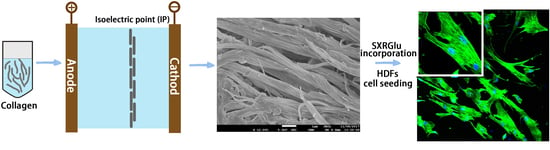
Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications
Abstract
:1. Introduction
2. Materials and Methods
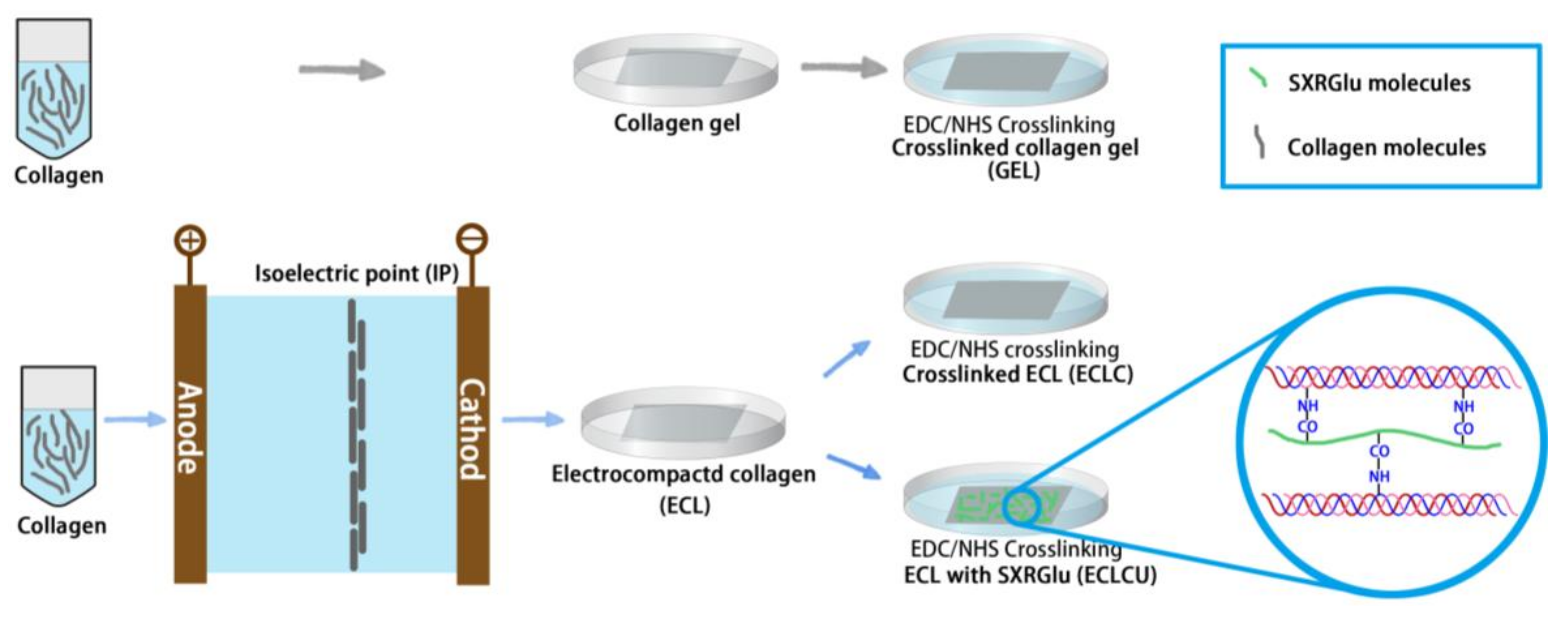
2.1. Synthesis of Electrocompacted Collagen Matrices
2.2. SXRGlu Conjugation to ECL Matrices
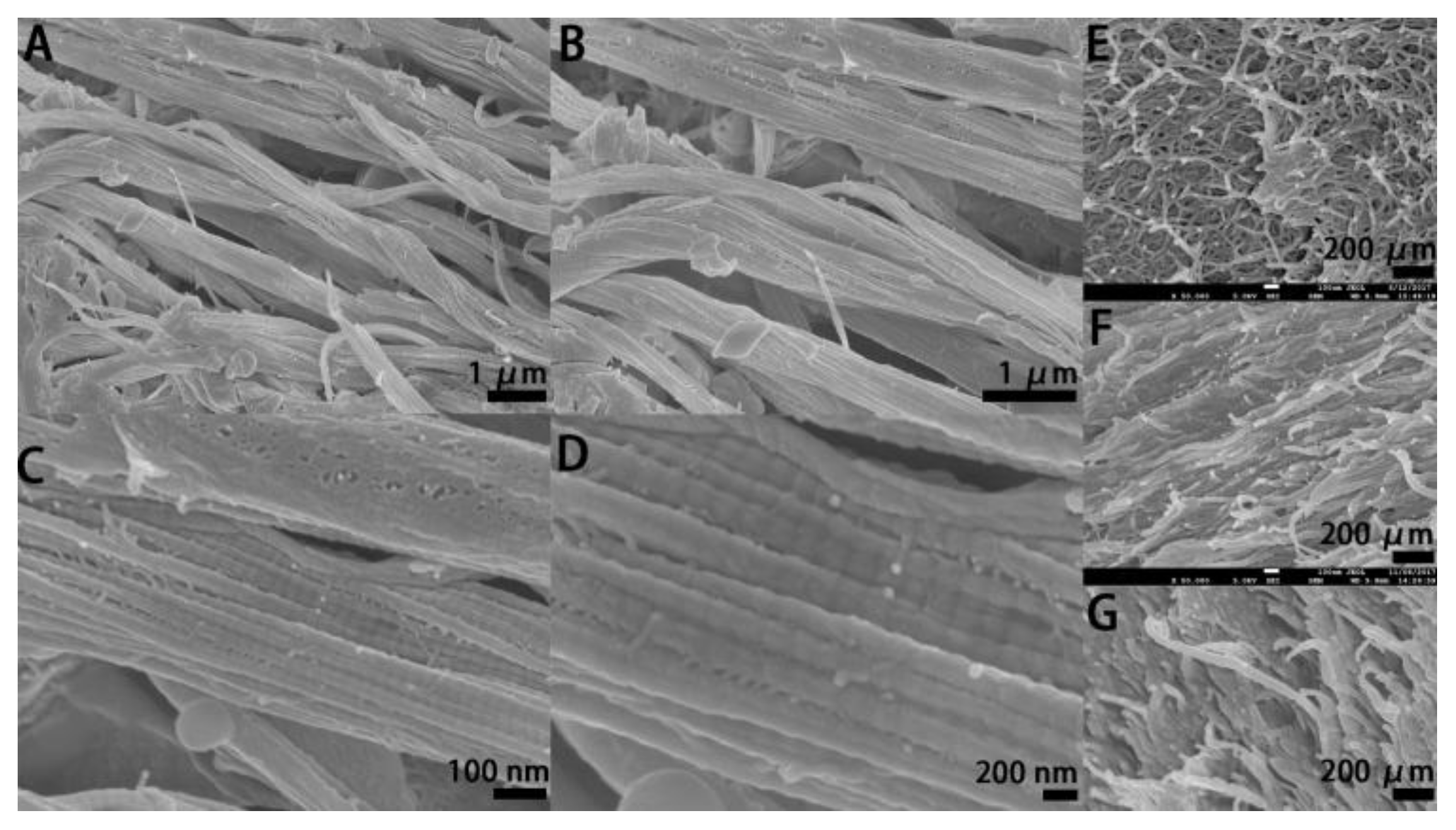
2.3. SEM Inspection of the Electrocompacted Collagen Matrices
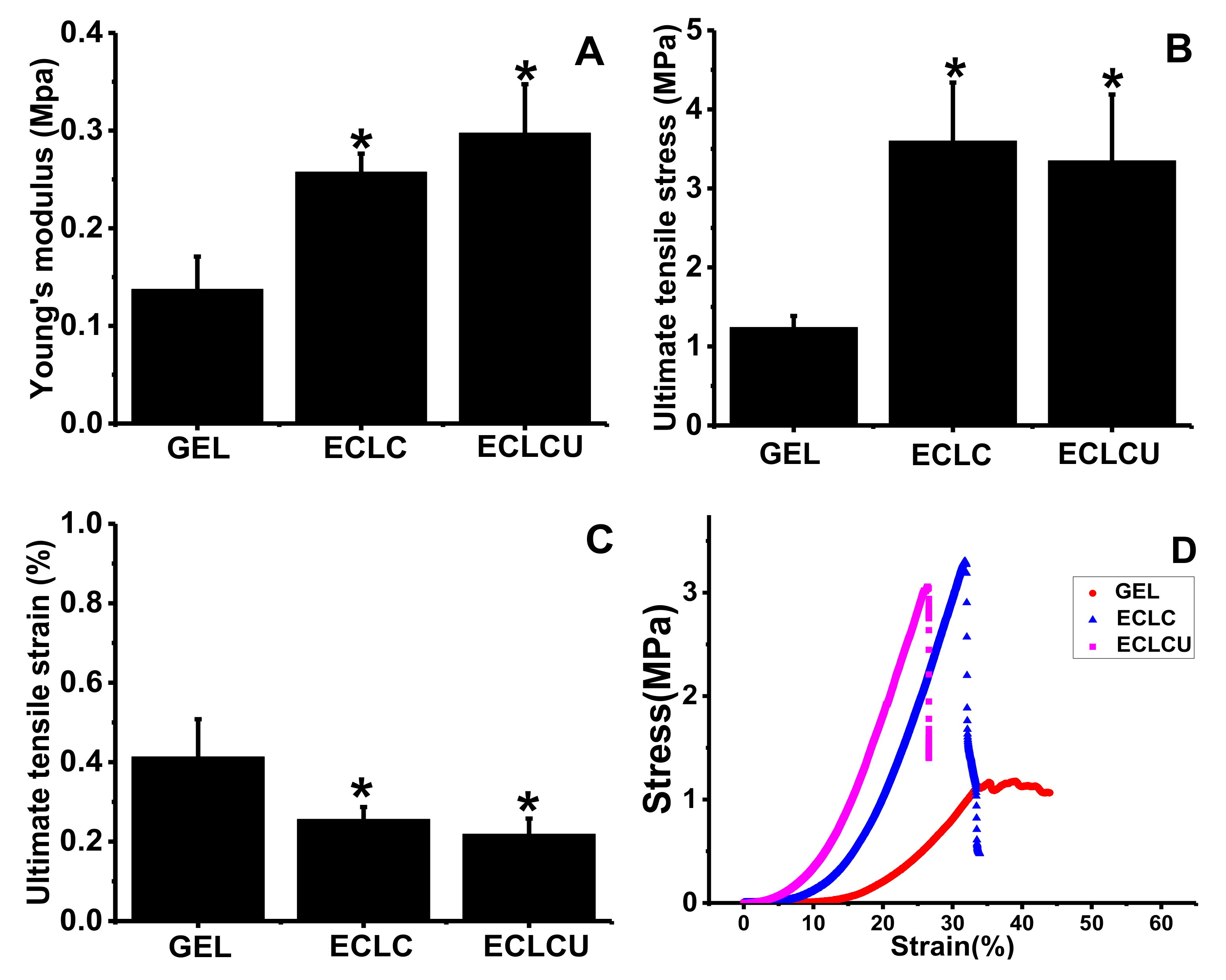
2.4. Mechanical Property
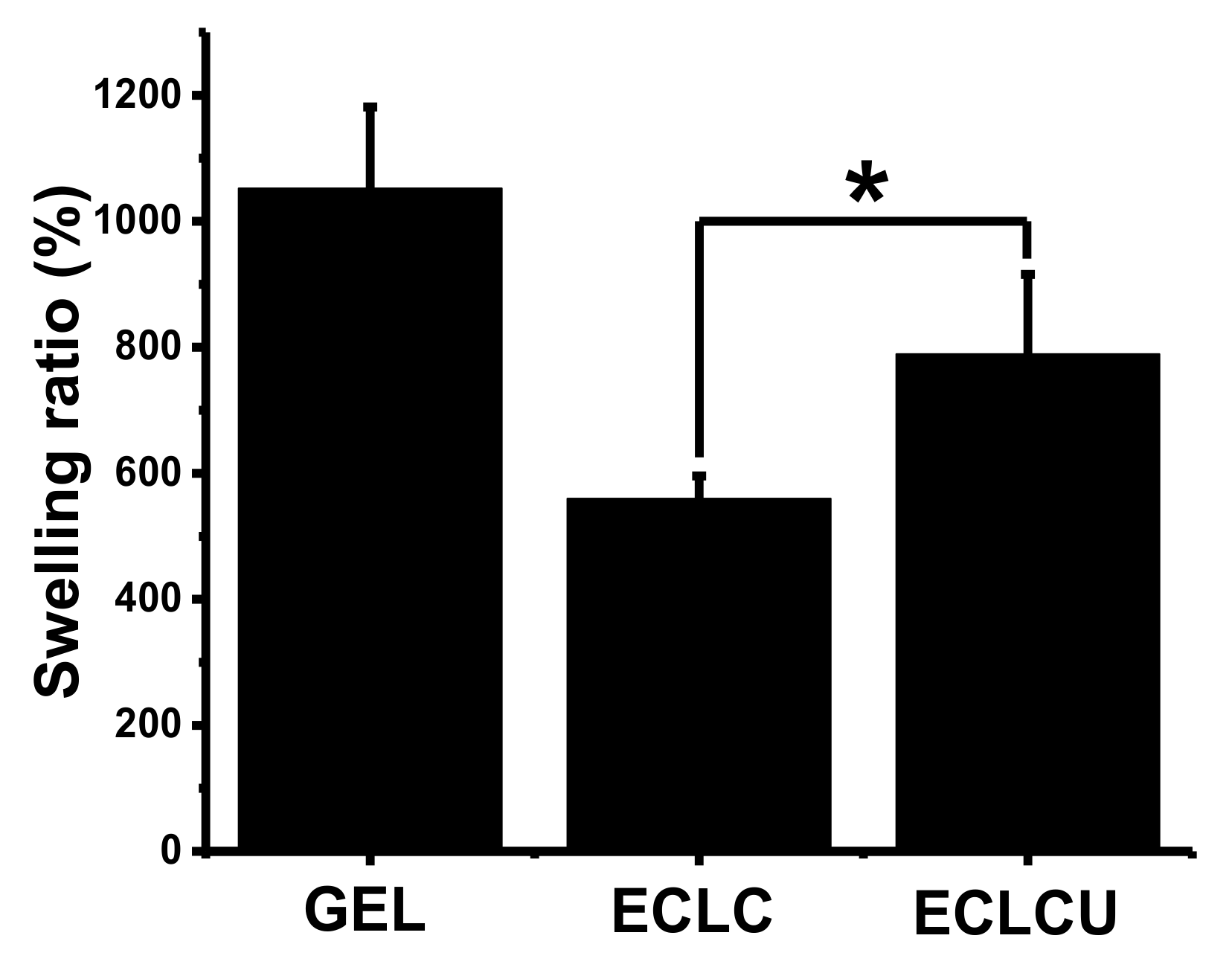
2.5. Swelling Ratio
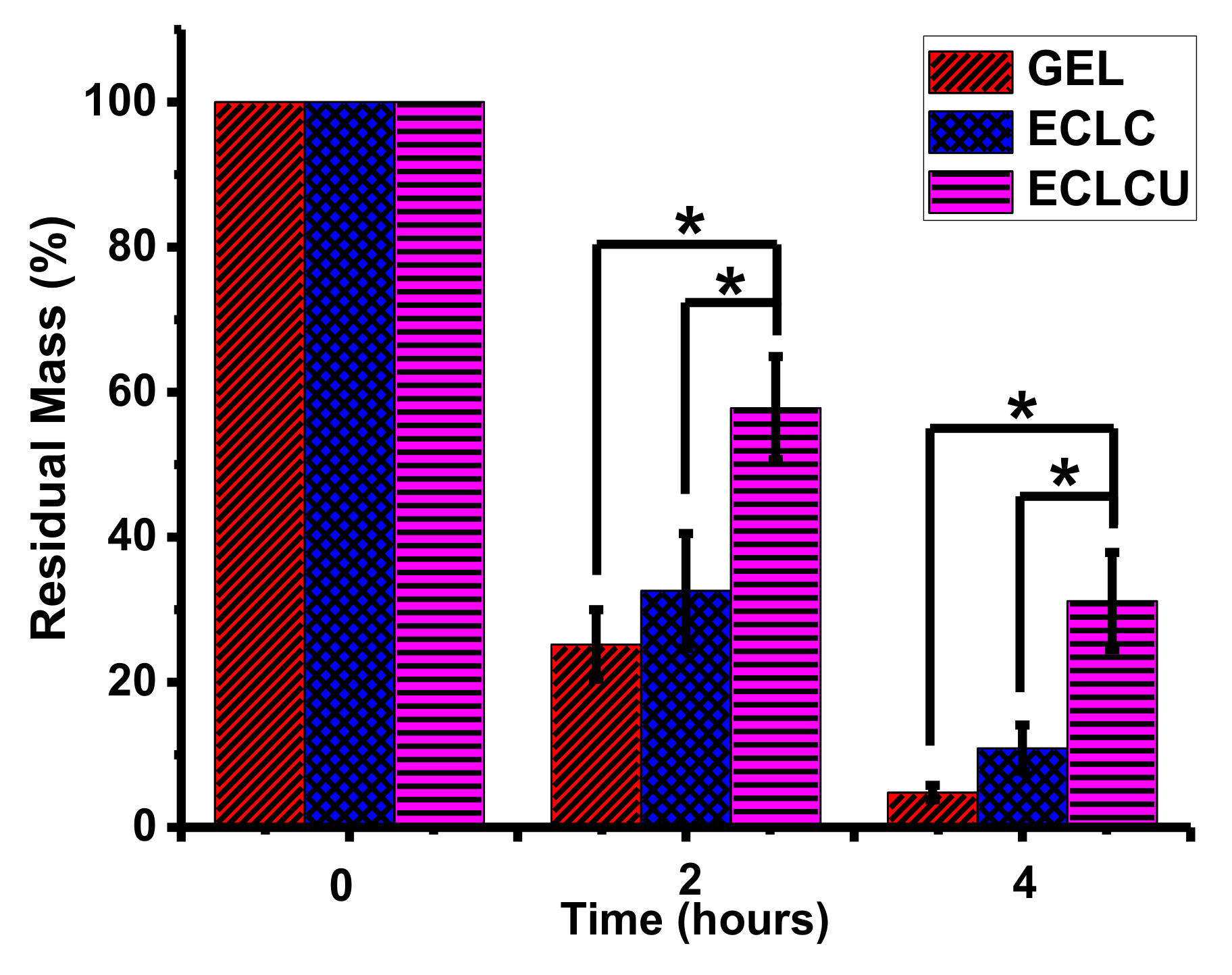
2.6. Degradability
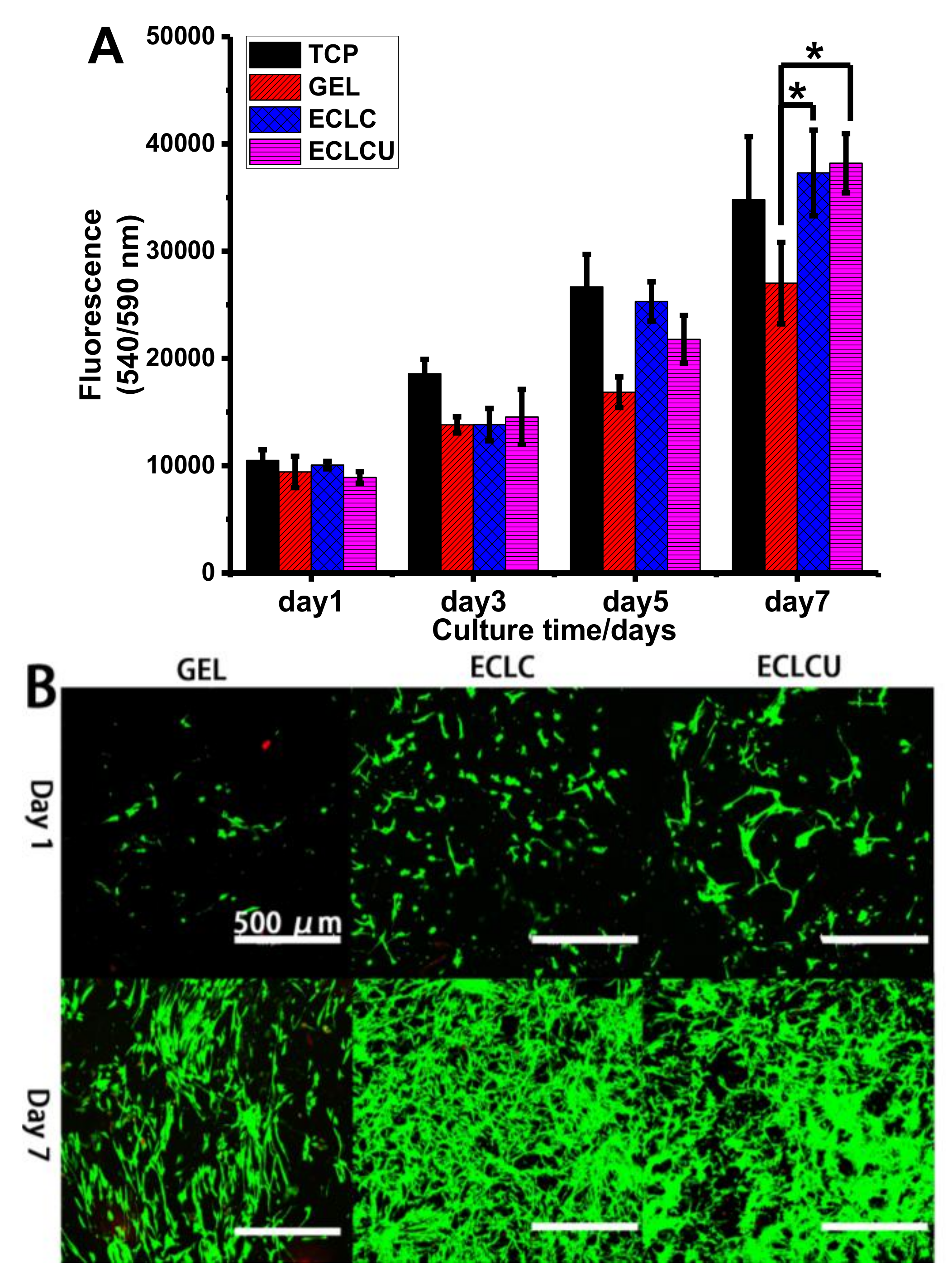
2.7. Human Dermal Fibroblast Cell Viability, Proliferation and Morphology on Electrocompacted Collagen Matrices
3. Results and Discussion
3.1. SEM Inspection
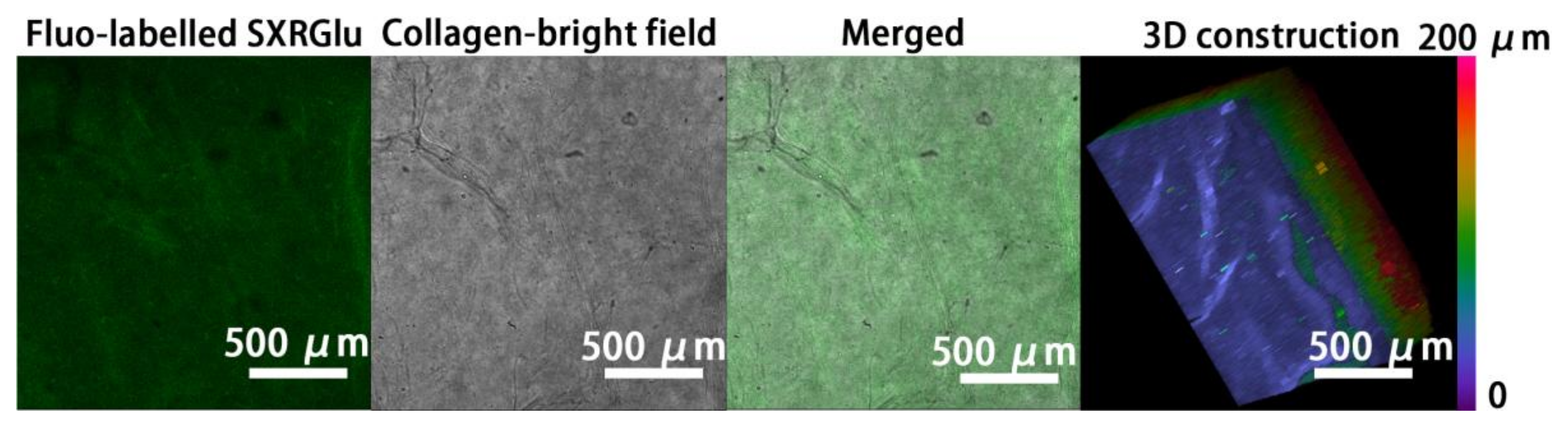
3.2. SXRGlu Content Quantification
3.3. Mechanical Property
3.4. Swelling Ratio
3.5. Degradability
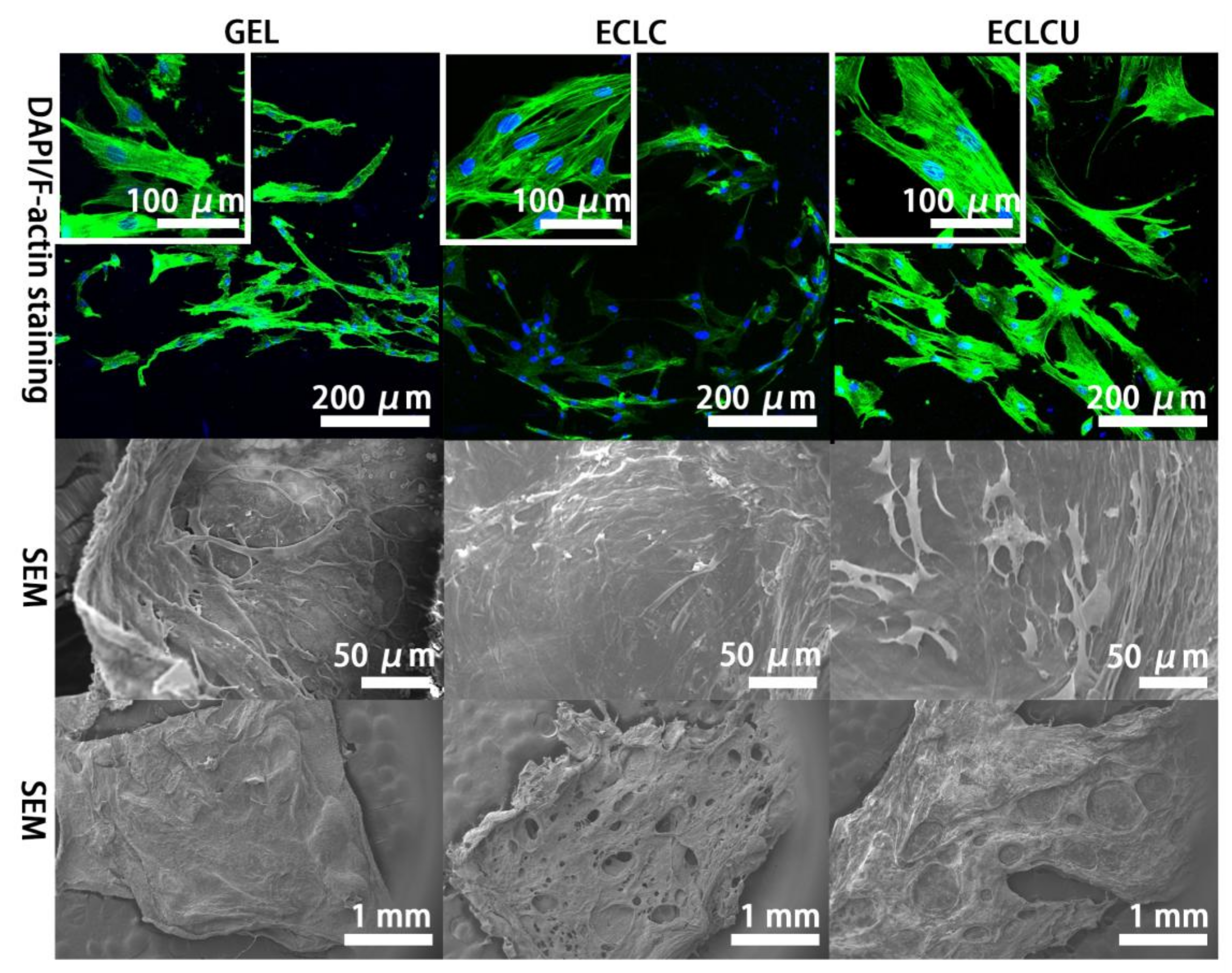
3.6. Human Dermal Fibroblast Cell Viability, Proliferation and Morphology on Electrocompacted Collagen Matrices
