Identification of Streptococcus pneumoniae Sortase A Inhibitors and the Interactive Mechanism
Abstract
1. Background
2. Methodology
2.1. Strain, Compounds and Reagents
2.2. Structure-Based Virtual Screening and Molecular Docking
2.3. Molecular Simulation
2.4. Parameter Analysis
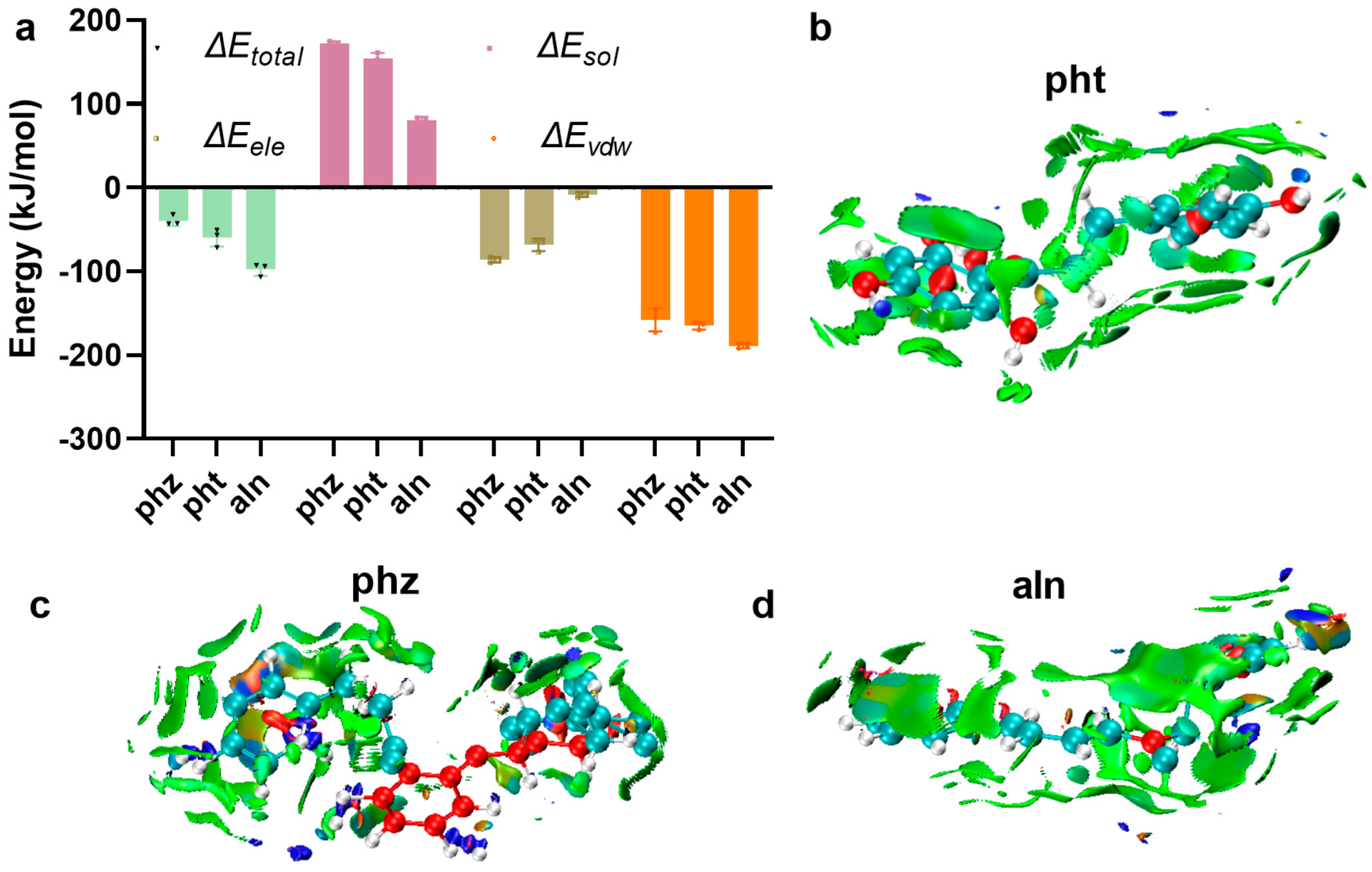
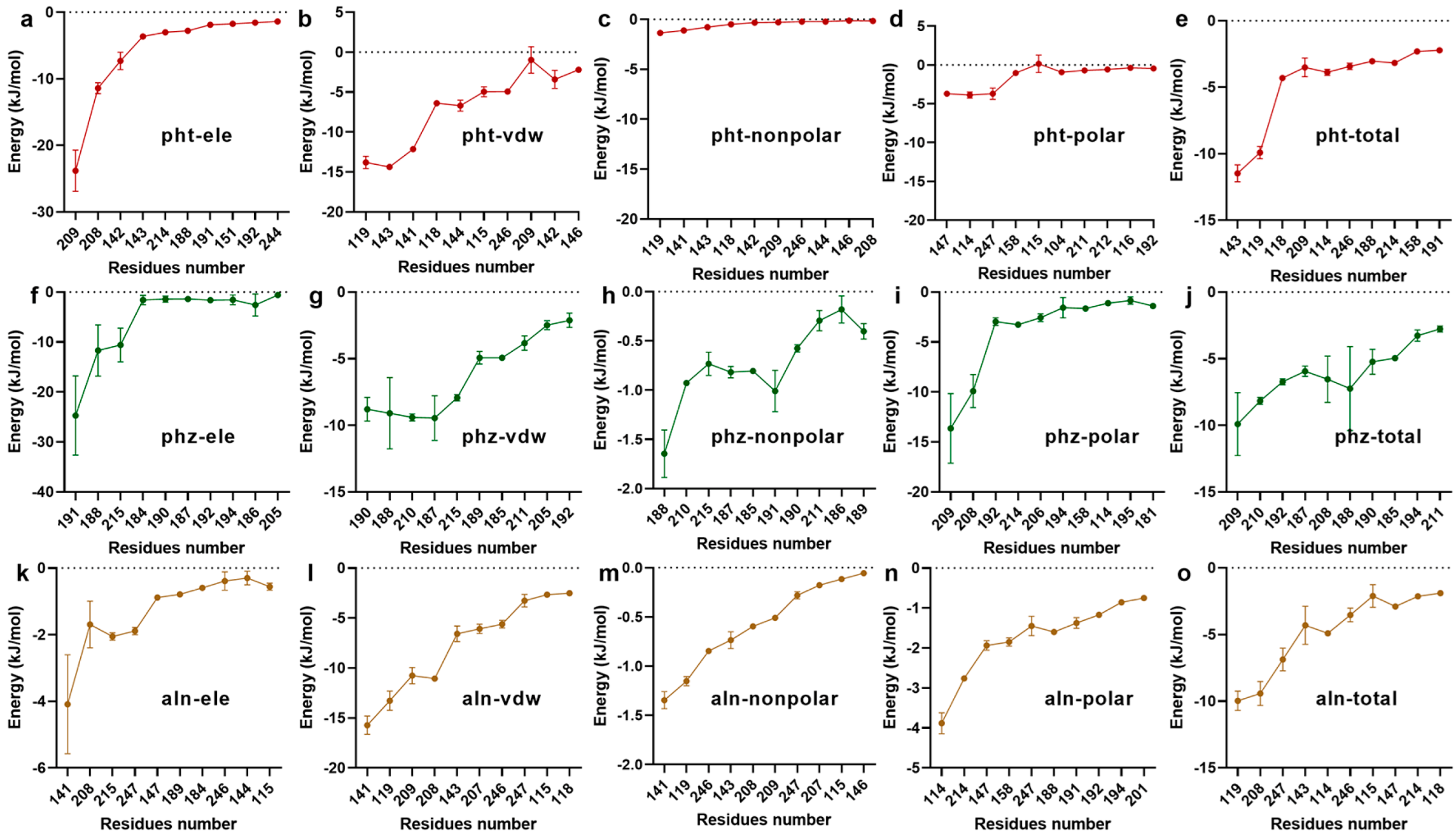
2.5. Binding Free Energy Analysis
2.6. Hydrogen Bond Analysis
2.7. Visualization of Weak Interaction Analysis
2.8. SrtA Activity Inhibition Assay
2.9. Growth Curve Assay
2.10. Adhesion Inhibition Assay
2.11. Bacterial Biofilm Assay
2.12. Statistical Analysis
3. Results
3.1. Analysis of the Results
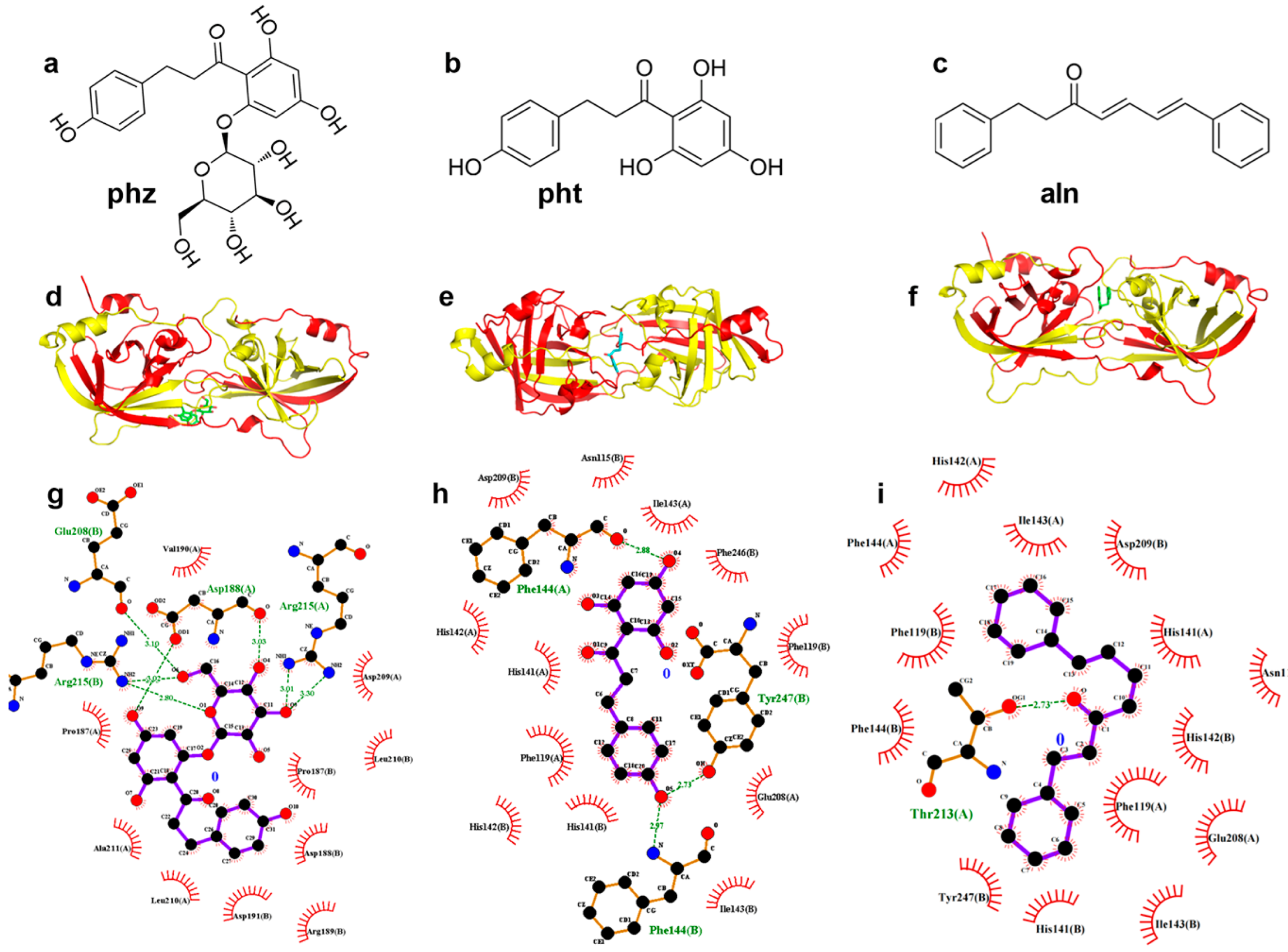
3.1.1. pht, phz, and aln Bind to the Active Center of SrtA
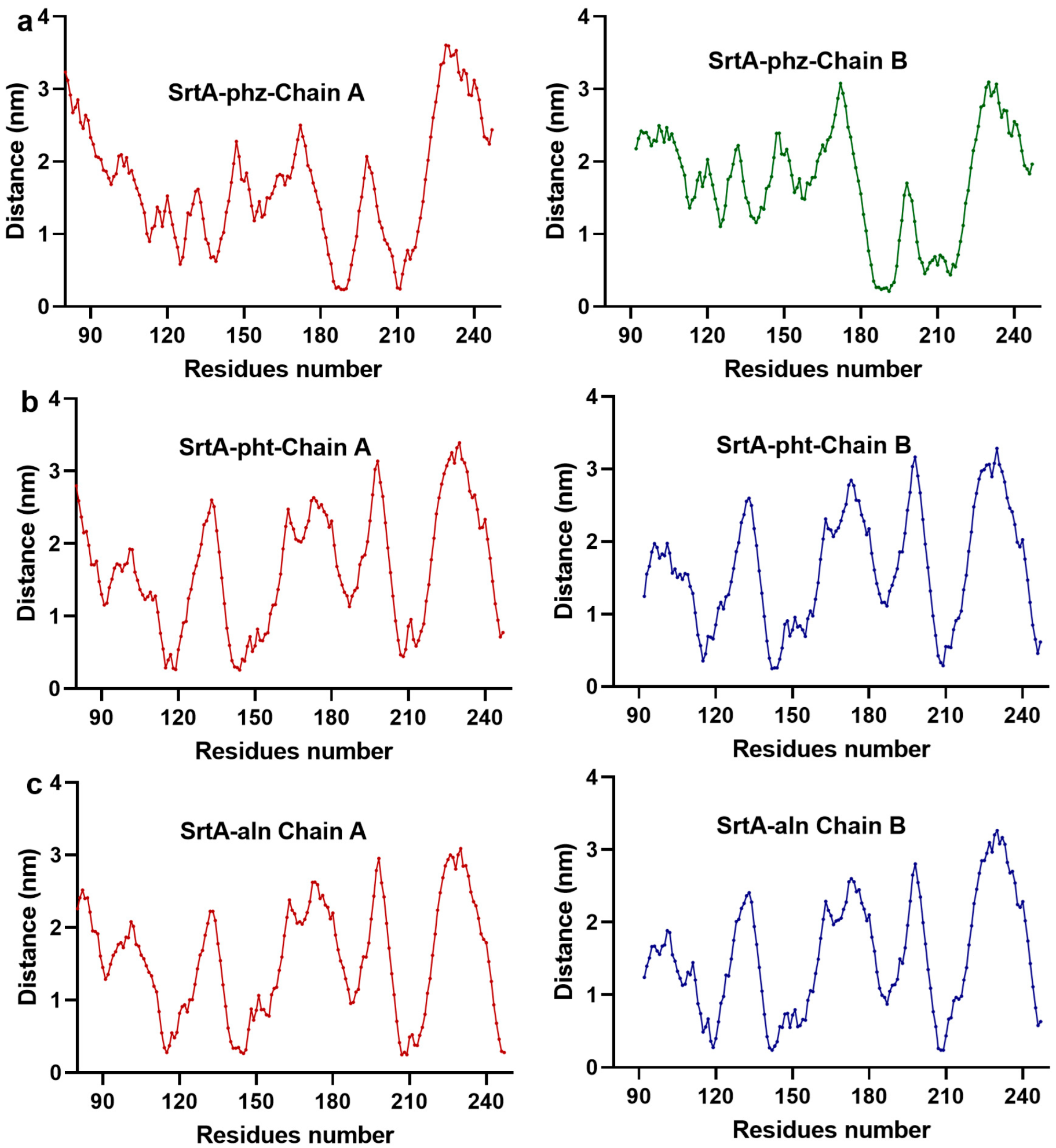
3.1.2. pht, phz, aln, and SrtA Maintained Stable Binding During the Simulation
3.1.3. Confirmation of the Interactive Residues
3.1.4. Hbonds Are Generated Between SrtA and pht, phz
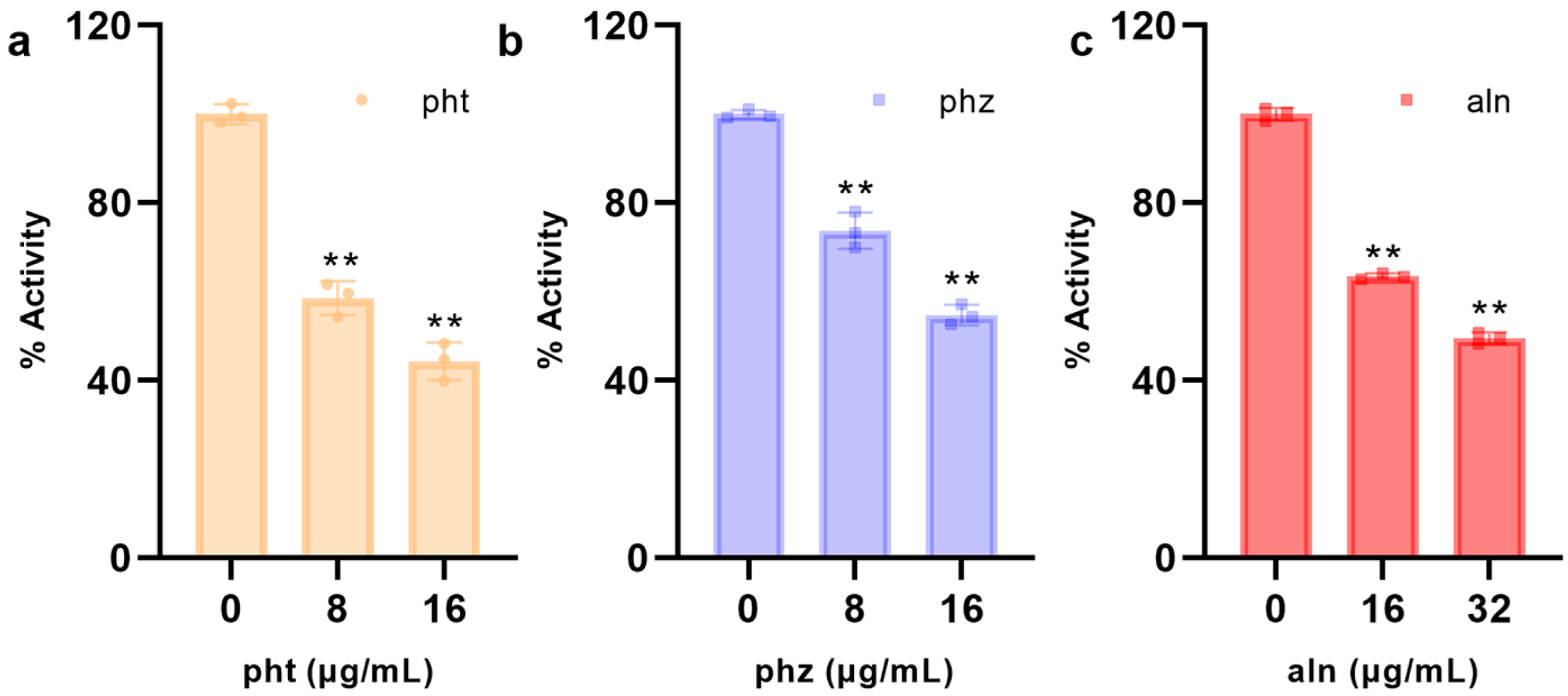
3.1.5. pht, phz, aln Inhibit the Activities of SrtA
3.1.6. pht, phz and aln Does Not Affect the Growth of S. pneumoniae
3.1.7. pht, phz and aln Inhibits the Adhesion of S. pneumoniae to A549 Cells
