An Investigation of the Intermolecular Interactions and Recognition Properties of Molecular Imprinted Polymers for Deltamethrin through Computational Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Computational Methods and Characterization
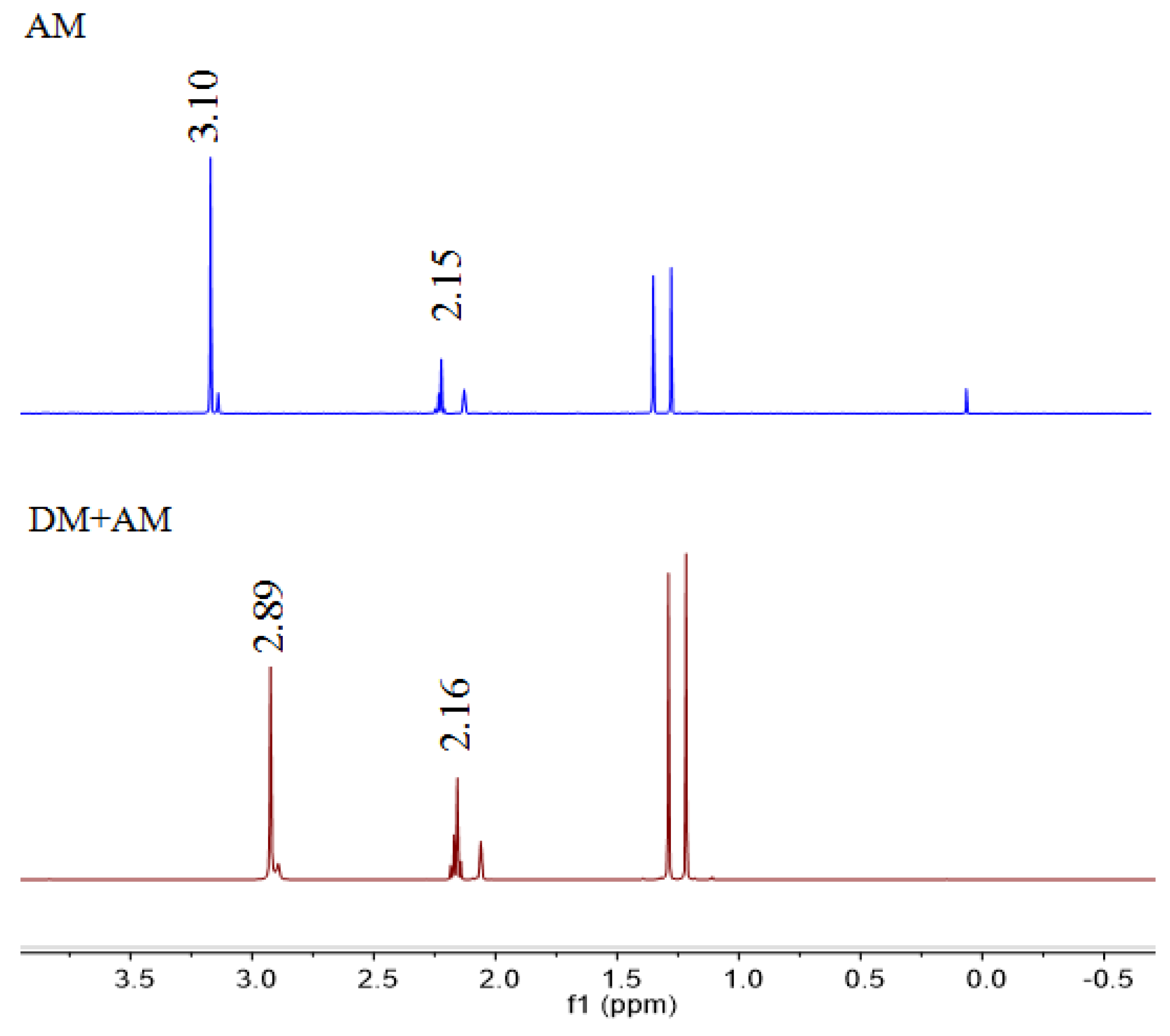
2.3. H-NMR Analysis
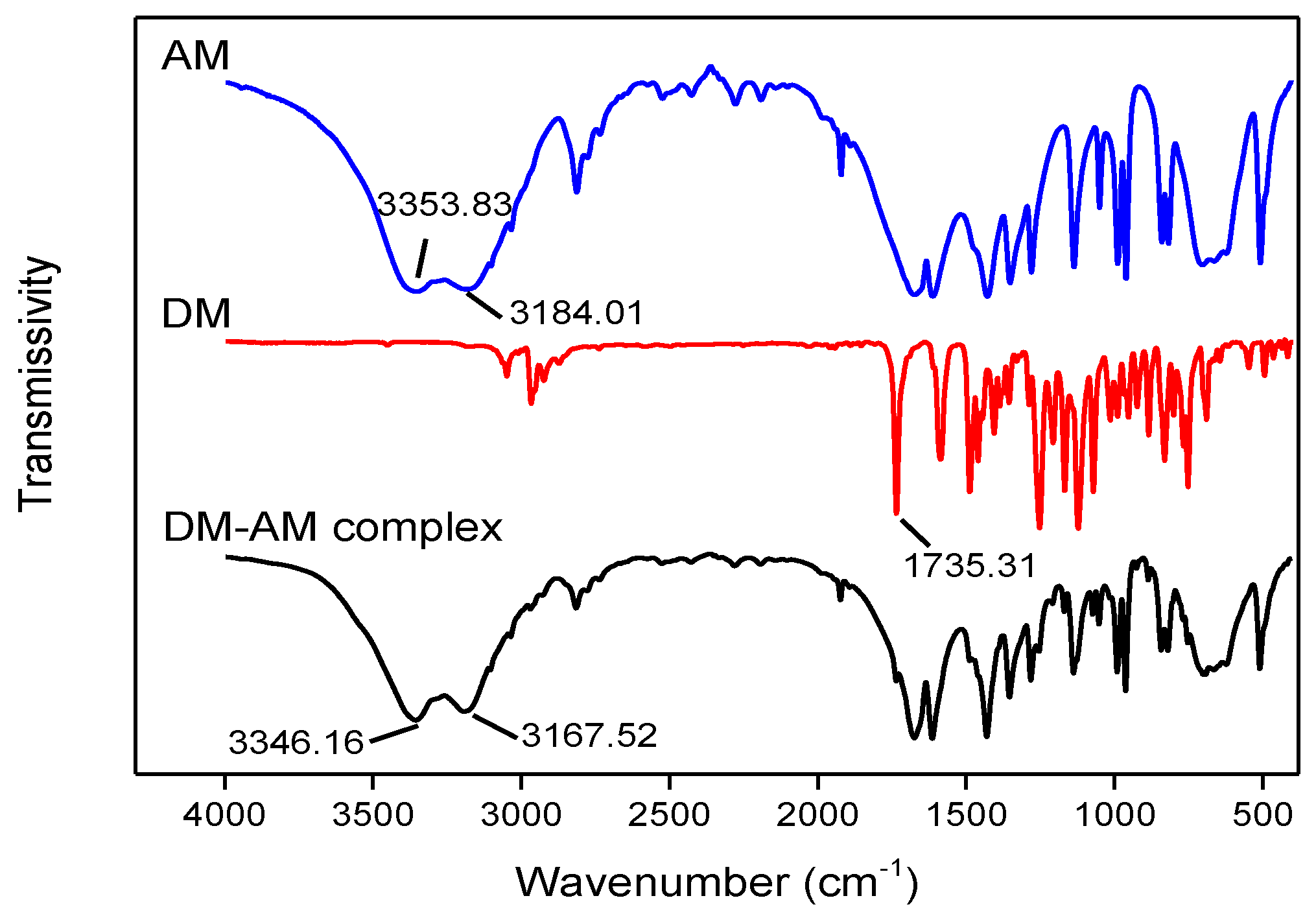
2.4. FTIR Analysis
2.5. Thermodynamic Properties
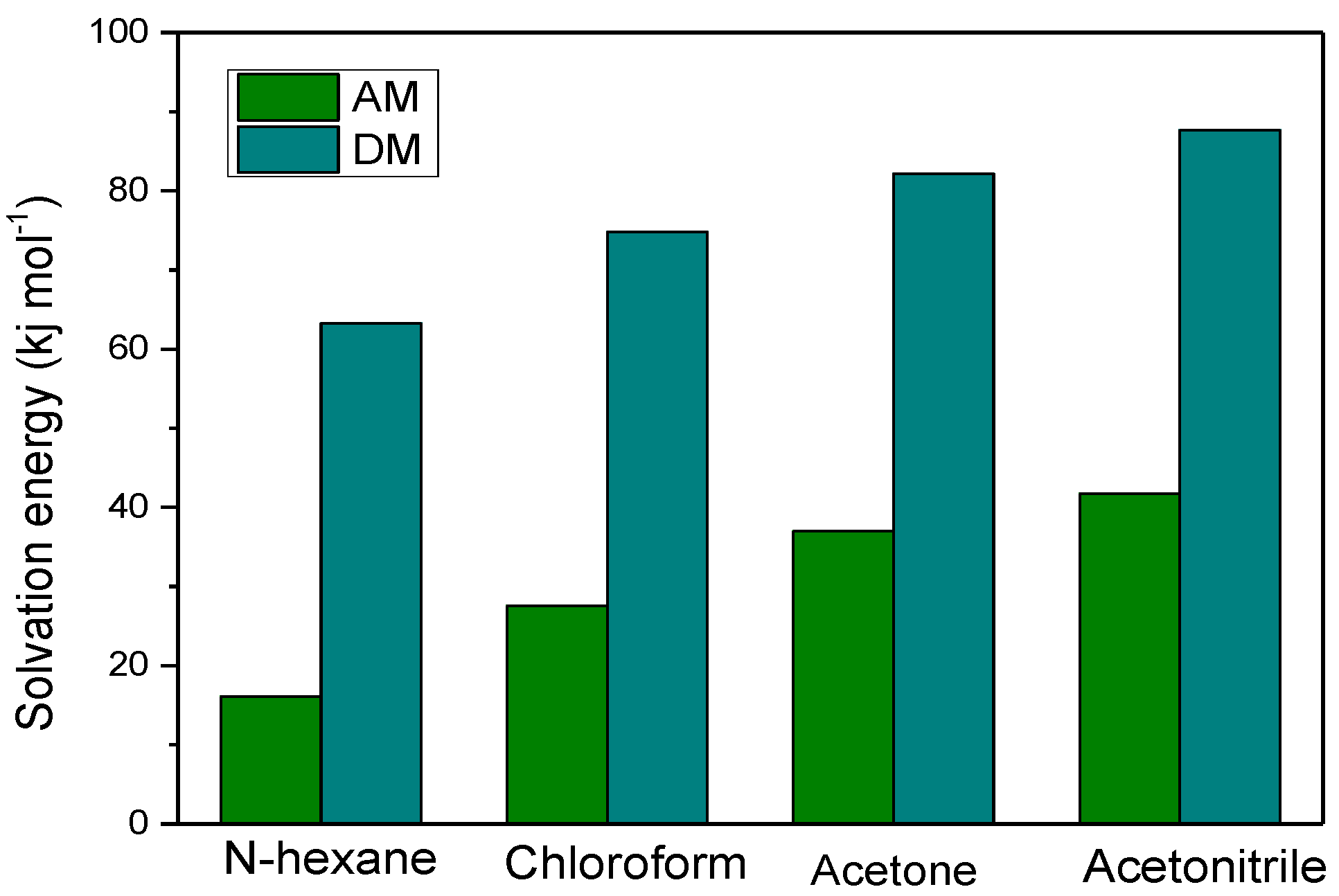
2.6. Solvation Analysis
2.7. Synthesis of DM MIPs
2.8. Evaluation of Specific Adsorption Characteristics
2.9. The Adsorption of DM in Different Teas by DM MIPs Solid-Phase Extraction
3. Results and Discussion
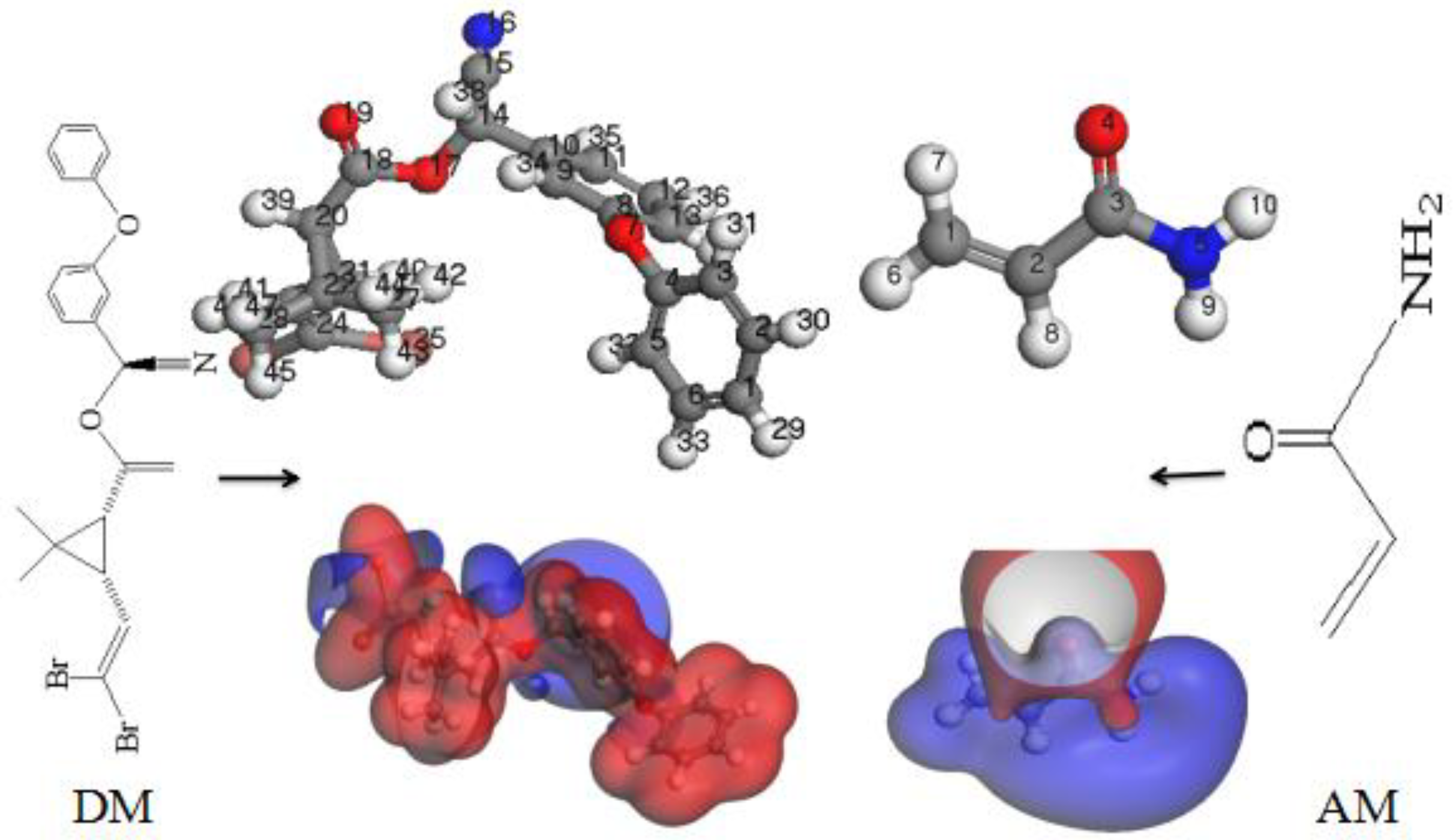
3.1. Active Sites for DM and AM Binding
3.1.1. Analysis of Frontier Molecular Orbitals
3.1.2. Analysis of Molecular Electrostatic Potentials
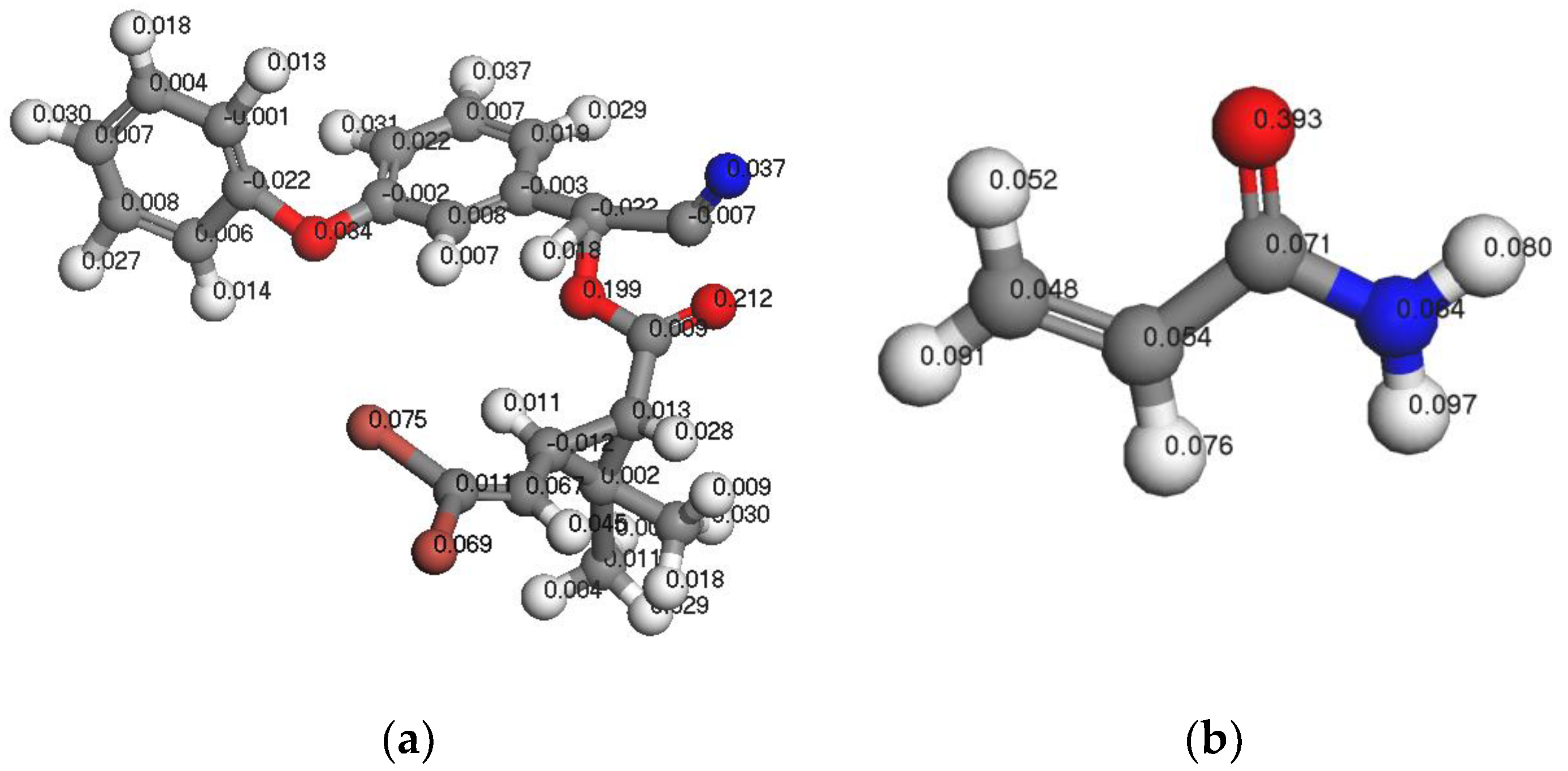
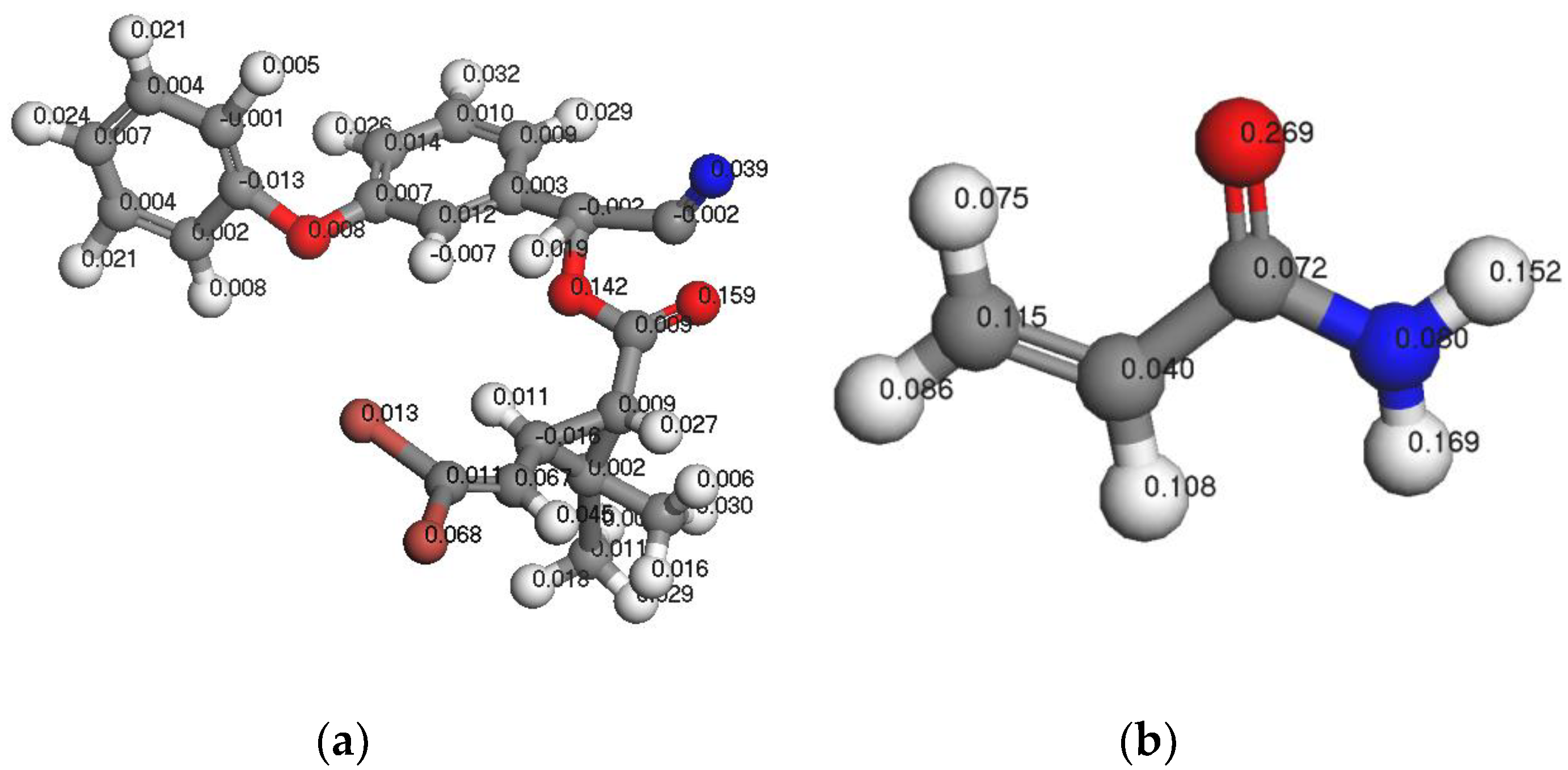
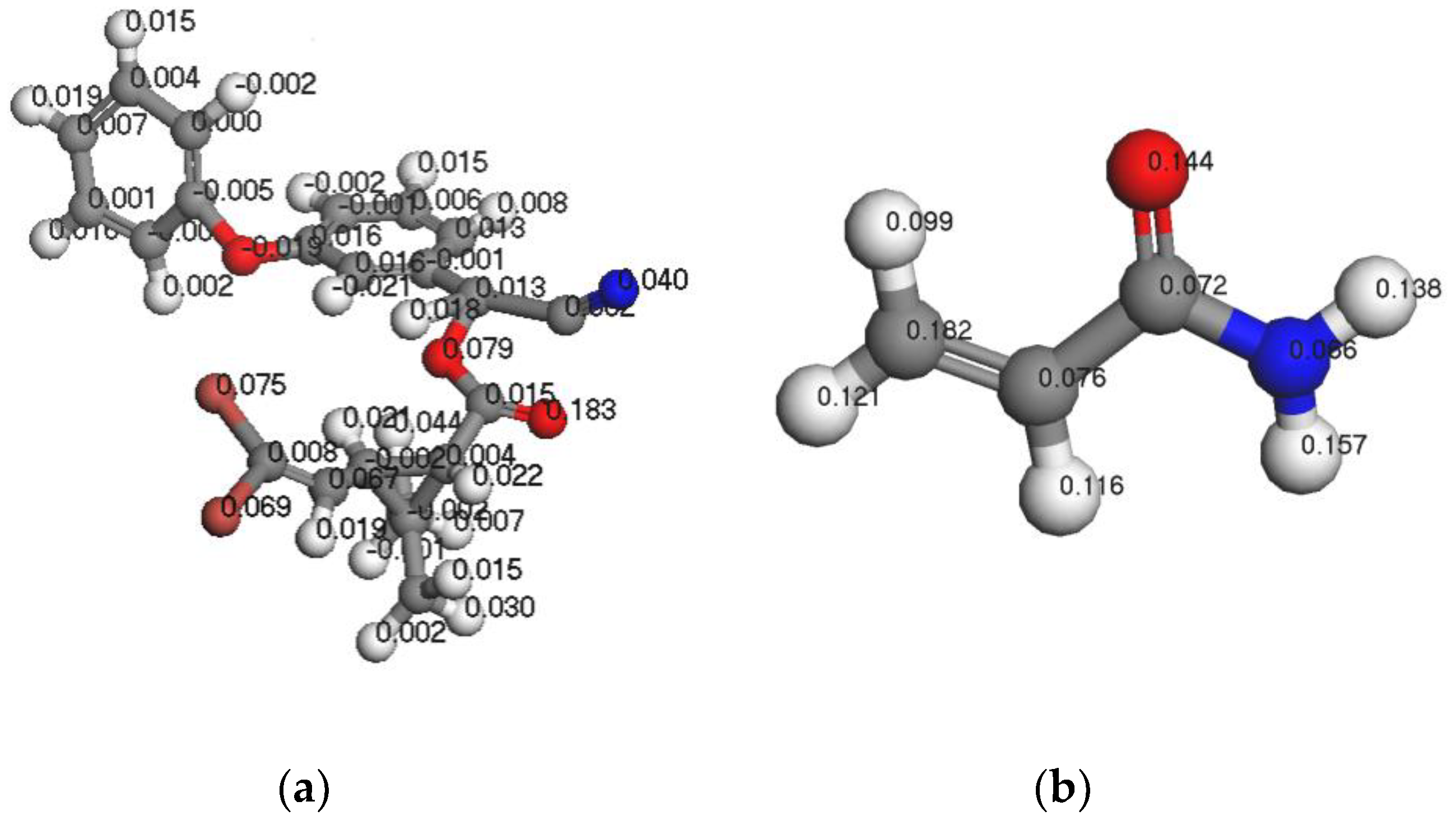
3.1.3. Analysis of Fukui Functions
3.2. H-NMR Analysis of a Prepolymerized System
3.3. FTIR Analysis
3.4. Adsorption Thermodynamics
3.5. The Effect of the Solvent on MIPs Molecular Recognition
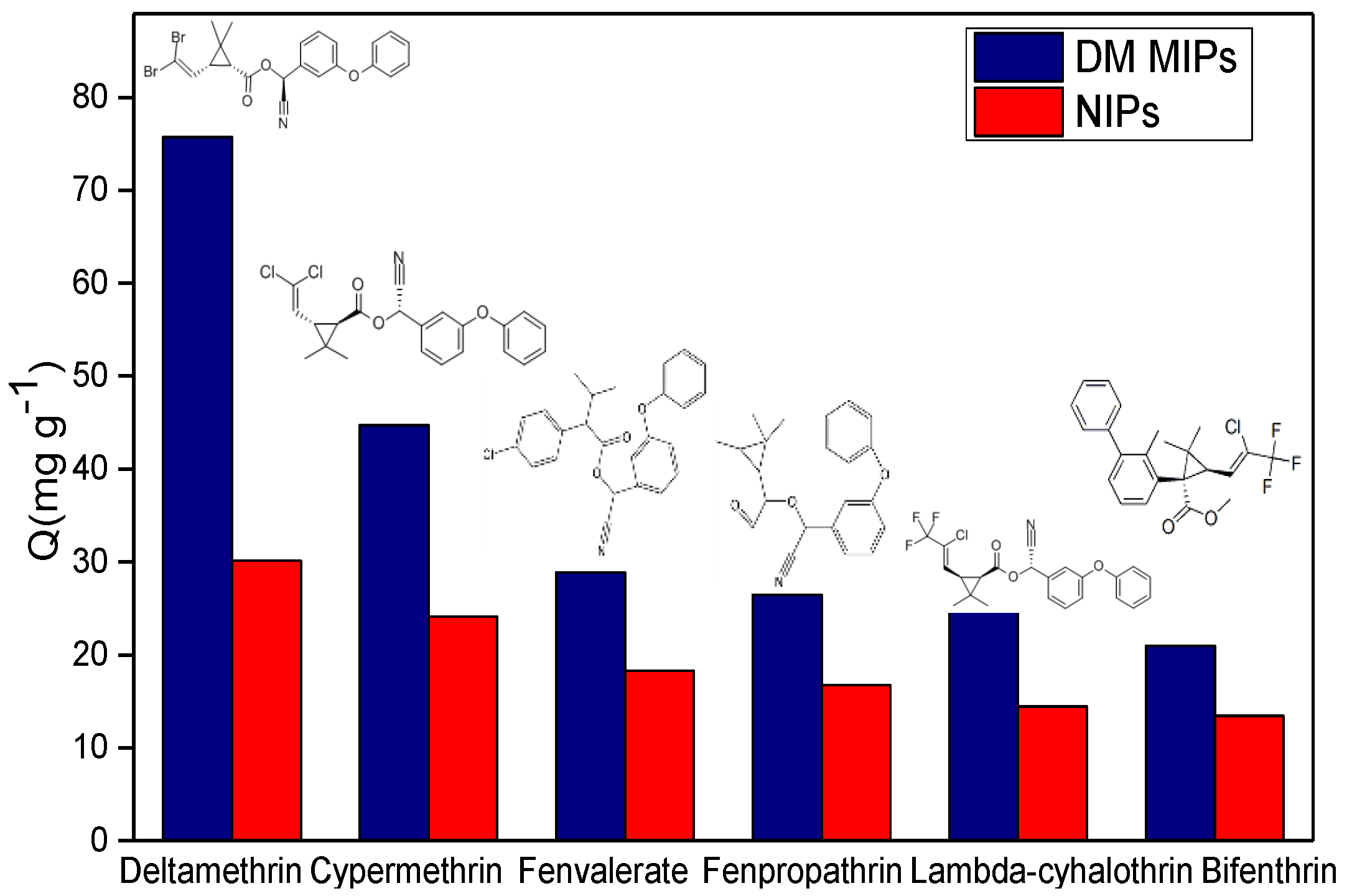
3.6. Specific Adsorption Characteristics of DM MIPs
3.7. Selective Extraction of DM in Different Tea Samples
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Liu, X.Y.; Zhang, Q.P.; Li, S.B.; Mi, P.; Chen, D.Y.; Zhao, X.; Feng, X.Z. Developmental toxicity and neurotoxicity of synthetic organic insecticides in zebrafish (Danio rerio): A comparative study of deltamethrin, acephate, and thiamethoxam. Chemosphere 2018, 199, 16–25. [Google Scholar] [CrossRef]