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pereira, R.F.; Barrias, C.C.; Granja, P.L.; Bartolo, P.J. Advanced biofabrication strategies for skin regeneration and repair. Nanomedicine 2013, 8, 603–621. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, A.D.; Ferguson, M.W. Tissue engineering of replacement skin: The crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. J. R. Soc. Interface 2007, 4, 413–437. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Yeong, W.Y.; Naing, M.W. Cellular approaches to tissue-engineering of skin: A review. J. Tissue Sci. Eng. 2015, 6. [Google Scholar] [CrossRef]
- Ng, W.L.; Wang, S.; Yeong, W.Y.; Naing, M.W. Skin bioprinting: Impending reality or fantasy? Trends Biotechnol. 2016, 34, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Tracy, L.E.; Minasian, R.A.; Caterson, E. Extracellular matrix and dermal fibroblast function in the healing wound. Adv. Wound Care 2016, 5, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Kahan, V.; Andersen, M.; Tomimori, J.; Tufik, S. Stress, immunity and skin collagen integrity: Evidence from animal models and clinical conditions. Brain Behav. Immun. 2009, 23, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Wojtowicz, A.M.; Shekaran, A.; Oest, M.E.; Dupont, K.M.; Templeman, K.L.; Hutmacher, D.W.; Guldberg, R.E.; García, A.J. Coating of biomaterial scaffolds with the collagen-mimetic peptide GFOGER for bone defect repair. Biomaterials 2010, 31, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Gurkan, U.A.; Dehen, C.J.; Tate, M.P.; Hillhouse, H.W.; Simpson, G.J.; Akkus, O. An electrochemical fabrication process for the assembly of anisotropically oriented collagen bundles. Biomaterials 2008, 29, 3278–3288. [Google Scholar] [CrossRef] [PubMed]
- Chiellini, F.; Morelli, A. Chapter Book: Ulvan: A versatile platform of biomaterials from renewable resources. In Biomaterials-Physics and Chemistry, I; Pignatello, R., Ed.; In Tech: Croatia, Balkans, 2011; pp. 75–97. [Google Scholar]
- Kishore, V.; Iyer, R.; Frandsen, A.; Nguyen, T.-U. In vitro characterization of electrochemically compacted collagen matrices for corneal applications. Biomed. Mater. 2016, 11, 055008. [Google Scholar] [CrossRef] [PubMed]
