detection with Cell Surface Imprinted Polymers. Adv. Mater. 2001, 13, 1480–1483. [Google Scholar] [CrossRef]
- Dabrowski, M.; Lach, P.; Cieplak, M.; Kutner, W. Nanostructured molecularly imprinted polymers for protein chemosensing. Biosens. Bioelectron. 2018, 102, 17–26. [Google Scholar] [CrossRef]
- Jetzschmann, K.J.; Zhang, X.; Yarman, A.; Wollenberger, U.; Scheller, F.W. Label-Free MIP Sensors for Protein Biomarkers. In Label-Free Biosensing; Springer: Cham, Switzerland, 2017; pp. 291–321. [Google Scholar]
- Bosserdt, M.; Erdőssy, J.; Lautner, G.; Witt, J.; Köhler, K.; Gajovic-Eichelmann, N.; Yarman, A.; Wittstock, G.; Scheller, F.W.; Gyurcsányi, R.E. Microelectrospotting as a new method for electrosynthesis of surface-imprinted polymer microarrays for protein recognition. Biosens. Bioelectron. 2015, 73, 123–129. [Google Scholar] [CrossRef]
- Poma, A.; Guerreiro, A.; Whitcombe, M.J.; Piletska, E.V.; Turner, A.P.F.; Piletsky, S.A. Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template—“Plastic Antibodies”. Adv. Funct. Mater. 2013, 23, 2821–2827. [Google Scholar] [CrossRef]
- Ambrosini, S.; Beyazit, S.; Haupt, K.; Tse Sum Bui, B. Solid-phase synthesis of molecularly imprinted nanoparticles for protein recognition. Chem. Commun. 2013, 49, 6746–6748. [Google Scholar] [CrossRef]
- Merkoçi, A.; Alegret, S. New materials for electrochemical sensing IV. Molecular imprinted polymers. TrAC Trends Anal. Chem. 2002, 21, 717–725. [Google Scholar] [CrossRef]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular Imprinting of Macromolecules for Sensor Applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef] [PubMed]
- Uzun, L.; Turner, A.P.F. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Gálvez, A.; Morales-Narváez, E.; Mayorga-Martinez, C.C.; Merkoçi, A. Nanomaterials connected to antibodies and molecularly imprinted polymers as bio/receptors for bio/sensor applications. Appl. Mater. Today 2017, 9, 387–401. [Google Scholar] [CrossRef]
- Kalow, W. Butyrylcholine esterase in the blood serum of man and animal. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1952, 215, 370–377. [Google Scholar] [PubMed]
- Rao, R.V.; Balasubramanian, A.S. Isolation of a galactose-free 20-kDa fragment exhibiting butyrylcholine esterase and aryl acylamidase activity from human serum butyrylcholine esterase by limited alpha-chymotrypsin digestion. Eur. J. Biochem. 1989, 179, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, A.; Svensson, A.L. Cholinesterase inhibitors in the treatment of Alzheimer’s disease. A comparison of tolerability and pharmacology. Drug Saf. 1998, 19, 465–480. [Google Scholar] [CrossRef]
- Stahl, S.M. Cholinesterase Inhibitors for Alzheimer’s Disease. Hosp. Pract. 1998, 33, 131–136. [Google Scholar] [CrossRef]
- Holmstedt, B. Pharmacology of organophosphorus cholinesterase inhibitors. Pharmacol. Rev. 1959, 11, 567–688. [Google Scholar]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Xu, X.; Cen, Y.; Xu, G.; Wei, F.; Shi, M.; Hu, Q. A ratiometric fluorescence probe based on carbon dots for discriminative and highly sensitive detection of acetylcholinesterase and butyrylcholinesterase in human whole blood. Biosens. Bioelectron. 2019, 131, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Bozal-Palabiyik, B.; Erkmen, C.; Uslu, B. Molecularly imprinted electrochemical sensors: Analytical and pharmaceutical applications based on ortho-phenylenediamine polymerization. Curr. Pharm. Anal. 2019, 15. [Google Scholar] [CrossRef]
- Yarman, A. Development of a molecularly imprinted polymer-based electrochemical sensor for tyrosinase. Turk. J. Chem. 2018, 42, 346–354. [Google Scholar] [CrossRef]
- Jann, M.W. Rivastigmine, a new-generation cholinesterase inhibitor for the treatment of Alzheimer’s disease. Pharmacotherapy 2000, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.; Minoura, N. Molecular Imprinting: Synthesis of Polymer Particles with Antibody-like Binding Characteristics for Glucose Oxidase. Biochem. Biophys. Res. Commun. 1996, 227, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Bossi, A.; Piletsky, S.A.; Piletska, E.V.; Righetti, P.G.; Turner, A.P.F. Surface-Grafted Molecularly Imprinted Polymers for Protein Recognition. Anal. Chem. 2001, 73, 5281–5286. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, R.; Guo, H.; Wei, Y.; Yang, W. A facile horseradish peroxidase electrochemical biosensor with surface molecular imprinting based on polyaniline nanotubes. J. Electroanal. Chem. 2018, 817, 184–194. [Google Scholar] [CrossRef]