- Rehman, H.; Ali, M.; Atif, F.; Kaur, M.; Bhatia, K.; Raisuddin, S. The modulatory effect of deltamethrin on antioxidants in mice. Clin. Chim. Acta. 2006, 369, 61–65. [Google Scholar] [CrossRef]
- Chen, L.X.; Wang, X.Y.; Lu, W.H.; Wu, X.Q.; Li, J.H. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 452, 137–2211. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.N. Study on the Multi-Residues Detection of Triazole Pesticides Based on Magnetic Nano Molecularly Imprinted Technology. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2017. [Google Scholar]
- Ge, S.H.; Lu, J.J.; Ge, L.; Yan, M.; Yu, J.H. Development of a novel deltamethrin sensor based on molecularly imprinted silica nanospheres embedded CdTe quantum dots. Spectrochima Acta Part A 2011, 79, 1704–1709. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.Z.; Liu, J.H.; Su, A.L.; Li, D.X.; Chen, J. Group-selective enrichment and determination of pyrethroid insecticides in aquaculture seawater via molecularly imprinted solid phase extraction coupled with gas chromatography-electron capture detection. J. Chromatogr. A 2012, 1227, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Juan, S.G.; Angeles, P.G.; Jesus, S.; Ana, M.B.; Pilar, B.B.; Antonio, M.P. NMR spectroscopy for assessing cocaine-functional monomer interactions when preparing molecularly imprinted polymers. Microchem. J. 2019, 147, 813–817. [Google Scholar]
- Wu, H.; Tian, Q.; Zheng, W.; Jiang, Y.; Xu, J.C.; Li, X.; Zhang, W.C.; Qiu, F.X. Non-enzymatic glucose sensor based on molecularly imprinted polymer: A theoretical, strategy fabrication and application. J. Solid State Electrchem. 2019, 23, 1379–1388. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, Y.H.; Wang, L.; Long, R.Q.; Chen, C.L. Preparation and application of magnetic molecularly imprinted polymers for the isolation of chelerythrine from Macleaya cordata. J. Sep. Sci. 2018, 41, 3318–3327. [Google Scholar] [CrossRef] [PubMed]
