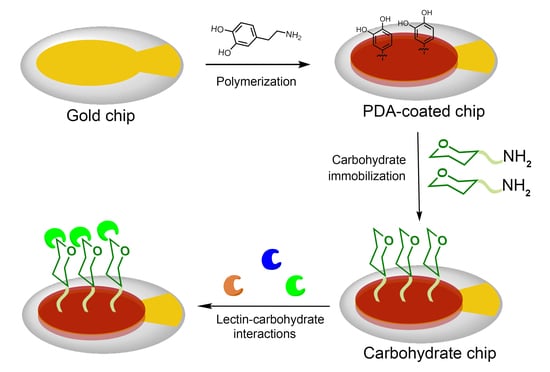
Fabrication of Carbohydrate Chips Based on Polydopamine for Real-Time Determination of Carbohydrate–Lectin Interactions by QCM Biosensor
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Gold Surface Functionalization by PDA Coating
2.3. Synthesis of Amino-Carbohydrate
2.4. Immobilization of Amino-Carbohydrate on the PDA-Coated Surface
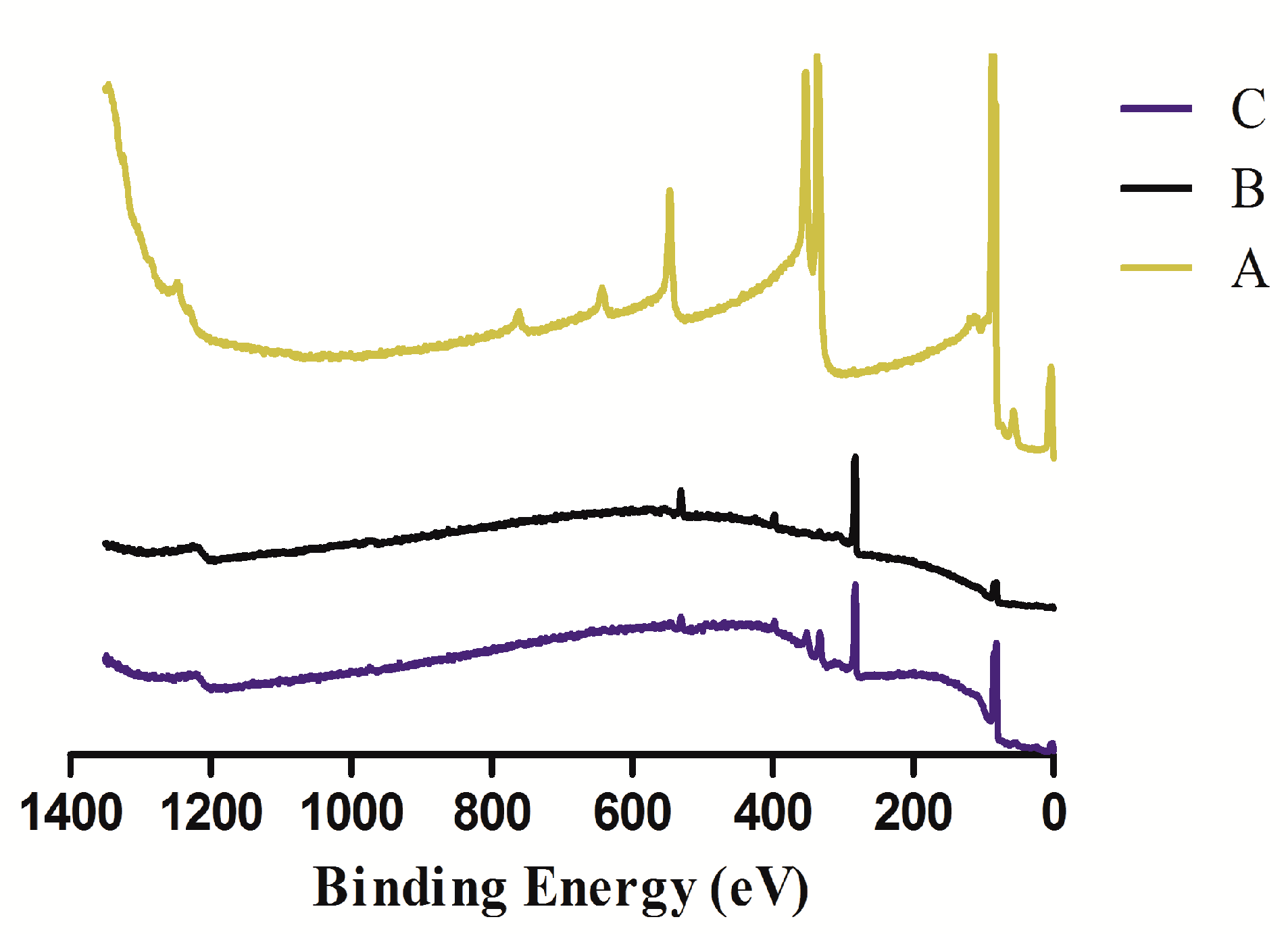
2.5. XPS Measurement
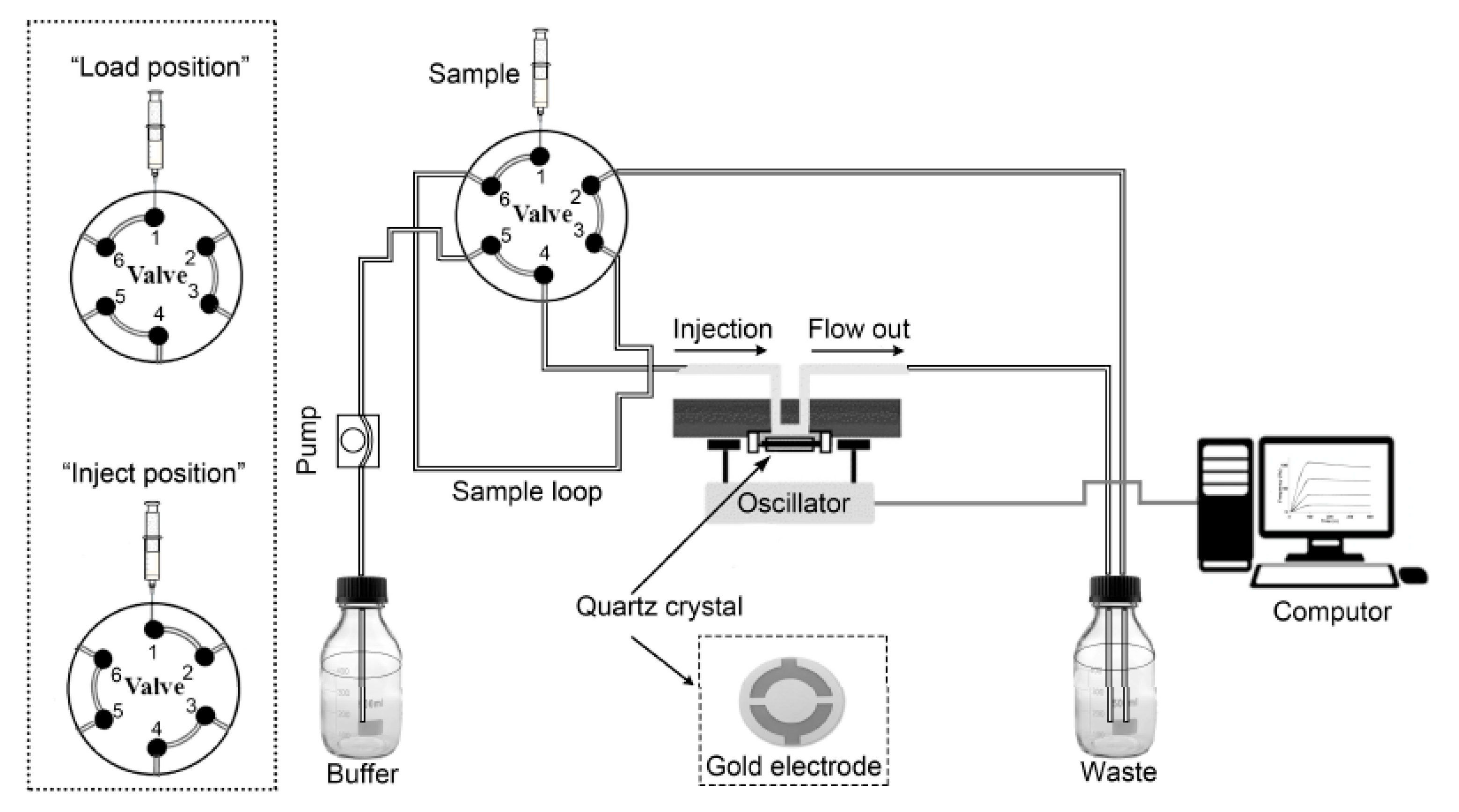
2.6. Analysis of Lectin-Carbohyhdrate Interactions by QCM
2.7. Reuse of the Sensor Chip
3. Results and Discussion
3.1. XPS Analysis of the Sensor Chip Surface
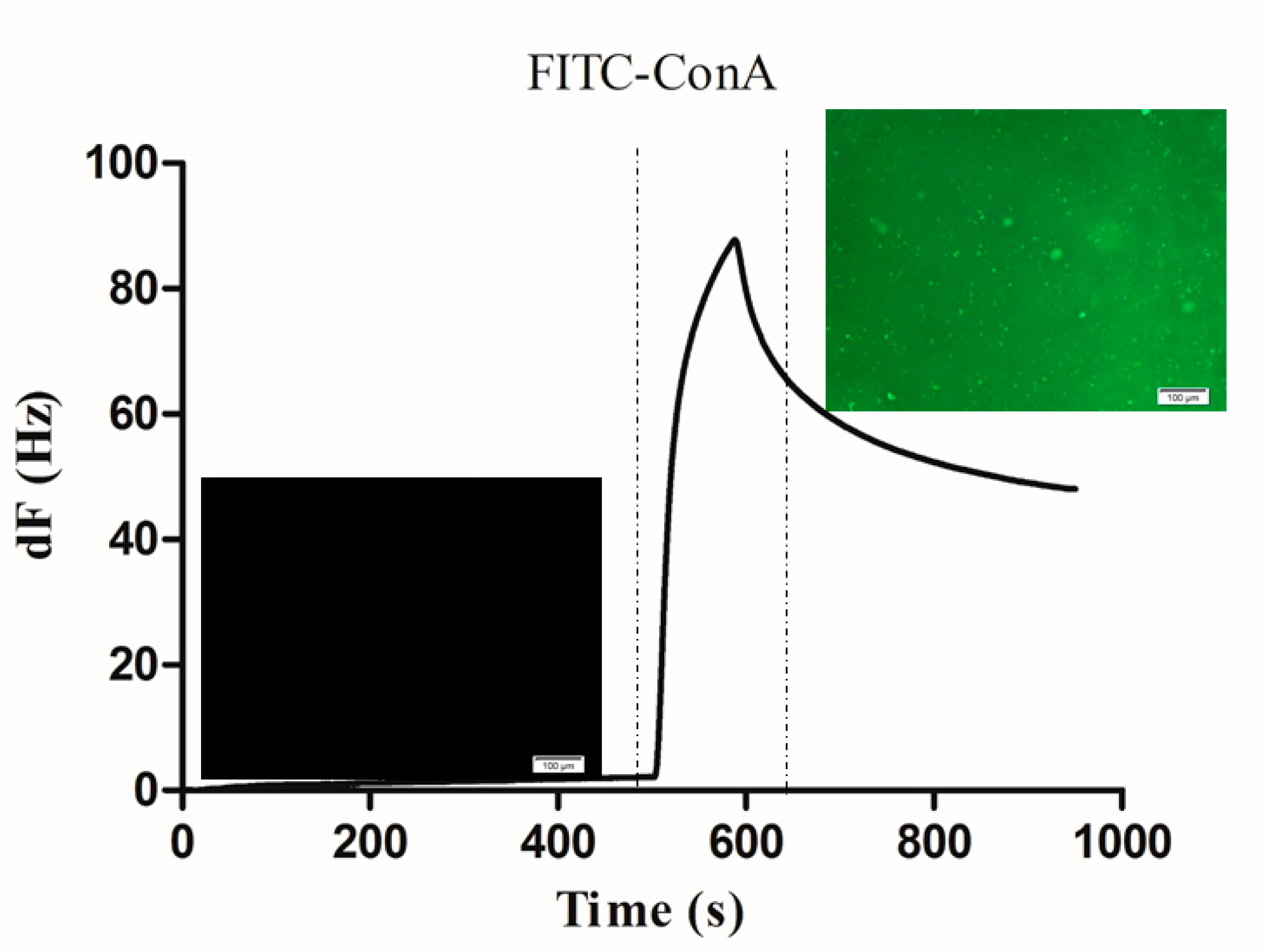
3.2. Binding of FITC-Con A on the PDA Coated Carbohydrate Chip Surface
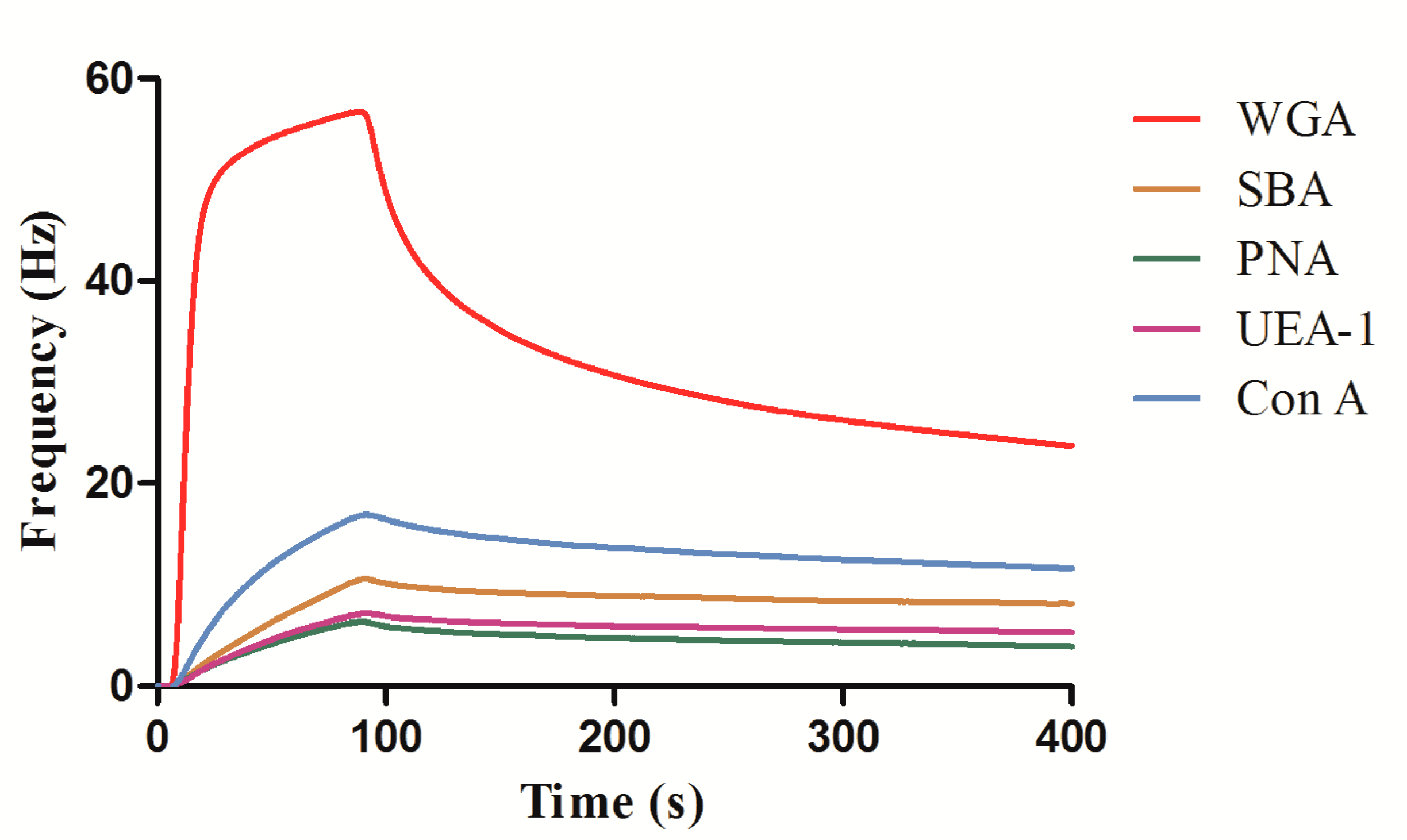
3.3. QCM Measurements of Lectin–Carbohyhdrate Interactions
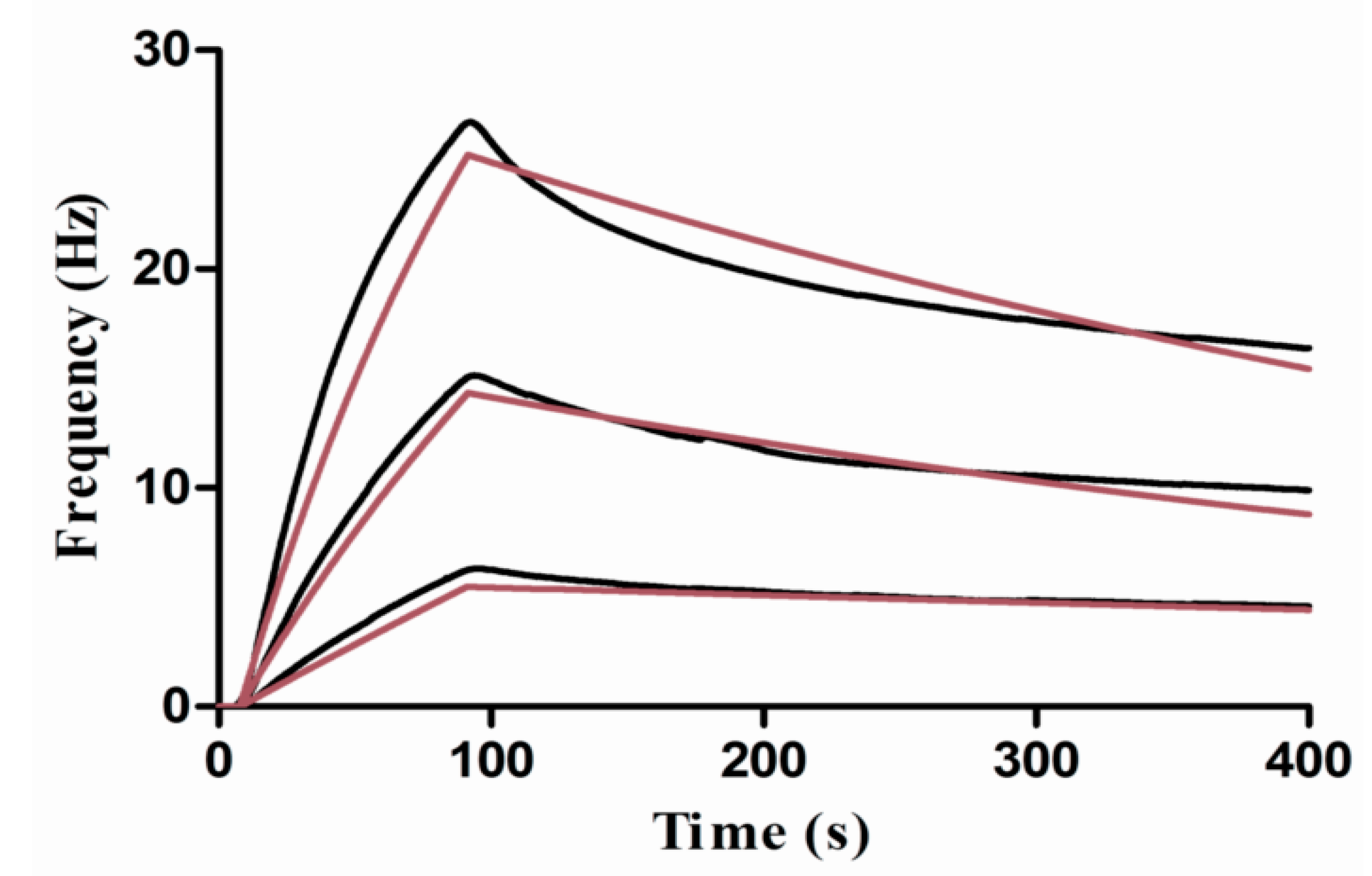
3.4. Kinetic Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Park, S.; Lee, M.R.; Pyo, S.J.; Shin, I. Carbohydrate chips for studying high-throughput carbohydrate-protein interactions. J. Am. Chem. Soc. 2004, 126, 4812–4819. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Wang, X.; Deng, L.; Ramström, O.; Yan, M. Photogenerated carbohydrate microarrays to study carbohydrate-protein interactions using surface plasmon resonance imaging. Biosens. Bioelectron. 2010, 26, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, J.; Liu, D.; Wang, Z. Studying the interaction of carbohydrate-protein on the dendrimer-modified solid support by microarray-based plasmon resonance light scattering assay. Analyst 2011, 136, 4301–4307. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Q.; Liu, D.J.; Wang, Z.X. Microarray-based study of carbohydrate-protein binding by gold nanoparticle probes. Anal. Chem. 2008, 80, 8822–8827. [Google Scholar] [CrossRef] [PubMed]
- Gruber, K.; Horlacher, T.; Castelli, R.; Mader, A.; Seeberger, P.H.; Hermann, B.A. Cantilever array sensors detect specific carbohydrate-protein interactions with picomolar sensitivity. ACS Nano 2011, 5, 3670–3678. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Hiromori, Y.; Nakamura, S.; Kawahara, K.; Fukakusa, S.; Maruno, T.; Noda, M.; Uchiyama, S.; Fukui, K.; Nishikawa, J. Structural basis for ppar gamma transactivation by endocrine-disrupting organotin compounds. Sci. Rep. 2015, 5, 8520. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Nakanishi, T.; Kami, K.; Arata, Y.; Shimada, I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 2000, 7, 220–223. [Google Scholar] [PubMed]
- Takeuchi, K.; Wagner, G. NMR studies of protein interactions. Curr. Opin. Struct. Biol. 2006, 16, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, T.; Blommers, M.J.J.; Meiler, J.; Jahnke, W.; Schupp, T.; Petersen, F.; Schinzer, D.; Altmann, K.H.; Griesinger, C. The high-resolution solution structure of epothilone a bound to tubulin: An understanding of the structure-activity relationships for a powerful class of antitumor agents. Angew. Chem. Int. Ed. 2003, 42, 2511–2515. [Google Scholar] [CrossRef] [PubMed]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug. Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Li, R.L.; Lin, C.W.; Shao, Y.L.; Chang, C.W.; Yao, F.K.; Kowal, M.D.; Wang, H.S.; Yeung, M.T.; Huang, S.C.; Kaner, R.B. Characterization of aniline tetramer by MALDI TOF mass spectrometry upon oxidative and reductive cycling. Polymers 2016, 8, 401. [Google Scholar] [CrossRef]
- Lee, C.S.; Muthusamy, A.; Abdul-Rahman, P.S.; Bhavanandan, V.P.; Hashim, O.H. An improved lectin-based method for the detection of mucin-type O-glycans in biological samples. Analyst 2013, 138, 3522–3529. [Google Scholar] [CrossRef] [PubMed]
- Funari, R.; Della, V.B.; Altucci, C.; Offenhäusser, A.; Mayer, D.; Velotta, R. Single molecule characterization of UV-activated antibodies on gold by atomic force microscopy. Langmuir 2016, 32, 8084–8091. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Aastrup, T.; Anderson, H.; Ramström, O. Redox-responsive and calcium-dependent switching of glycosyldisulfide interactions with concanavalin A. Bioorg. Med. Chem. Lett. 2005, 15, 2707–2710. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Lu, Y.; Li, X.; Cao, S.; Pei, Y.; Aastrup, T.; Pei, Z. Optimization of 3D surfaces of dextran with different molecule weights for real-time detection of biomolecular interactions by a QCM biosensor. Polymers 2017, 9, 409. [Google Scholar] [CrossRef]
- Pei, Z.; Larsson, R.; Aastrup, T.; Anderson, H.; Lehn, J.M.; Ramström, O. Quartz crystal microbalance bioaffinity sensor for rapid identification of glycosyldisulfide lectin inhibitors from a dynamic combinatorial library. Biosens. Bioelectron. 