3.1.8. pht, phz and aln Inhibits S. pneumoniae Biofilm Formation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, X.; Kimmey, J.M.; Matarazzo, L.; de Bakker, V.; Van Maele, L.; Sirard, J.-C.; Nizet, V.; Veening, J.-W. Exploration of Bacterial Bottlenecks and Streptococcus pneumoniae Pathogenesis by CRISPRi-Seq. Cell Host Microbe 2021, 29, 107–120.e106. [Google Scholar] [CrossRef] [PubMed]
- Gil, E.; Noursadeghi, M.; Brown, J.S. Streptococcus pneumoniae interactions with the complement system. Front. Cell. Infect. Microbiol. 2022, 12, 929483. [Google Scholar] [CrossRef]
- Obaro, S.; Adegbola, R. The pneumococcus: Carriage, disease and conjugate vaccines. J. Med. Microbiol. 2002, 51, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.; Sá-Leão, R.; Carriço, J.; Alves, C.R.; Mato, R.; Avô, A.B.; Saldanha, J.; Almeida, J.S.; Sanches, I.S.; de Lencastre, H. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: A summary of 4 years of annual surveillance. J. Clin. Microbiol. 2005, 43, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, J.; Yu, Z.; Li, M.; Zhang, W.; Sun, H. Epidemiological characteristics and antibiotic resistance mechanisms of Streptococcus pneumoniae: An updated review. Microbiol. Res. 2023, 266, 127221. [Google Scholar] [CrossRef]