- El Nashar, R.M.; Abdel Ghani, N.T.; El Gohary, N.A. Molecularly imprinted polymers based biomimetic sensors for mosapride citrate detection in biological fluids. Mater. Sci. Eng: C 2017, 76, 123–129. [Google Scholar] [CrossRef]
- Latorre, A.L.; Pérez, M.C.; Fernández, S.F.; Vilariño, J.L.; Rodríguez, M.G. Selective removal of ATP degradation products from food matrices I: Design and characterization of a dummy molecularly imprinted specific sorbent for hypoxanthine. React. Funct. Polym. 2015, 91, 51–61. [Google Scholar] [CrossRef]
- Dai, Z.Q.; Liu, J.B.; Tang, S.S.; Wang, Y.; Wang, Y.M.; Jin, R.F. Optimization of enrofloxacin-imprinted polymers by computer-aided design. J. Mol. Model. 2015, 21, 290. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Yang, B.W.; Zhu, Q.J. Configurational Simulations and Theoretical Calculations of Molecularly Imprinted Polymers of Histamine and 2-(Trifluoromethyl)acrylic Acid Based on Computational Chemistry. J. Chin. Chem. Soc. 2017, 64, 434–439. [Google Scholar] [CrossRef]
- Nezhadali, A.; Senobari, S.; Mojarrab, M. 1,4-dihydroxyanthraquinone electrochemical sensor based on molecularly imprinted polymer using multi-walled carbon nanotubes and multivariate optimization method. Talanta 2016, 146, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Gholivand, M.B.; Khodadadian, M.; Ahmadi, F. Computer aided-molecular design and synthesis of a high selective molecularly imprinted polymer for solid-phase extraction of furosemide from human plasma. Anal. Chim. Acta 2010, 658, 225–232. [Google Scholar] [CrossRef]