2006, 22, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, S.; Shuai, Q.; Pei, Y.; Aastrup, T.; Pei, Y.; Pei, Z. Real-time and label-free analysis of binding thermodynamics of carbohydrate-protein interactions on unfixed cancer cell surfaces using a QCM biosensor. Sci. Rep. 2015, 5, 14066. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pei, Y.; Zhang, R.; Shuai, Q.; Wang, F.; Aastrup, T.; Pei, Z. A suspension-cell biosensor for real-time determination of binding kinetics of protein-carbohydrate interactions on cancer cell surfaces. Chem. Commun. 2013, 49, 9908–9910. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Saint-Guirons, J.; Käck, C.; Ingemarsson, B.; Aastrup, T. Real-time analysis of the carbohydrates on cell surfaces using a QCM biosensor: A lectin-based approach. Biosens. Bioelectron. 2012, 35, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Yu, H.; Pei, Z.; Theurer, M.; Ammer, C.; André, S.; Gabius, H.J.; Yan, M.; Ramström, O. Photoderivatized polymer thin films at quartz crystal microbalance surfaces: Sensors for carbohydrate-protein interactions. Anal. Chem. 2007, 79, 6897–6902. [Google Scholar] [CrossRef] [PubMed]
- Norberg, O.; Lee, I.H.; Aastrup, T.; Yan, M.; Ramström, O. Photogenerated lectin sensors produced by thiol-ene/yne photo-click chemistry in aqueous solution. Biosens. Bioelectron. 2012, 34, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Song, S.; Hou, C.; Pang, S.; Li, X.; Wu, X.; Shao, C.; Pei, Y.; Pei, Z. Facile fabrication of branched-chain carbohydrate chips for studying carbohydrate-protein interactions by QCM biosensor. Chin. Chem. Lett. 2018, 29, 65–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, S.; Tang, Y.; Yu, L.; Hou, K.Y.; Cheng, J.P.; Zeng, X.; Wang, P.G. Carbohydrate-protein