Optimization of Vapor Diffusion Conditions for Anti-CD20 Crystallization and Scale-Up to Meso Batch
Abstract
1. Introduction
2. Materials and Methods
2.1. Anti-CD20 Crystallization Experiments
2.2. Analysis of Anti-CD20 Crystals
2.3. Mass Spectroscopy
2.4. X-ray Diffraction Analysis
3. Results
3.1. Anti-CD20 Crystallization and Crystal Analysis
3.2. Crystallographic Analysis of Anti-CD20 Crystals
3.3. Crystal Morphological Analysis
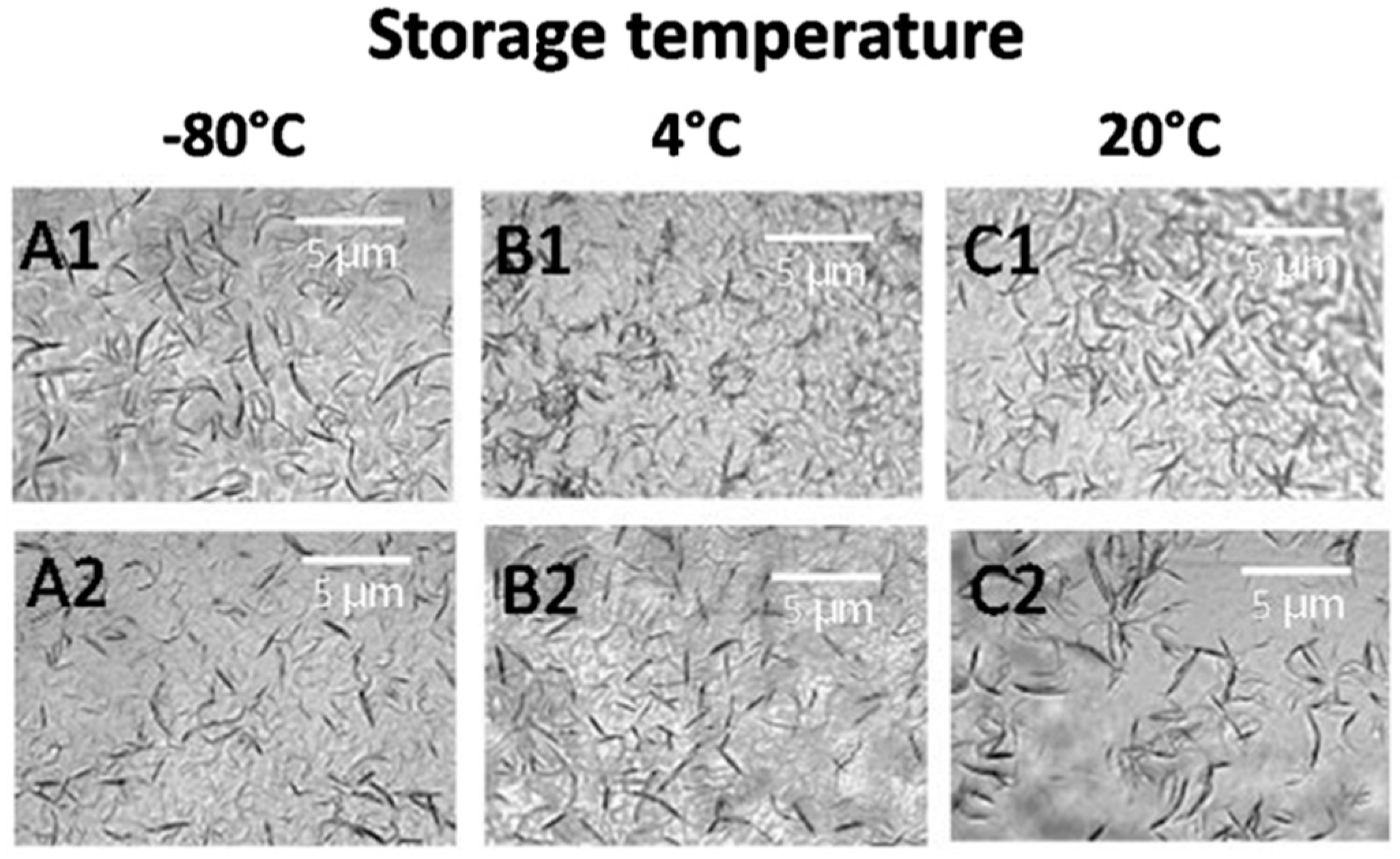
3.4. Effect of Storage Conditions on Anti-CD20 Crystallization
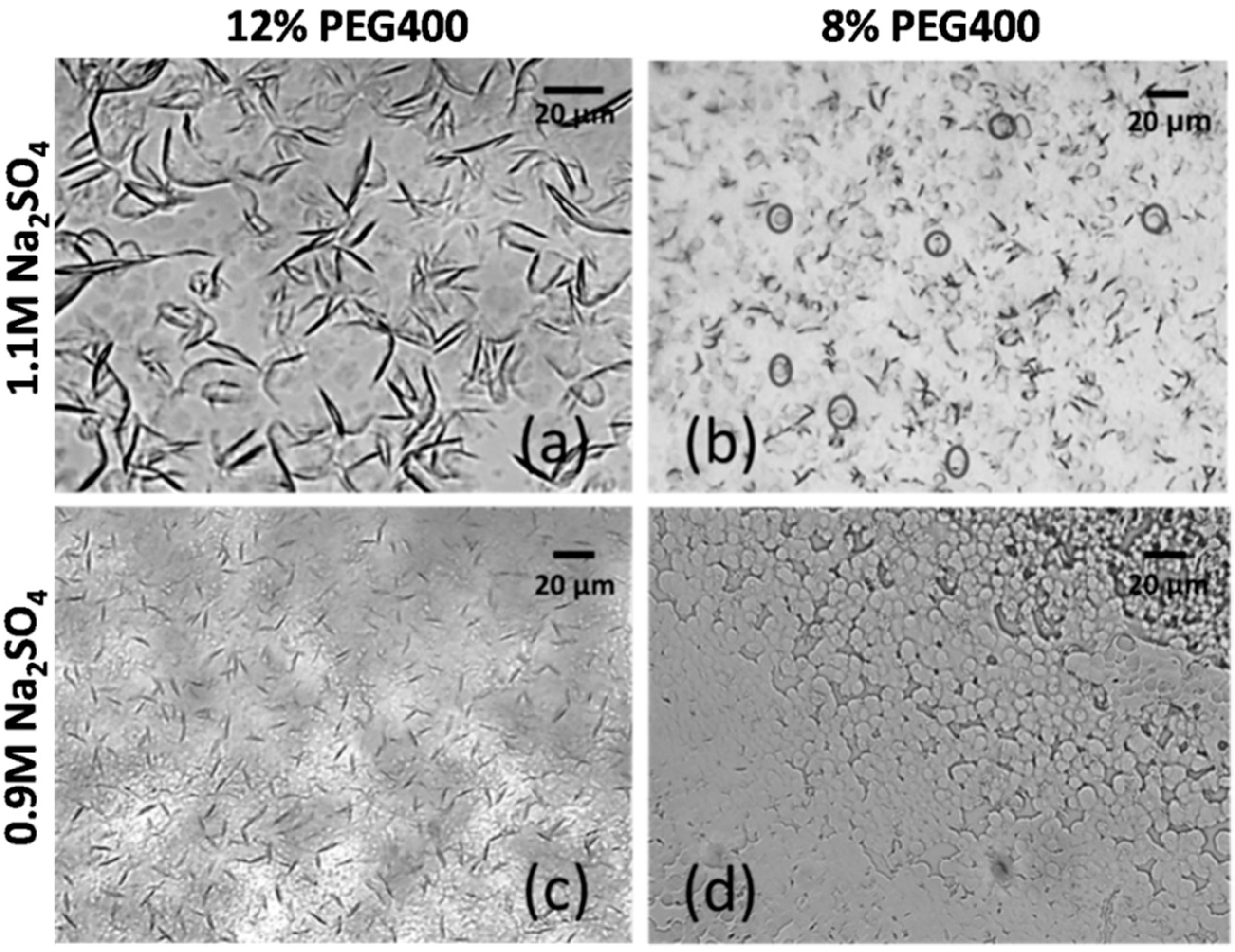
3.5. Influence of Ions on mAb Crystallization
3.6. Meso Batch Crystallization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Urquhart, L. Market watch: Top drugs and companies by sales in 2017. Nat. Rev. Drug Discovery 2018, 232, 17. [Google Scholar] [CrossRef] [PubMed]
- Kelley, B. Industrialization of mAb production technology the bioprocessing industry at a crossroads. Mabs 2009, 1, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Pagkaliwangan, M.; Gjoka, X.; Davidovits, T.; Stock, R.; Ransohoff, T.; Gantier, R.; Schofield, M. Modeling the Downstream Processing of Monoclonal Antibodies Reveals Cost Advantages for Continuous Methods for a Broad Range of Manufacturing Scales. Biotechnol. J. 2018, 14, 1700665. [Google Scholar] [CrossRef] [PubMed]
- Soetaert, W.; Vandamme, E. The impact of industrial biotechnology. Biotechnol. J. 2006, 1, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rasmuson, Å.C. Investigation of batch cooling crystallization in a liquid–liquid separating system by PAT. Organ. Process Res. Develop. 2012, 16, 1212–1224. [Google Scholar] [CrossRef]
- Yang, H.; Chen, W.; Peczulis, P.; Heng, J.Y.Y. Development and Workflow of a Continuous Protein Crystallization Process: A Case of Lysozyme. Cryst. Growth Des. 2019, 19, 983–991. [Google Scholar] [CrossRef]
- Govada, L.; Chayen, N.E. Choosing the Method of Crystallization to Obtain Optimal Results. Crystals 2019, 9, 106. [Google Scholar] [CrossRef]