- Dong, C.; Li, X.; Guo, Z.; Qi, J. Development of a model for the rational design of molecular imprinted polymer: Computational approach for combined molecular dynamics/quantum mechanics calculations. Anal. Chim. Acta 2009, 647, 117–124. [Google Scholar] [CrossRef]
- Li, L.; Zhou, L.M.; Xie, X.A.; Li, Y. Screening of molecularly imprinted pre-assembly system for detection of deltamethrin pesticide residues and its specific adsorption properties. Trans. Chin. Soc. Agric. Eng. 2019, 35, 269–277. [Google Scholar]
- Zhang, C.L.; Hu, J.L.; Chen, S.J.; Ji, F.L. Theoretical study of hydrogen bonding interactions on MDI-based polyurethane. J. Mol. Model. 2010, 16, 1391–1399. [Google Scholar] [CrossRef]
- Torkashvand, M.; Gholivand, M.B.; Taherkhani, F. Fabrication of an electrochemical sensor based on computationally designed molecularly imprinted polymer for the determination of mesalamine in real samples. Mater. Sci. Eng. C 2015, 55, 209–217. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.Z.; Yang, L.W.; Fan, Y.M.; Liu, Y.S. Molecular structure and spectral characteristics of hyperoside and analysis of its molecular imprinting adsorption properties based on density functional theory. J. Mol. Graph. Model. 2019, 88, 228–236. [Google Scholar] [CrossRef]
- Khan, M.S.; Pal, S. Quantum mechanical studies on dioxin-imprinted polymer precursor composites: Fundamental insights to enhance the binding strength and selectivity of biomarkers. J. Mol. Recognit. 2018, 31, e2736. [Google Scholar] [CrossRef]
- Li, L.; Gao, Y.; Li, H.; Zhao, Y.; Pei, Y.; Chen, Z.F.; Zeng, X.C. CO oxidation on TiO2 (110) supported subnanometer gold clusters: Size and shape effects. J. Am. Chem. Soc. 2013, 135, 19336–19346. [Google Scholar] [CrossRef]