- Tran-Quang, K.; Nguyen-Thi-Dieu, T.; Tran-Do, H.; Pham-Hung, V.; Nguyen-Vu, T.; Tran-Xuan, B.; Larsson, M.; Duong-Quy, S. Antibiotic resistance of Streptococcus pneumoniae in Vietnamese children with severe pneumonia: A cross-sectional study. Front. Public Health 2023, 11, 1110903. [Google Scholar] [CrossRef]
- Kharat, A.S.; Tomasz, A. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 2003, 71, 2758–2765. [Google Scholar] [CrossRef]
- Lalioui, L.; Pellegrini, E.; Dramsi, S.; Baptista, M.; Bourgeois, N.; Doucet-Populaire, F.; Rusniok, C.; Zouine, M.; Glaser, P.; Kunst, F.; et al. The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 2005, 73, 3342–3350. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Ton-That, H.; Schneewind, O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 2001, 40, 1049–1057. [Google Scholar] [CrossRef]
- Paterson, G.K.; Mitchell, T.J. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004, 12, 89–95. [Google Scholar] [CrossRef]
- Paterson, G.K.; Mitchell, T.J. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 2006, 8, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Aceil, J.; Avci, F.Y. Pneumococcal Surface Proteins as Virulence Factors, Immunogens, and Conserved Vaccine Targets. Front. Cell. Infect. Microbiol. 2022, 12, 832254. [Google Scholar] [CrossRef]
