Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level
Abstract
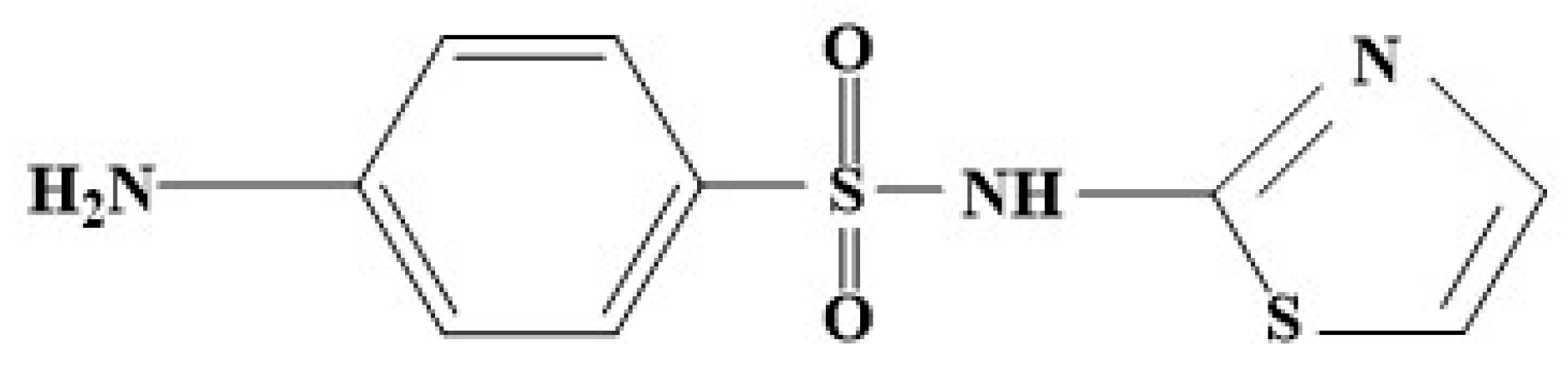
1. Introduction
2. Methods
3. Results and Discussion
3.1. Gibbs Free Energy Calculations
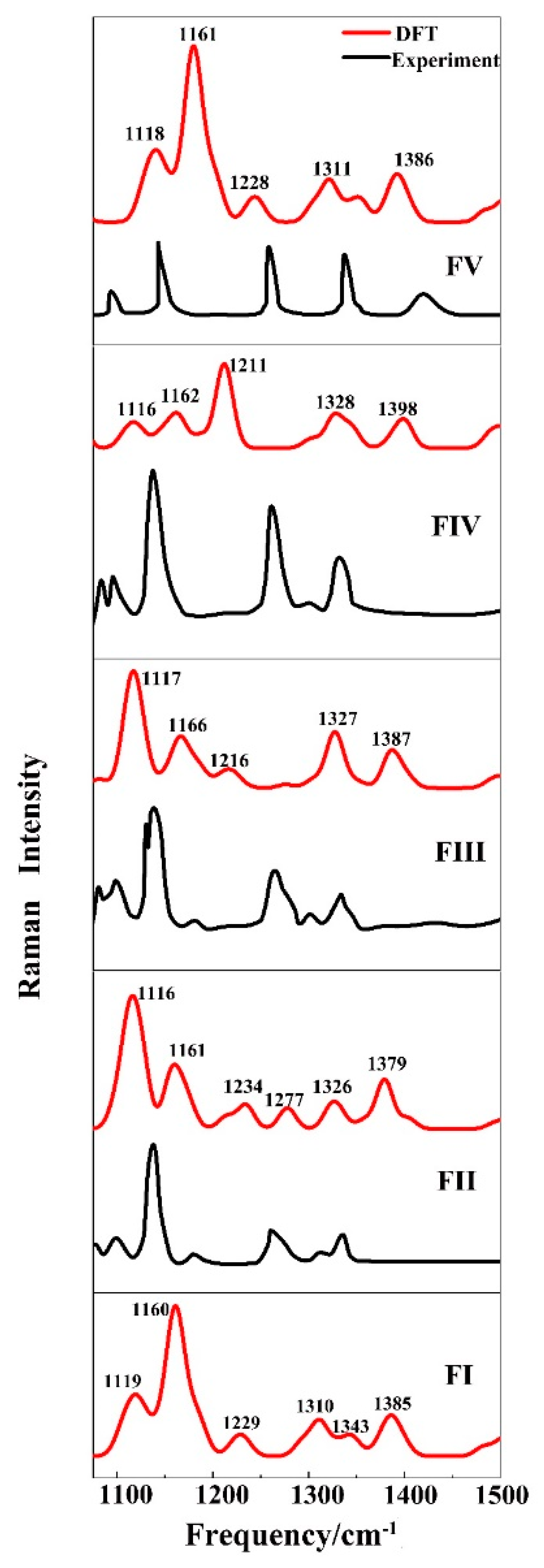
3.2. Vibrational Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Craven, R.J.; Lencki, R.W. Polymorphism of Acylglycerols: A Stereochemical Perspective. Chem. Rev. 2013, 113, 7402–7420. [Google Scholar] [CrossRef]
- Cruz-Cabeza, A.J.; Bernstein, J. Conformational Polymorphism. Chem. Rev. 2014, 114, 2170–2191. [Google Scholar] [CrossRef]
- Neumann, M.A.; de Streek, J.V.; Fabbiani, F.P.A.; Hidber, P.; Grassmann, O. Combined crystal structure prediction and high-pressure crystallization in rational pharmaceutical polymorph screening. Nat. Commun. 2015, 6, 7. [Google Scholar] [CrossRef]
- Hoser, A.A.; Sovago, I.; Lanzac, A.; Madsen, A.O. A crystal structure prediction enigma solved: The gallic acid monohydrate system—Surprises at 10 K. Chem. Commun. 2017, 53, 925–928. [Google Scholar] [CrossRef]
- Terayama, K.; Yamashita, T.; Oguchi, T.; Tsuda, K. Fine-grained optimization method for crystal structure prediction. NPJ Comput. Mater. 2018, 4, 8. [Google Scholar] [CrossRef]
- Ryan, K.; Lengyel, J.; Shatruk, M. Crystal Structure Prediction via Deep Learning. J. Am. Chem. Soc. 2018, 140, 10158–10168. [Google Scholar] [CrossRef] [PubMed]
- Mitchell-Koch, K.R.; Matzger, A.J. Evaluating computational predictions of the relative stabilities of polymorphic pharmaceuticals. J. Pharm. Sci. 2008, 97, 2121–2129. [Google Scholar] [CrossRef]
- Drebushchak, T.N.; Boldyreva, E.V.; Mikhailenko, M.A. Crystal structures of sulfathiazole polymorphs in the temperature range 100-295 K: A comparative analysis. J. Struct. Chem. 2008, 49, 84–94. [Google Scholar] [CrossRef]
- Abramov, Y.A. QTAIM Application in Drug Development: Prediction of Relative Stability of Drug Polymorphs from Experimental Crystal Structures. J. Phys. Chem. A 2011, 115, 12809–12817. [Google Scholar] [CrossRef]