- Inukai, J.; Tryk, D.A.; Abe, T.; Wakisaka, M.; Uchida, H.; Watanabe, M. Direct STM elucidation of the effects of atomic-level structure on Pt (111) electrodes for dissolved CO oxidation. J. Am. Chem. Soc. 2013, 135, 1476–1490. [Google Scholar] [CrossRef] [PubMed]
- Eşme, A.; Sağdınç, S.G. Molecular structures, spectroscopic (FT–IR, NMR, UV) studies, NBO analysis and NLO properties for tautomeric forms of 1,3–dimethyl–5–(phenylazo)–6–aminouracil by density functional method. Spectrochim. Acta Part A 2018, 188, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhou, L.M.; Li, L.; Xin, X.A.; Li, Y. Preparation and adsorption selectivity of deltamethrin molecularly imprinted polymers by two-step seed swelling method. J. Appl. Polym. Sci. 2019, 136, 47415. [Google Scholar] [CrossRef]
- Halim, S.A.; Ibrahim, M.A. Synthesis, DFT calculations, electronic structure, electronic absorption spectra, natural bond orbital (NBO) and nonlinear optical (NLO) analysis of the novel 5-methyl-8H-benzo[h]chromeno[2,3-b] [1,6] naphthyridine-6(5H),8-dione (MBCND). J. Mol. Struct. 2017, 1130, 543–558. [Google Scholar] [CrossRef]
- James, C.; Raj, A.A.; Reghunathan, R.; Jayakumar, V.S.; Joe, I.H. Structural conformation and vibrational spectroscopic studies of 2,6-bis-N, N-dimethyl benzylidene cyclohexanone using density functional theory. J. Raman Spectrosc. 2006, 37, 1381–1392. [Google Scholar] [CrossRef]
- Shakila, G.; Saleem, H. Sundaraganesan FT–IR, FT–Raman, NMR and U–V Spectral investigation: Computation of vibrational frequency, chemical shifts and electronic structure calculations of 1–bromo–4–nitrobenzene. World Sci. News 2017, 61, 150–185. [Google Scholar]