- Nanev, C.N. Peculiarities of Protein Crystal Nucleation and Growth. Crystals 2018, 8, 422. [Google Scholar] [CrossRef]
- Nanev, C.N. Recent Insights into Protein Crystal Nucleation. Crystals 2018, 8, 219. [Google Scholar] [CrossRef]
- Koizumi, H.; Uda, S.; Fujiwara, K.; Okada, J.; Nozawa, J. Effect of an External Electric Field on the Kinetics of Dislocation-Free Growth of Tetragonal Hen Egg White Lysozyme Crystals. Crystals 2017, 7, 170. [Google Scholar] [CrossRef]
- Hekmat, D. Large-scale crystallization of proteins for purification and formulation. Bioprocess Biosyst. Eng. 2015, 38, 1209–1231. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Larson, S.B.; Hasel, K.W.; McPherson, A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 1997, 36, 1581–1597. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Skaletsky, E.; McPherson, A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 1998, 275, 861–872. [Google Scholar] [CrossRef]
- Saphire, E.O.; Parren, P.W.H.I.; Barbas, C.F.; Burton, D.R.; Wilson, I.A. Crystallization and preliminary structure determination of an intact human immunoglobulin, b12: An antibody that broadly neutralizes primary isolates of HIV-1. Acta Crystallogr. D 2001, 57, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Saphire, E.O.; Parren, P.W.H.I.; Pantophlet, R.; Zwick, M.B.; Morris, G.M.; Rudd, P.M.; Dwek, R.A.; Stanfield, R.L.; Burton, D.R.; Wilson, I.A. Crystal structure of a neutralizing human IgG against HIV-1: A template for vaccine design. Science 2001, 293, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Wendling, J.; He, F.; Trilisky, E.; Stevenson, R.; Franey, H.; Kinderman, F.; Li, G.; Piedmonte, D.M.; Osslund, T.; et al. In Vivo Crystallization of Human IgG in the Endoplasmic Reticulum of Engineered Chinese Hamster Ovary (CHO) Cells. J. Biol. Chem. 2011, 286, 19917–19931. [Google Scholar] [CrossRef] [PubMed]
- Smejkal, B.; Agrawal, N.J.; Helk, B.; Schulz, H.; Giffard, M.; Mechelke, M.; Ortner, F.; Heckmeier, P.; Trout, B.L.; Hekmat, D. Fast and Scalable Purification of a Therapeutic Full-Length Antibody Based on Process Crystallization. Biotech. Bioeng. 2013, 110, 2452–2461. [Google Scholar] [CrossRef]