interactions by “clicked” carbohydrate self-assembled monolayers. Anal. Chem. 2006, 78, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, H.; Kamiya, S.; Minoura, N.; Dohi, H.; Nishida, Y.; Taguchi, K.; Yokoyama, S.; Mori, H.; Shimizu, T.; Kobayashi, K. A quartz crystal microbalance method for rapid detection and differentiation of shiga toxins by applying a monoalkyl globobioside as the toxin ligand. Biomacromolecules 2002, 3, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Anderson, H.; Aastrup, T.; Ramström, O. Study of real-time lectin-carbohydrate interactions on the surface of a quartz crystal microbalance. Biosens. Bioelectron. 2005, 21, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Seto, H.; Ogata, Y.; Murakami, T.; Yu, H.; Miura, Y. Selective protein separation using siliceous materials with a trimethoxysilane-containing glycopolymer. ACS Appl. Mater. Interfaces 2012, 4, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Norberg, O.; Deng, L.; Aastrup, T.; Yan, M.; Ramström, O. Photo-click immobilization on quartz crystal microbalance sensors for selective carbohydrate-protein interaction analyses. Anal. Chem. 2011, 83, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Norberg, O.; Deng, L.; Yan, M.; Ramström, O. Photo-click immobilization of carbohydrates on polymeric surfaces—A quick method to functionalize surfaces for biomolecular recognition studies. Bioconjug. Chem. 2009, 20, 2364–2370. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ai, K.; Lu, L. Polydopamine and its derivative materials: Synthesis and promising applications in energy, environmental, and biomedical fields. Chem. Rev. 2014, 114, 5057–5115. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Dong, Y.; Zhang, W.; Zhang, S.; Li, J. Preparation of stable superhydrophobic coatings on wood substrate surfaces via mussel-Inspired polydopamine and electroless deposition methods. Polymers 2017, 9, 218. [Google Scholar] [CrossRef]
- Luo, R.; Tang, L.; Wang, J.; Zhao, Y.; Tu, Q.; Weng, Y.; Shen, R.; Huang, N. Improved immobilization of biomolecules to quinone-rich polydopamine for efficient surface functionalization. Colloids Surf. B Biointerfaces 2013, 106, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Rho, J.; Messersmith, P.B. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Adv. Mater. 2009, 21, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Chen, D.; Hu, W. Patterning of metal films on arbitrary substrates by using polydopamine as a UV-sensitive catalytic layer for electroless deposition. Langmuir 2016, 32, 5285–5290. [Google Scholar]
- Frick, C.P.; Merkel, D.R.; Laursen, C.M.; Brinckmann, S.A.; Yakacki, C.M. Copper-coated liquid-crystalline elastomer via bioinspired polydopamine adhesion and electroless deposition. Macromol. Rapid Commun. 2016, 37, 1912–1917. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Li, X.; Song, S.; Pei, Y.; Guo, L.; Pei, Z.; Wu, C.; Li, X.; Song, S.; Pei, Y. QCM biosensor based on polydopamine surface for real-time analysis of the binding kinetics of protein-protein interactions. Polymers 2017, 9, 482. [Google Scholar] [CrossRef]