- Sovago, I.; Gutmann, M.J.; Hill, J.G.; Senn, H.M.; Thomas, L.H.; Wilson, C.C.; Farrugia, L.J. Experimental Electron Density and Neutron Diffraction Studies on the Polymorphs of Sulfathiazole. Cryst. Growth Des. 2014, 14, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Shtukenberg, A.G.; Zhu, Q.; Carter, D.J.; Vogt, L.; Hoja, J.; Schneider, E.; Song, H.; Pokroy, B.; Polishchuk, I.; Tkatchenko, A.; et al. Powder diffraction and crystal structure prediction identify four new coumarin polymorphs. Chem. Sci. 2017, 8, 4926–4940. [Google Scholar] [CrossRef]
- Price, S.L. Predicting crystal structures of organic compounds. Chem. Soc. Rev. 2014, 43, 2098–2111. [Google Scholar] [CrossRef]
- Price, S.L.; Braun, D.E.; Reutzel-Edens, S.M. Can computed crystal energy landscapes help understand pharmaceutical solids? Chem. Commun. 2016, 52, 7065–7077. [Google Scholar] [CrossRef]
- Li, J.J.; Sode, O.; Voth, G.A.; Hirata, S. A solid-solid phase transition in carbon dioxide at high pressures and intermediate temperatures. Nat. Commun. 2013, 4, 141–155. [Google Scholar] [CrossRef]
- Li, J.J.; Sode, O.; Hirata, S. Second-Order Many-Body Perturbation Study on Thermal Expansion of Solid Carbon Dioxide. J. Chem. Theory Comput. 2015, 11, 224–229. [Google Scholar] [CrossRef]
- Bingham, A.L.; Hughes, D.S.; Hursthouse, M.B.; Lancaster, R.W.; Tavener, S.; Threlfall, T.L. Over one hundred solvates of sulfathiazole. Chem. Commun. 2001, 7, 603–604. [Google Scholar] [CrossRef]
- Gelbrich, T.; Hughes, D.S.; Hursthouse, M.B.; Threlfall, T.L. Packing similarity in polymorphs of sulfathiazole. CrystEngComm 2008, 10, 1328–1334. [Google Scholar] [CrossRef]
- Parmar, M.M.; Khan, O.; Seton, L.; Ford, J.L. Polymorph selection with morphology control using solvents. Cryst. Growth Des. 2007, 7, 1635–1642. [Google Scholar] [CrossRef]
- Lee, I.S.; Lee, A.Y.; Myerson, A.S. Concomitant polymorphism in confined environment. Pharm. Res. 2008, 25, 960–968. [Google Scholar] [CrossRef]