- Smith, M.R. Rituximab (monoclonal anti-CD20 antibody): Mechanisms of action and resistance. Oncogene 2003, 22, 7359–7368. [Google Scholar] [CrossRef]
- Appelbaum, F. Treatment of aggressive non-Hodgkin’s lymphoma with marrow transplantation. Marrow Transplant. Rev. 1993, 3, 1993–1996. [Google Scholar]
- Pescovitz, M.D. Rituximab, an anti-CD20 monoclonal antibody: History and mechanism of action. Am. J. Transplant. 2006, 6, 859–866. [Google Scholar] [CrossRef]
- Kelley, B. Very large scale monoclonal antibody purification: The case for conventional unit operations. Biotechnol. Progr. 2007, 23, 995–1008. [Google Scholar] [CrossRef]
- Utturkar, A.; Gillette, K.; Sun, C.Y.; Pagkaliwangan, M.; Quesenberry, R.; Schofield, M. A Direct Approach for Process Development Using Single Column Experiments Results in Predictable Streamlined Multi-Column Chromatography Bioprocesses. Biotechnol. J. 2019, 14, 1800243. [Google Scholar] [CrossRef]
- Grilo, A.L.; Mateus, M.; Aires-Barros, M.R.; Azevedo, A.M. Monoclonal Antibodies Production Platforms: An Opportunity Study of a Non-Protein-A Chromatographic Platform Based on Process Economics. Biotechnol. J. 2017, 12, 1700260. [Google Scholar] [CrossRef]
- Franzreb, M.; Müller, E.; Vajda, J. Cost estimation for protein A chromatography. Bioprocess Int. 2014, 12, 44–52. [Google Scholar]
- Yang, M.X.; Shenoy, B.; Disttler, M.; Patel, R.; McGrath, M.; Pechenov, S.; Margolin, A.L. Crystalline monoclonal antibodies for subcutaneous delivery. Proc. Natl. Acad. Sci. USA 2003, 100, 6934–6939. [Google Scholar] [CrossRef]
- Wilkins, J.A.; Oshodi, S.A.; Lobo, B.; Breece, T. Crystallization of Anti-cd20 Antibodies. U.S. Patent US20110020322A1, 27 January 2011. [Google Scholar]