- Susmitha, A.; Bajaj, H.; Madhavan Nampoothiri, K. The divergent roles of sortase in the biology of Gram-positive bacteria. Cell Surf. 2021, 7, 100055. [Google Scholar] [CrossRef]
- Maggio, B.; Raffa, D.; Raimondi, M.V.; Cascioferro, S.; Plescia, F.; Schillaci, D.; Cusimano, M.G.; Leonchiks, A.; Zhulenkovs, D.; Basile, L.; et al. Discovery of a New Class of Sortase A Transpeptidase Inhibitors to Tackle Gram-Positive Pathogens: 2-(2-Phenylhydrazinylidene)alkanoic Acids and Related Derivatives. Molecules 2016, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chhimwal, J.; Kumar, S.; Singh, R.; Patial, V.; Purohit, R.; Padwad, Y.S. Phloretin and phlorizin mitigates inflammatory stress and alleviate adipose and hepatic insulin resistance by abrogating PPARγ S273-Cdk5 interaction in type 2 diabetic mice. Life Sci. 2023, 322, 121668. [Google Scholar] [CrossRef] [PubMed]
- Ongay, K.K.; Granato, D.; Barreto, G.E. Comparison of Antioxidant Capacity and Network Pharmacology of Phloretin and Phlorizin against Neuroinflammation in Traumatic Brain Injury. Molecules 2023, 28, 919. [Google Scholar] [CrossRef]