- Munroe, A.; Rasmuson, A.C.; Hodnett, B.K.; Croker, D.M. Relative Stabilities of the Five Polymorphs of Sulfathiazole. Cryst. Growth Des. 2012, 12, 2825–2835. [Google Scholar] [CrossRef]
- Kamiya, M.; Hirata, S.; Valiev, M. Fast electron correlation methods for molecular clusters without basis set superposition errors. J. Chem. Phys. 2008, 128, 074103. [Google Scholar] [CrossRef]
- Sode, O.; Hirata, S. Second-order many-body perturbation study of solid hydrogen fluoride under pressure. Phys. Chem. Chem. Phys. 2012, 14, 7765–7779. [Google Scholar] [CrossRef]
- Beran, G.J.O. Modeling Polymorphic Molecular Crystals with Electronic Structure Theory. Chem. Rev. 2016, 116, 5567–5613. [Google Scholar] [CrossRef]
- Dennis, J.E., Jr.; More, J.J. Quasi-Newton methods, motivation and theory. SIAM Rev. 1977, 19, 46–89. [Google Scholar] [CrossRef]
- Head, J.D.; Zerner, M.C. A Broyden-Fletcher-Goldfarb-Shanno optimization procedure for molecular geometries. Chem. Phys. Lett. 1985, 122, 264–270. [Google Scholar] [CrossRef]
- Blagden, N.; Davey, R.J.; Lieberman, H.F.; Williams, L.; Payne, R.; Roberts, R.; Rowe, R.; Docherty, R. Crystal chemistry and solvent effects in polymorphic systems sulfathiazole. J. Chem. Soc.-Faraday Trans. 1998, 94, 1035–1044. [Google Scholar] [CrossRef]
- Chan, F.C.; Anwar, J.; Cernik, R.; Barnes, P.; Wilson, R.M. Ab initio structure determination of sulfathiazole polymorph V from synchrotron X-ray powder diffraction data. J. Appl. Crystallogr. 1999, 32, 436–441. [Google Scholar] [CrossRef]
- Davey, R.J.; Allen, K.; Blagden, N.; Cross, W.I.; Lieberman, H.F.; Quayle, M.J.; Righini, S.; Seton, L.; Tiddy, G.J.T. Crystal engineering—Nucleation, the key step. CrystEngComm 2002, 4, 257–264. [Google Scholar] [CrossRef]
- He, X.; Sode, O.; Xantheas, S.S.; Hirata, S. Second-order many-body perturbation study of ice Ih. J. Chem. Phys. 2012, 137, 204505. [Google Scholar] [CrossRef]
- He, X.; Zhu, T.; Wang, X.W.; Liu, J.F.; Zhang, J.Z.H. Fragment Quantum Mechanical Calculation of Proteins and Its Applications. Accounts Chem. Res. 2014, 47, 2748–2757. [Google Scholar] [CrossRef]