- Basham, M.; Filik, J.; Wharmby, M.T.; Chang, P.C.; El Kassaby, B.; Gerring, M.; Aishima, J.; Levik, K.; Pulford, B.C.; Sikharulidze, I. Data analysis workbench (DAWN). J. Synchrotron Rad. 2015, 22, 853–858. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0-new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Caliandro, R.; Belviso, D.B. RootProf: Software for multivariate analysis of unidimensional profiles. J. Appl. Crystallogr. 2014, 47, 1087–1096. [Google Scholar] [CrossRef]
- Shenoy, B.; Govardhan, C.P.; Yang, M.X.; Margolin, A.L. Crystals of whole Antibodies and Fragments Thereof and Methods for Making and Using Them. U.S. Patent US7833525B2, 16 November 2010. [Google Scholar]
- Scapin, G.; Yang, X.Y.; Prosise, W.W.; Mccoy, M.; Reichert, P.; Johnston, J.M.; Kashi, R.S.; Strickland, C. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab. Nat. Struct. Mol. Biol. 2015, 22, 953–958. [Google Scholar] [CrossRef]
- Voak, D.; Sacks, S.; Alderson, T.; Takei, F.; Lennox, E.; Jarvis, J.; Milstein, C.; Darnborough, J. Monoclonal anti-A from a hybrid-myeloma: evaluation as a blood grouping reagent. Vox Sanguinis 1980, 39, 134–140. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Janis, L.J. Influence of pH, buffer species, and storage temperature on physicochemical stability of a humanized monoclonal antibody LA298. Int. J. Pharm. 2006, 308, 46–51. [Google Scholar] [CrossRef]
- Wang, W.; Nema, S.; Teagarden, D. Protein aggregation-Pathways and influencing factors. Int. J. Pharm. 2010, 390, 89–99. [Google Scholar] [CrossRef]
- Ng, J.D.; Lorber, B.; Witz, J.; TheobaldDietrich, A.; Kern, D.; Giege, R. The crystallization of biological macromolecules from precipitates: Evidence for Ostwald ripening. J. Cryst. Growth 1996, 168, 50–62. [Google Scholar] [CrossRef]
- Mullin, J.W. Crystallization; Butterworth Heinemann: Oxford, UK, 2001. [Google Scholar]
- Biscans, B.; Laguerie, C. Determination of Induction Time of Lysozyme Crystals by Laser Diffraction. J. Phys. D Appl. Phys. 1993, 26, B118–B122. [Google Scholar] [CrossRef]
- EPA. Estimation Programs Interface Suite™ for Microsoft® Windows, V 4.11; United States Environmental Protection Agency: Washington, DC, USA, 2012.