- Chang, W.T.; Huang, W.C.; Liou, C.J. Evaluation of the anti-inflammatory effects of phloretin and phlorizin in lipopolysaccharide-stimulated mouse macrophages. Food Chem. 2012, 134, 972–979. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, L.; Ma, L.; Ahmad Farooqi, A.; Qiao, G.; Zhang, Y.; Ye, H.; Liu, M.; Huang, J.; Yang, X.; et al. Alnustone inhibits the growth of hepatocellular carcinoma via ROS- mediated PI3K/Akt/mTOR/p70S6K axis. Phytother. Res. PTR 2022, 36, 525–542. [Google Scholar] [CrossRef]
- Zhang, C.; Deng, Y.; Wang, X.; Shi, L.; Zhan, B.; Hou, N.; Liu, S.; Bao, M.; Chi, G.; Fang, T. Alnustone inhibits Streptococcus pneumoniae virulence by targeting pneumolysin and sortase A. Fitoterapia 2022, 162, 105261. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock Vina Adopts More Accurate Binding Poses but Autodock4 Forms Better Binding Affinity. J. Chem. Inf. Model. 2020, 60, 204–211. [Google Scholar] [CrossRef]
- Gaillard, T. Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J. Chem. Inf. Model. 2018, 58, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Chen, R.; Lin, M.; Lin, Q.; Zhu, Y.; Ding, J.; Hu, H.; Ling, M.; Wu, J. Accelerating AutoDock Vina with GPUs. Molecules 2022, 27, 3041. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, X.; Gao, Y.; Wang, H.; Niu, X. Insight into the Dual inhibitory Mechanism of verbascoside targeting serine/threonine phosphatase Stp1 against Staphylococcus aureus. Eur. J. Pharm. Sci. 2021, 157, 105628. [Google Scholar] [CrossRef]
- Dong, J.; Qiu, J.; Zhang, Y.; Lu, C.; Dai, X.; Wang, J.; Li, H.; Wang, X.; Tan, W.; Luo, M.; et al. Oroxylin A inhibits hemolysis via hindering the self-assembly of α-hemolysin heptameric transmembrane pore. PLoS Comput. Biol. 2013, 9, e1002869. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Liu, S.; Li, G.; Shi, L.; Dong, J.; Li, W.; Deng, X.; Niu, X. Morin hydrate attenuates Staphylococcus aureus virulence by inhibiting the self-assembly of α-hemolysin. J. Appl. Microbiol. 2015, 118, 753–763. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, L.; Xu, Y.; Jiang, S.; Shao, Y. Deciphering the binding behavior of flavonoids to the cyclin dependent kinase 6/cyclin D complex. PLoS ONE 2018, 13, e0196651. [Google Scholar] [CrossRef]
- Lu, T.; Chen, Q. Visualization Analysis of Weak Interactions in Chemical Systems. In Comprehensive Computational Chemistry; Elsevier: Amsterdam, The Netherlands, 2024; pp. 240–264. [Google Scholar]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38, 27–28. [Google Scholar] [CrossRef]