- Zhang, B.C.; Fan, X.; Zhao, D.Y. Computer-aided design of molecularly imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Polymers 2019, 11, 17. [Google Scholar] [CrossRef]
- Dong, W.G.; Yan, M.; Liu, Z.; Wu, G.S.; Li, Y.M. Effects of solvents on the adsorption selectivity of molecularly imprinted polymers: Molecular simulation and experimental validation. Sep. Purif. Technol. 2007, 53, 183–188. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; Wang, N.N.; Luo, J.J.; Zhou, Y.; Yu, Z.W. Hydrogen-bonding interactions between [BMIM][BF4] and acetonitrile. Phys. Chem. Chem. Phys. 2013, 15, 18055–18064. [Google Scholar] [CrossRef]
- Liu, S.C.; Pan, J.M.; Zhu, H.J.; Pan, G.Q.; Qiu, F.X.; Meng, M.J.; Yao, J.; Yuan, D. Graphene oxide based molecularly imprinted polymers with double recognition abilities: The combination of covalent boronic acid and traditional non-covalent monomers. Chem. Eng. J. 2016, 290, 220–231. [Google Scholar] [CrossRef]
- Alireza, M.; Majid, A.; Payam, Z. Isosorbide dinitrate template-based molecularly imprinted poly (methacrylic acid) nanoparticles: Effect of initiator concentration on morphology and physicochemical properties. Chem. Pap. 2018, 72, 3005–3016. [Google Scholar]
- She, Y.X.; Wang, M.; Shi, X.M.; Liu, J.J.; Lu, X.L.; Xia, H.; Cao, W.Q.; Wang, J. Spectroscopy study of binding mechanisms and molecular recognition of class-specific molecularly imprinted polymer beads. Spectrosc. Spect. Anal. 2010, 30, 3052–3055. [Google Scholar]
- Zhang, P.P.; Ji, X.Y.; Zhang, H.; Xia, B.H. Quantum investigation into intermolecular interactions between bisphenol A and 2-vinyl/4-vinylpyridine: Theoretical insight into molecular imprinting complexes. Comput. Theor. Chem. 2017, 1108, 76–85. [Google Scholar] [CrossRef]