- Arakawa, T.; Timasheff, S.N. Mechanism of Poly(Ethylene Glycol) Interaction with Proteins. Biochemistry 1985, 24, 6756–6762. [Google Scholar] [CrossRef]
- Timasheff, S.N. Protein-solvent preferential interactions, protein hydration, and the modulation of biochemical reactions by solvent components. Proc. Natl. Acad. Sci. USA 2002, 99, 9721–9726. [Google Scholar] [CrossRef]
- Hildebrandt, C.; Mathaes, R.; Saedler, R.; Winter, G. Origin of Aggregate Formation in Antibody Crystal Suspensions Containing PEG. J. Pharm. Sci. 2016, 105, 1059–1065. [Google Scholar] [CrossRef]
- Rakel, N.; Galm, L.; Bauer, K.C.; Hubbuch, J. Influence of Macromolecular Precipitants on Phase Behavior of Monoclonal Antibodies. Biotechnol. Progr. 2015, 31, 145–153. [Google Scholar] [CrossRef]
- Ten Wolde, P.R.; Frenkel, D. Enhancement of protein crystal nucleation by critical density fluctuations. Science 1997, 277, 1975–1978. [Google Scholar] [CrossRef]
- Hagewiesche, A.M.C.; Fukami, J.; Cromwell, M.E.; Dinges, R.B. Crystallization of Antibody or Fragment Thereof. U.S. Patent US8008447B2, 30 August 2011. [Google Scholar]
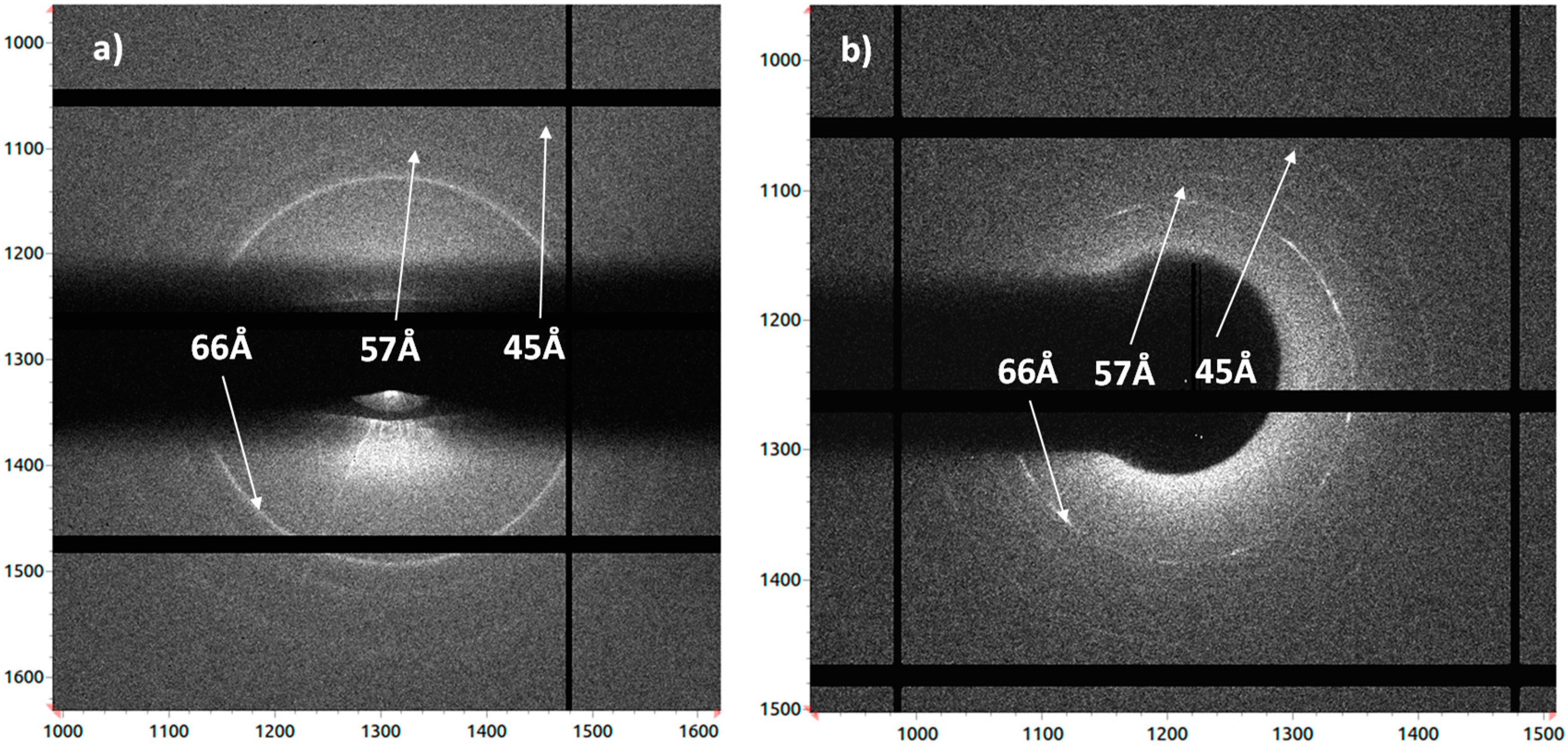
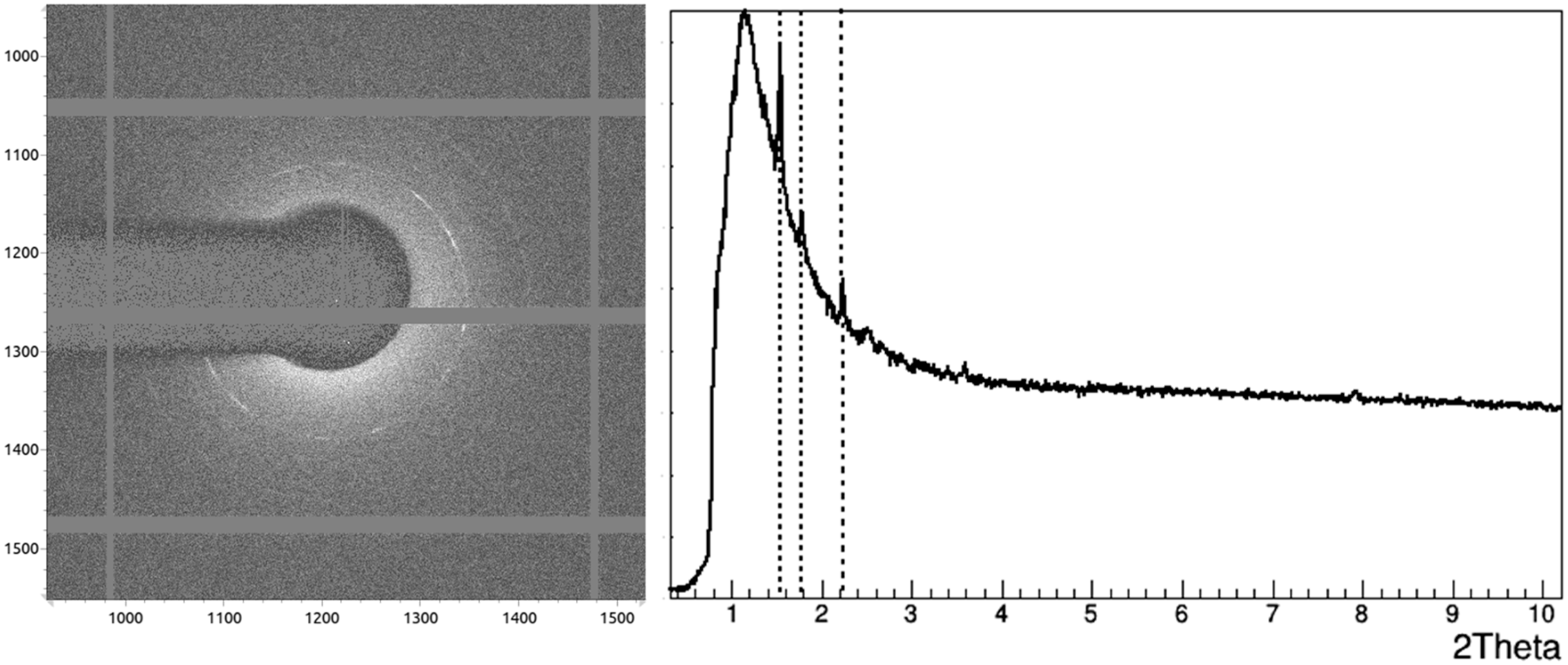
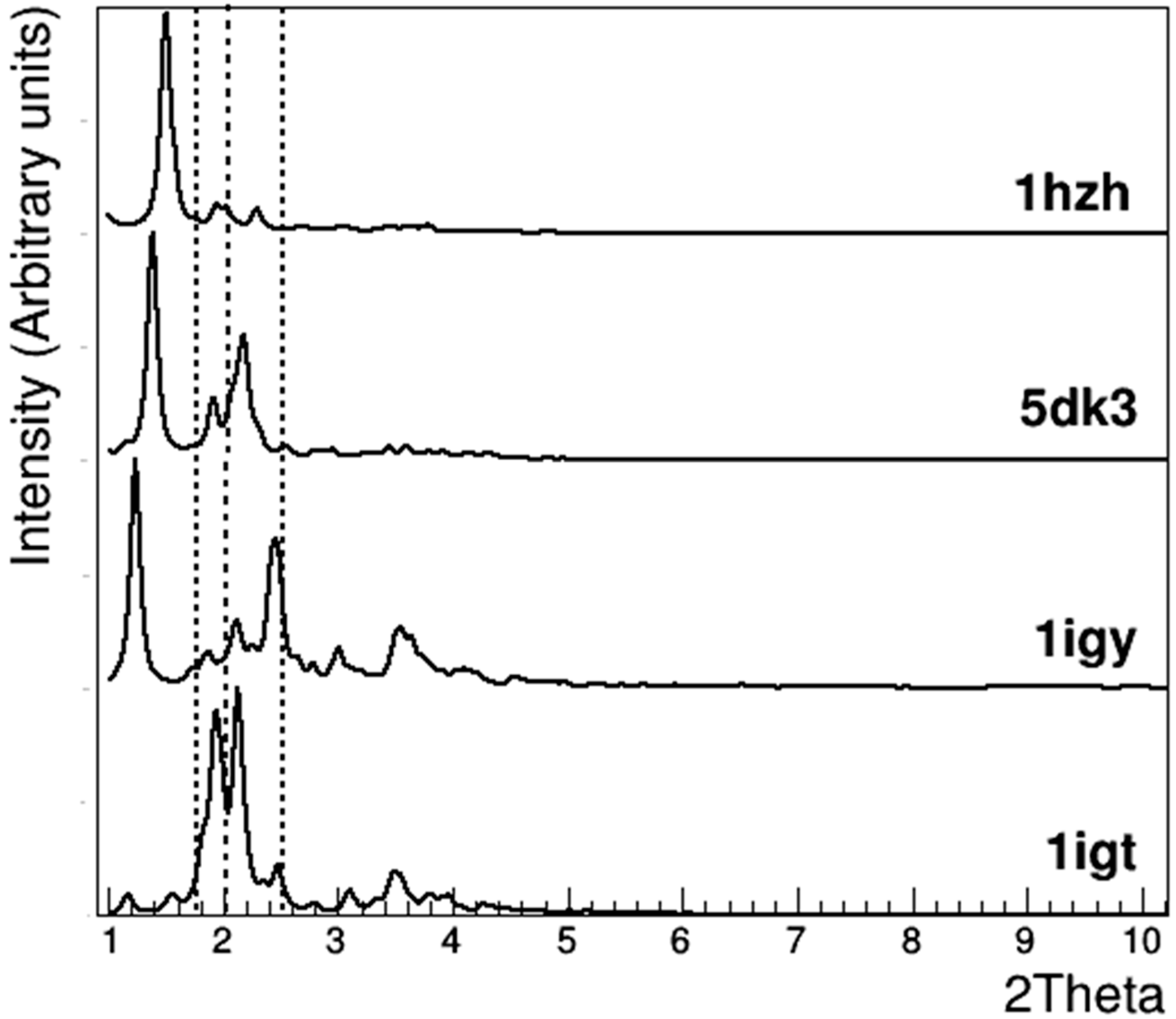

| 1IGT [12] | 1IGY [13] | 5DK3 [31] | 1HZH [14,15] | |
|---|---|---|---|---|
| mAb type | IgG1 | IgG1 | IgG4 | IgG1 |
| Organism | Mus musculus | Mus musculus | Human | Human |
| Sequence identity with anti-CD20 (%) | 60 | 64 | 83 | 87 |
| Space group | P 1 | P 21 | P 212121 | H 3 2 |
| Crystal cell parameters (Å) | 65.8, 76.8, 100.6 | 66.7, 190.7, 73.1 | 63.8, 110.8, 265.0 | 271.3, 271.3, 175.2 |