- Younesi, M.; Donmez, B.O.; Islam, A.; Akkus, O. Heparinized collagen sutures for sustained delivery of PDGF-BB: Delivery profile and effects on tendon-derived cells In Vitro. Acta Biomater. 2016, 41, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.Y.; Chow, K.L. SEM sample preparation for cells on 3D scaffolds by freeze-drying and HMDS. Scanning 2012, 34, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Sherman, V.R.; Tang, Y.; Zhao, S.; Yang, W.; Meyers, M.A. Structural characterization and viscoelastic constitutive modeling of skin. Acta Biomater. 2017, 53, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Loh, Q.L.; Choong, C. Three-dimensional scaffolds for tissue engineering applications: Role of porosity and pore size. Tissue Eng. Part B Rev. 2013, 19, 485–502. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E.; Lalia, B.S.; Hashaikeh, R. A review on electrospinning for membrane fabrication: Challenges and applications. Desalination 2015, 356, 15–30. [Google Scholar] [CrossRef]
- Mikos, A.G.; Temenoff, J.S. Formation of highly porous biodegradable scaffolds for tissue engineering. Electron. J. Biotechnol. 2000, 3, 23–24. [Google Scholar] [CrossRef]
- Schugens, C.; Maquet, V.; Grandfils, C.; Jérôme, R.; Teyssie, P. Polylactide macroporous biodegradable implants for cell transplantation. II. Preparation of polylactide foams by liquid-liquid phase separation. J. Biomed. Mater. Res. Part A 1996, 30, 449–461. [Google Scholar] [CrossRef]
- O’Brien, F.J.; Harley, B.A.; Yannas, I.V.; Gibson, L. Influence of freezing rate on pore structure in freeze-dried collagen-GAG scaffolds. Biomaterials 2004, 25, 1077–1086. [Google Scholar] [CrossRef]
- Cudjoe, E.; Younesi, M.; Cudjoe, E.; Akkus, O.; Rowan, S.J. Synthesis and Fabrication of Nanocomposite Fibers of Collagen-Cellulose Nanocrystals by Coelectrocompaction. Biomacromolecules 2017, 18, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Boppart, S.A. Biomechanical properties of in vivo human skin from dynamic optical coherence elastography. IEEE Trans. Biomed. Eng. 2010, 57, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Benavides, S.; Villalobos-Carvajal, R.; Reyes, J.E. Physical, mechanical