- Song, M.; Teng, Z.; Li, M.; Niu, X.; Wang, J.; Deng, X. Epigallocatechin gallate inhibits Streptococcus pneumoniae virulence by simultaneously targeting pneumolysin and sortase A. J. Cell Mol. Med. 2017, 21, 2586–2598. [Google Scholar] [CrossRef]
- Biswas, T.; Misra, A.; Das, S.; Yadav, P.; Ramakumar, S.; Roy, R. Interrogation of 3D-swapped structure and functional attributes of quintessential Sortase A from Streptococcus pneumoniae. Biochem. J. 2020, 477, 4711–4728. [Google Scholar] [CrossRef]
- Wang, J.; Song, M.; Pan, J.; Shen, X.; Liu, W.; Zhang, X.; Li, H.; Deng, X. Quercetin impairs Streptococcus pneumoniae biofilm formation by inhibiting sortase A activity. J. Cell Mol. Med. 2018, 22, 6228–6237. [Google Scholar] [CrossRef]
- Gu, K.; Ding, L.; Wang, Z.; Sun, Y.; Sun, X.; Yang, W.; Sun, H.; Tian, Y.; Wang, Z.; Sun, L. Wogonin attenuates the pathogenicity of Streptococcus pneumoniae by double-target inhibition of Pneumolysin and Sortase A. J. Cell Mol. Med. 2023, 27, 563–575. [Google Scholar] [CrossRef]
- Alharthi, S.; Alavi, S.E.; Moyle, P.M.; Ziora, Z.M. Sortase A (SrtA) inhibitors as an alternative treatment for superbug infections. Drug Discov. Today 2021, 26, 2164–2172. [Google Scholar] [CrossRef]