- Wu, Y.L.; Ma, Y.; Pan, J.M.; Gu, R.X.; Luo, J.L. Porous and Magnetic Molecularly Imprinted Polymers via Pickering High Internal Phase Emulsions Polymerization for Selective Adsorption of λ-Cyhalothrin. Front. Chem. 2017, 5, 18. [Google Scholar] [CrossRef]
- Yu, Q.; Deng, S.; Yu, G. Selective removal of perflfluorooctane sulfonate from aqueous solution using chitosan-based molecularly imprinted polymer adsorbents. Water Res. 2018, 42, 3089–3097. [Google Scholar] [CrossRef]
- Machicote, R.G.; Pacheco, M.E.; Bruzzone, L. Binding of several benzodiazepines to bovine serum albumin: Fluorescence study. Spectrochim. Acta A 2010, 77, 466–472. [Google Scholar] [CrossRef]
- Zhao, X.C.; Liu, R.T.; Chi, Z.X.; Teng, Y.; Qin, P.F. New insights into the behavior of bovine serum albumin adsorbed onto carbon nanotubes: Comprehensive spectroscopic studies. J. Phys. Chem. B 2010, 114, 5625–5631. [Google Scholar] [CrossRef]
- Gao, B.J.; Li, Y.B.; Zhang, Z.G. Preparation and recognition performance of creatinine-imprinted material prepared with novel surface-imprinting technique. J. Chromatogr. B 2010, 878, 2077–2086. [Google Scholar] [CrossRef]
- Mazouz, Z.; Rahali, S.; Fourati, N.; Zerrouki, C.; Aloui, N.; Seydou, M.; Yaakoubi, N.; Chehimi, M.M.; Othmane, A.; Kalfat, R. Highly Selective Polypyrrole MIP-Based Gravimetric and Electrochemical Sensors for Picomolar Detection of Glyphosate. Sensors 2017, 17, 2586. [Google Scholar] [CrossRef] [Green Version]
- Nezhadali, A.; Mojarrab, M. Computational study and multivariate optimization of hydrochlorothiazide analysis using molecularly imprinted polymer electrochemical sensor based on carbon nanotube/polypyrrole fifilm. Sens. Actuators B Chem. 2014, 190, 829–837. [Google Scholar] [CrossRef]