- Yang, S.; Bai, C.; Teng, Y.; Zhang, J.; Peng, J.; Fang, Z.; Xu, W. Study of horseradish peroxidase and hydrogen peroxide bi-analyte sensor with boronate affinity-based molecularly imprinted film. Can. J. Chem. 2019, 97, 833–839. [Google Scholar] [CrossRef]
- Peng, L.; Yarman, A.; Jetzschmann, K.J.; Jeoung, J.-H.; Schad, D.; Dobbek, H.; Wollenberger, U.; Scheller, F.W. Molecularly Imprinted Electropolymer for a Hexameric Heme Protein with Direct Electron Transfer and Peroxide Electrocatalysis. Sensors 2016, 16, 272. [Google Scholar] [CrossRef]
- Jetzschmann, K.J.; Yarman, A.; Rustam, L.; Kielb, P.; Urlacher, V.B.; Fischer, A.; Weidinger, I.M.; Wollenberger, U.; Scheller, F.W. Molecular LEGO by domain-imprinting of cytochrome P450 BM3. Colloids Surf. B Biointerfaces 2018, 164, 240–246. [Google Scholar] [CrossRef]
- Yarman, A. Electrosynthesized Molecularly Imprinted Polymer for Laccase Using the Inactivated Enzyme as the Target. Bull. Korean Chem. Soc. 2018, 39, 483–488. [Google Scholar] [CrossRef]
- Dolak, I.; Canpolat, G.; Ersöz, A.; Say, R. Metal chelate based site recognition of ceruloplasmin using molecularly imprinted polymer/cryogel system. Sep. Sci. Technol. 2019, 1–10. [Google Scholar] [CrossRef]
- Takeuchi, T.; Hishiya, T. Molecular imprinting of proteins emerging as a tool for protein recognition. Org. Biomol. Chem. 2008, 6, 2459–2467. [Google Scholar] [CrossRef]
- Kempe, M.; Glad, M.; Mosbach, K. An approach towards surface imprinting using the enzyme ribonuclease A. J. Mol. Recognit. 1995, 8, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhou, D.; Guo, T. Construction of a novel macroporous imprinted biosensor based on quartz crystal microbalance for ribonuclease Adetection. Biosens. Bioelectron. 2013, 42, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Cutivet, A.; Schembri, C.; Kovensky, J.; Haupt, K. Molecularly Imprinted Microgels as Enzyme Inhibitors. J. Am. Chem. Soc. 2009, 131, 14699–14702. [Google Scholar] [CrossRef] [PubMed]
- Karaseva, N.A.; Pluhar, B.; Beliaeva, E.A.; Ermolaeva, T.N.; Mizaikoff, B. Synthesis and application of molecularly imprinted polymers for trypsin piezoelectric sensors. Sens. Actuators B Chem. 2019, 280, 272–279. [Google Scholar] [CrossRef]
- Lee, M.H.; Chen, Y.C.; Ho, M.H.; Lin, H.Y. Optical recognition of salivary proteins by use of molecularly imprinted poly(ethylene-co-vinyl alcohol)/quantum dot composite nanoparticles. Anal. Bioanal. Chem. 2010, 397, 1457–1466. [Google Scholar] [CrossRef]
- Chen, X.; Yang, Z.; Si, S. Potentiometric urea biosensor based on immobilization of urease onto molecularly imprinted TiO2 film. J. Electroanal. Chem. 2009, 635, 1–6. [Google Scholar] [CrossRef]
- Jetzschmann, K.J.; Jágerszki, G.; Dechtrirat, D.; Yarman, A.; Gajovic-Eichelmann, N.; Gilsing, H.-D.; Schulz, B.; Gyurcsányi, R.E.; Scheller, F.W. Vectorially Imprinted Hybrid Nanofilm for Acetylcholinesterase Recognition. Adv. Funct. Mater. 2015, 25, 5178–5183. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Chen, Y.-C.; Sheu, D.-C.; Chou, T.C. Molecularly imprinted polymers for the recognition of sodium dodecyl sulfate denatured creatine kinase. J. Taiwan Inst. Chem. Eng. 2012, 43, 188–194. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozcelikay, G.; Kurbanoglu, S.; Zhang, X.; Kosak Soz, C.; Wollenberger, U.; Ozkan, S.A.; Yarman, A.; Scheller, F.W. Electrochemical MIP Sensor for Butyrylcholinesterase. Polymers 2019, 11, 1970. https://doi.org/10.3390/polym11121970
Ozcelikay G, Kurbanoglu S, Zhang X, Kosak Soz C, Wollenberger U, Ozkan SA, Yarman A, Scheller FW. Electrochemical MIP Sensor for Butyrylcholinesterase. Polymers. 2019; 11(12):1970. https://doi.org/10.3390/polym11121970
Chicago/Turabian StyleOzcelikay, Goksu, Sevinc Kurbanoglu, Xiaorong Zhang, Cagla Kosak Soz, Ulla Wollenberger, Sibel A. Ozkan, Aysu Yarman, and Frieder W. Scheller. 2019. "Electrochemical MIP Sensor for Butyrylcholinesterase" Polymers 11, no. 12: 1970. https://doi.org/10.3390/polym11121970
APA StyleOzcelikay, G., Kurbanoglu, S., Zhang, X., Kosak Soz, C., Wollenberger, U., Ozkan, S. A., Yarman, A., & Scheller, F. W. (2019). Electrochemical MIP Sensor for Butyrylcholinesterase. Polymers, 11(12), 1970. https://doi.org/10.3390/polym11121970