- Hirata, S.; Gilliard, K.; He, X.; Li, J.J.; Sode, O. Ab Initio Molecular Crystal Structures, Spectra, and Phase Diagrams. Accounts Chem. Res. 2014, 47, 2721–2730. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Qi, L.W.; Zhang, J.Z.H.; He, X. Fragment Quantum Mechanical Method for Large-Sized Ion-Water Clusters. J. Chem. Theory Comput. 2017, 13, 2021–2034. [Google Scholar] [CrossRef]
- Liu, J.F.; He, X. Accurate prediction of energetic properties of ionic liquid clusters using a fragment-based quantum mechanical method. Phys. Chem. Chem. Phys. 2017, 19, 20657–20666. [Google Scholar] [CrossRef]
- Liu, J.F.; He, X.; Zhang, J.Z.H. Structure of liquid water—A dynamical mixture of tetrahedral and ‘ring-and-chain’ like structures. Phys. Chem. Chem. Phys. 2017, 19, 11931–11936. [Google Scholar] [CrossRef]
- Liu, J.F.; He, X.; Zhang, J.Z.H.; Qi, L.W. Hydrogen-bond structure dynamics in bulk water: Insights from ab initio simulations with coupled cluster theory. Chem. Sci. 2018, 9, 2065–2073. [Google Scholar] [CrossRef]
- Liu, J.F.; Zhang, J.Z.H.; He, X. Probing the Ion-Specific Effects at the Water/Air Interface and Water-Mediated Ion Pairing in Sodium Halide Solution with Ab Initio Molecular Dynamics. J. Phys. Chem. B 2018, 122, 10202–10209. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef]
- Khoshkhoo, S.; Anwar, J. Crystallization of polymorphs: The effect of solvent. J. Phys. D Appl. Phys. 1993, 26, B90–B93. [Google Scholar] [CrossRef]
- Ueno, S.; Nishida, T.; Sato, K. Synchrotron radiation microbeam X-ray analysis of microstructures and the polymorphic transformation of spherulite crystals of trilaurin. Cryst. Growth Des. 2008, 8, 751–754. [Google Scholar] [CrossRef]
- Hedoux, A.; Guinet, Y.; Descamps, M. The contribution of Raman spectroscopy to the analysis of phase transformations in pharmaceutical compounds. Int. J. Pharm. 2011, 417, 17–31. [Google Scholar] [CrossRef]