- Qiang, Z.; Akolawala, S.A.; Wang, M. Simultaneous in-film polymer synthesis and self-assembly for hierarchical nanopatterns. ACS Macro Lett. 2018, 7, 566–571. [Google Scholar] [CrossRef]
- Zhou, J.; Butchosa, N.; Jayawardena, H.S.; Zhou, Q.; Yan, M.; Ramström, O. Glycan-functionalized fluorescent chitin nanocrystals for biorecognition applications. Bioconjug. Chem. 2014, 25, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Uzawa, H.; Nagatsuka, T.; Kondo, S.; Sato, K.; Ohsawa, I.; Kanamorikataoka, M.; Takei, Y.; Ota, S.; Furuno, M. Preparation and evaluation of lactose-modified monoliths for the adsorption and decontamination of plant toxins and lectins. Carbohydr. Res. 2011, 346, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Otman, O.; Boullanger, P.; Lafont, D.; Hamaide, T. New amphiphilic glycopolymers based on a polycaprolactone-maleic anhydride copolymer backbone: characterization by 15N NMR and application to colloidal stabilization of nanoparticles. Macromol. Chem. Phys. 2008, 209, 2410–2422. [Google Scholar] [CrossRef]
- Del Frari, D.; Bour, J.; Ball, V.; Toniazzo, V.; Ruch, D. Degradation of polydopamine coatings by sodiumhypochlorite: A process depending on the substrate and the film synthesis method. Polym. Degrad. Stab. 2012, 97, 1844–1849. [Google Scholar] [CrossRef]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.; Creavin, A.; O’Connell, M.; O’Connor, B.; Clarke, P. Optimization of the enzyme-linked lectin assay for enhanced glycoprotein and glycoconjugate analysis. Anal. Biochem. 