| Crystal cell volume (Å3) | 503,852 | 874,767 | 1,874,401 | 11,168,087 |
| Volume per atom (Å3) | 48.3 | 42.7 | 43.6 | 58.3 |
| Salt | Salt Concentration | ||
|---|---|---|---|
| 0.5 M | 1.0 M | 1.5 M | |
| Clear drop | Needle-like crystals | Needle-like crystals | |
| Clear drop | Needle-like crystals | Needle-like crystals | |
| Clear drop | Clear drop | Needle-like crystals | |
| Clear drop | Clear drop | Clear drop | |
| Clear drop | Clear drop | Clear drop | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Belviso, B.D.; Li, X.; Chen, W.; Mastropietro, T.F.; Di Profio, G.; Caliandro, R.; Heng, J.Y.Y. Optimization of Vapor Diffusion Conditions for Anti-CD20 Crystallization and Scale-Up to Meso Batch. Crystals 2019, 9, 230. https://doi.org/10.3390/cryst9050230
Yang H, Belviso BD, Li X, Chen W, Mastropietro TF, Di Profio G, Caliandro R, Heng JYY. Optimization of Vapor Diffusion Conditions for Anti-CD20 Crystallization and Scale-Up to Meso Batch. Crystals. 2019; 9(5):230. https://doi.org/10.3390/cryst9050230
Chicago/Turabian StyleYang, Huaiyu, Benny Danilo Belviso, Xiaoyu Li, Wenqian Chen, Teresa Fina Mastropietro, Gianluca Di Profio, Rocco Caliandro, and Jerry Y. Y. Heng. 2019. "Optimization of Vapor Diffusion Conditions for Anti-CD20 Crystallization and Scale-Up to Meso Batch" Crystals 9, no. 5: 230. https://doi.org/10.3390/cryst9050230
APA StyleYang, H., Belviso, B. D., Li, X., Chen, W., Mastropietro, T. F., Di Profio, G., Caliandro, R., & Heng, J. Y. Y. (2019). Optimization of Vapor Diffusion Conditions for Anti-CD20 Crystallization and Scale-Up to Meso Batch. Crystals, 9(5), 230. https://doi.org/10.3390/cryst9050230