and antibacterial properties of alginate film: Effect of the crosslinking degree and oregano essential oil concentration. J. Food Eng. 2012, 110, 232–239. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, G.D.; Boucek, R.J. Collagen and elastin of human dermis. J. Investig. Dermatol. 1960, 35, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Oxlund, H.; Manschot, J.; Viidik, A. The role of elastin in the mechanical properties of skin. J. Biomech. 1988, 21, 213–218. [Google Scholar] [CrossRef]
- Senior, R.M.; Griffin, G.L.; Mecham, R.P.; Wrenn, D.S.; Prasad, K.U.; Urry, D.W. Val-Gly-Val-Ala-Pro-Gly, a repeating peptide in elastin, is chemotactic for fibroblasts and monocytes. J. Cell Biol. 1984, 99, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Wachi, H.; Uemura, Y.; Okamoto, K. Modulation by elastin peptide VGVAPG of cell proliferation and elastin expression in human skin fibroblasts. Arch. Dermatol. Res. 1997, 289, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Järveläinen, H.; Sainio, A.; Koulu, M.; Wight, T.N.; Penttinen, R. Extracellular matrix molecules: Potential targets in pharmacotherapy. Pharmacol. Rev. 2009, 61, 198–223. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, P.; Sage, E.H. Matricellular proteins: Extracellular modulators of cell function. Curr. Opin. Cell Biol. 2002, 14, 608–616. [Google Scholar] [CrossRef]
- Okamoto, O.; Fujiwara, S. Dermatopontin, a novel player in the biology of the extracellular matrix. Connect. Tissue Res. 2006, 47, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Xia, H.; He, W.; Li, Z.; Zhao, J.; Liu, B.; Wang, Y.; Lei, Q.; Kong, Y.; Bai, Y. Controlled water vapor transmission rate promotes wound-healing via wound re-epithelialization and contraction enhancement. Sci. Rep. 2016, 6, 24596. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Mih, J.D.; Shea, B.S.; Kho, A.T.; Sharif, A.S.; Tager, A.M.; Tschumperlin, D.J. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J. Cell Biol. 2010, 190, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Andres, E.; Molinari, J.; Péterszegi, G.; Mariko, B.; Ruszova, E.; Velebny, V.; Faury, G.; Robert, L. Pharmacological properties of rhamnose-rich polysaccharides, potential interest in age-dependent alterations of connectives tissues. Pathol. Biol. 2006, 54, 420–425. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, L.; Liu, X.; Yue, Z.; Chen, Z.; Baker, C.; Winberg, P.C.; Wallace, G.G. Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications. Polymers 2018, 10, 415. https://doi.org/10.3390/polym10040415
Kang L, Liu X, Yue Z, Chen Z, Baker C, Winberg PC, Wallace GG. Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications. Polymers. 2018; 10(4):415. https://doi.org/10.3390/polym10040415
Chicago/Turabian StyleKang, Lingzhi, Xiao Liu, Zhilian Yue, Zhi Chen, Chris Baker, Pia C. Winberg, and Gordon G. Wallace. 2018. "Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications" Polymers 10, no. 4: 415. https://doi.org/10.3390/polym10040415
APA StyleKang, L., Liu, X., Yue, Z., Chen, Z., Baker, C., Winberg, P. C., & Wallace, G. G. (2018). Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications. Polymers, 10(4), 415. https://doi.org/10.3390/polym10040415