2011, 413, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Lienemann, M.; Paananen, A.; Boer, H.; de la Fuente, J.M.; García, I.; Penadés, S.; Koivula, A. Characterization of the wheat germ agglutinin binding to self-assembled monolayers of neoglycoconjugates by AFM and SPR. Glycobiology 2009, 19, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Vila-Perelló, M.; Gutiérrez Gallego, R.; Andreu, D. A simple approach to well-defined sugar-coated surfaces for interaction studies. ChemBioChem 2005, 6, 1831–1838. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, K.; Song, S.; Cheng, Y.; Guo, L.; Pei, Y.; Lv, X.; Aastrup, T.; Pei, Z. Fabrication of Carbohydrate Chips Based on Polydopamine for Real-Time Determination of Carbohydrate–Lectin Interactions by QCM Biosensor. Polymers 2018, 10, 1275. https://doi.org/10.3390/polym10111275
Shang K, Song S, Cheng Y, Guo L, Pei Y, Lv X, Aastrup T, Pei Z. Fabrication of Carbohydrate Chips Based on Polydopamine for Real-Time Determination of Carbohydrate–Lectin Interactions by QCM Biosensor. Polymers. 2018; 10(11):1275. https://doi.org/10.3390/polym10111275
Chicago/Turabian StyleShang, Kun, Siyu Song, Yaping Cheng, Lili Guo, Yuxin Pei, Xiaomeng Lv, Teodor Aastrup, and Zhichao Pei. 2018. "Fabrication of Carbohydrate Chips Based on Polydopamine for Real-Time Determination of Carbohydrate–Lectin Interactions by QCM Biosensor" Polymers 10, no. 11: 1275. https://doi.org/10.3390/polym10111275
APA StyleShang, K., Song, S., Cheng, Y., Guo, L., Pei, Y., Lv, X., Aastrup, T., & Pei, Z. (2018). Fabrication of Carbohydrate Chips Based on Polydopamine for Real-Time Determination of Carbohydrate–Lectin Interactions by QCM Biosensor. Polymers, 10(11), 1275. https://doi.org/10.3390/polym10111275