- Van Eerdenbrugh, B.; Taylor, L.S. Application of mid-IR spectroscopy for the characterization of pharmaceutical systems. Int. J. Pharm. 2011, 417, 3–16. [Google Scholar] [CrossRef]
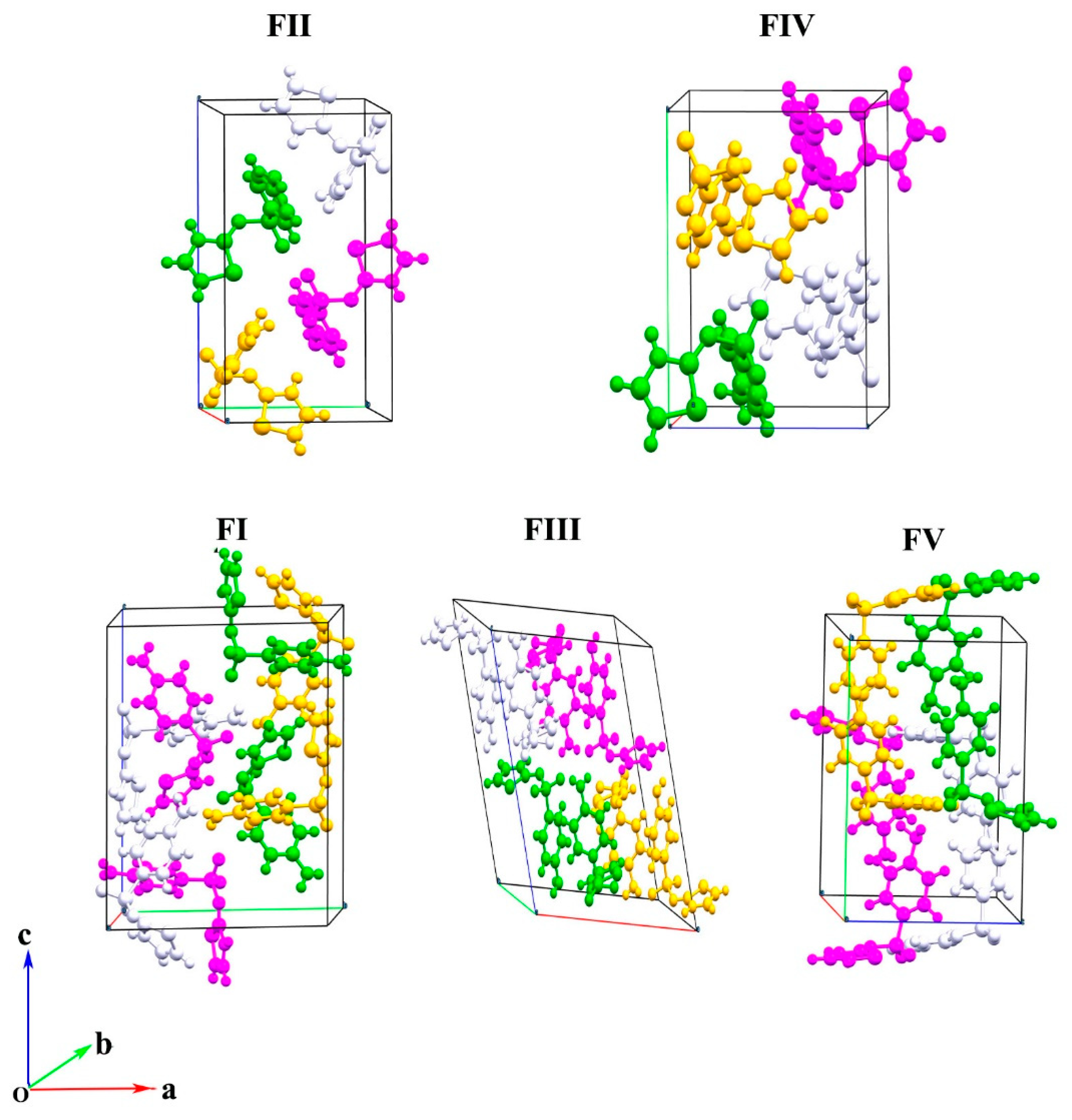

| Polymorph | FI | FII | FIII | FIV | FV |
|---|---|---|---|---|---|
| a (Å) | 10.554 | 8.235 | 17.570 | 10.867 | 14.330 |
| b (Å) | 13.220 | 8.550 | 8.574 | 8.543 | 15.273 |
| c (Å) | 17.050 | 15.558 | 15.583 | 11.456 | 10.443 |
| α (degree) | 90 | 90 | 90 | 90 | 90 |
| β (degree) | 108.06 | 93.67 | 112.93 | 88.13 | 91.05 |
| γ (degree) | 90 | 90 | 90 | 90 | 90 |
| Space group | P21/c | P21/c | P21/c | P21/n | P21/n |
| Cell volume(Å3) | 2261.68 | 1093.18 | 2162.01 | 1062.97 | 2285.13 |
| CCDC ref code | SUTHAZ01 | SUTHAZ | SUTHAZ02 | SUTHAZ04 | SUTHAZ06 |
| Parameters | FI | FII | FIII | FIV | FV | |
|---|---|---|---|---|---|---|
| a/Å | Exp. | 10.554 | 8235 | 17.570 | 10.867 | 14.330 |
| Cal. | 10.355 | 8119 | 17.333 | 10.643 | 14.096 | |
| b/Å | Exp. | 13.220 | 8550 | 8574 | 8543 | 15.273 |
| Cal. | 13.185 | 8419 | 8306 | 8251 | 14.854 | |
| c/Å | Exp. | 17.050 | 15.558 | 15.583 | 11.456 | 10.443 |
| Cal. | 16.842 | 15.472 | 15.550 | 11.398 | 10.319 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, X.; Liu, J.; Luo, H.; Han, Y.; Hu, W.; Liu, J.; Li, J.; He, X. Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level. Crystals 2019, 9, 256. https://doi.org/10.3390/cryst9050256
Hao X, Liu J, Luo H, Han Y, Hu W, Liu J, Li J, He X. Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level. Crystals. 2019; 9(5):256. https://doi.org/10.3390/cryst9050256
Chicago/Turabian StyleHao, Xuan, Jinfeng Liu, Hongyuan Luo, Yanqiang Han, Wenxin Hu, Jinyun Liu, Jinjin Li, and Xiao He. 2019. "Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level" Crystals 9, no. 5: 256. https://doi.org/10.3390/cryst9050256
APA StyleHao, X., Liu, J., Luo, H., Han, Y., Hu, W., Liu, J., Li, J., & He, X. (2019). Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level. Crystals, 9(5), 256. https://doi.org/10.3390/cryst9050256