| ΔH/kJ mol−1 | ΔS/J mol−1 K | ΔG/kJ mol−1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 293 K | 303 K | 313 K | 323 K | 293 K | 303 K | 313 K | 323 K | ||
| AM Monomer | 185.351 | 0.627 | 0.606 | 0.586 | 0.568 | −1.593 | −1.677 | −1.681 | 1.686 |
| DM MIPs | −61.00 | 188.9 | 188.0 | 185.1 | 181.2 | −5.655 | −4.046 | −3.052 | 2.489 |
| Tea type | Recovery (%) | RSD (%) |
|---|---|---|
| Black tea | 83.68 | 2.13 |
| Green tea | 107.55 | 0.98 |
| Tieguanyin | 94.38 | 1.79 |
| Black tea beverage | 85.82 | 0.35 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Xiao, N.; Li, L.; Xie, X.; Li, Y. An Investigation of the Intermolecular Interactions and Recognition Properties of Molecular Imprinted Polymers for Deltamethrin through Computational Strategies. Polymers 2019, 11, 1872. https://doi.org/10.3390/polym11111872
Xie L, Xiao N, Li L, Xie X, Li Y. An Investigation of the Intermolecular Interactions and Recognition Properties of Molecular Imprinted Polymers for Deltamethrin through Computational Strategies. Polymers. 2019; 11(11):1872. https://doi.org/10.3390/polym11111872
Chicago/Turabian StyleXie, Lei, Nan Xiao, Lu Li, Xinan Xie, and Yan Li. 2019. "An Investigation of the Intermolecular Interactions and Recognition Properties of Molecular Imprinted Polymers for Deltamethrin through Computational Strategies" Polymers 11, no. 11: 1872. https://doi.org/10.3390/polym11111872
APA StyleXie, L., Xiao, N., Li, L., Xie, X., & Li, Y. (2019). An Investigation of the Intermolecular Interactions and Recognition Properties of Molecular Imprinted Polymers for Deltamethrin through Computational Strategies. Polymers, 11(11), 1872. https://doi.org/10.3390/polym11111872