- Cascioferro, S.; Totsika, M.; Schillaci, D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014, 77, 105–112. [Google Scholar] [CrossRef]
- Bhardwaj, S.; Mehra, P.; Dhanjal, D.S.; Sharma, P.; Sharma, V.; Singh, R.; Nepovimova, E.; Chopra, C.; Kuča, K. Antibiotics and Antibiotic Resistance- Flipsides of the Same Coin. Curr. Pharm. Des. 2022, 28, 2312–2329. [Google Scholar] [CrossRef]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance-A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef] [PubMed]
- Krueger, E.; Brown, A.C. Inhibition of bacterial toxin recognition of membrane components as an anti-virulence strategy. J. Biol. Eng. 2019, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.L.; Mullally, C.A.; Haese, E.C.; Kibble, E.A.; McCluskey, N.R.; Mikucki, E.C.; Thai, V.C.; Stubbs, K.A.; Sarkar-Tyson, M.; Kahler, C.M. Anti-Virulence Therapeutic Approaches for Neisseria gonorrhoeae. Antibiotics 2021, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, J.; Pieper, F.; Göbel, J.; Knispel, H.; McCarthy, M.; Goncalves, M.; Turner, M.; Merrill, A.R.; Genersch, E. Anti-Virulence Strategy against the Honey Bee Pathogenic Bacterium Paenibacillus larvae via Small Molecule Inhibitors of the Bacterial Toxin Plx2A. Toxins 2021, 13, 607. [Google Scholar] [CrossRef]



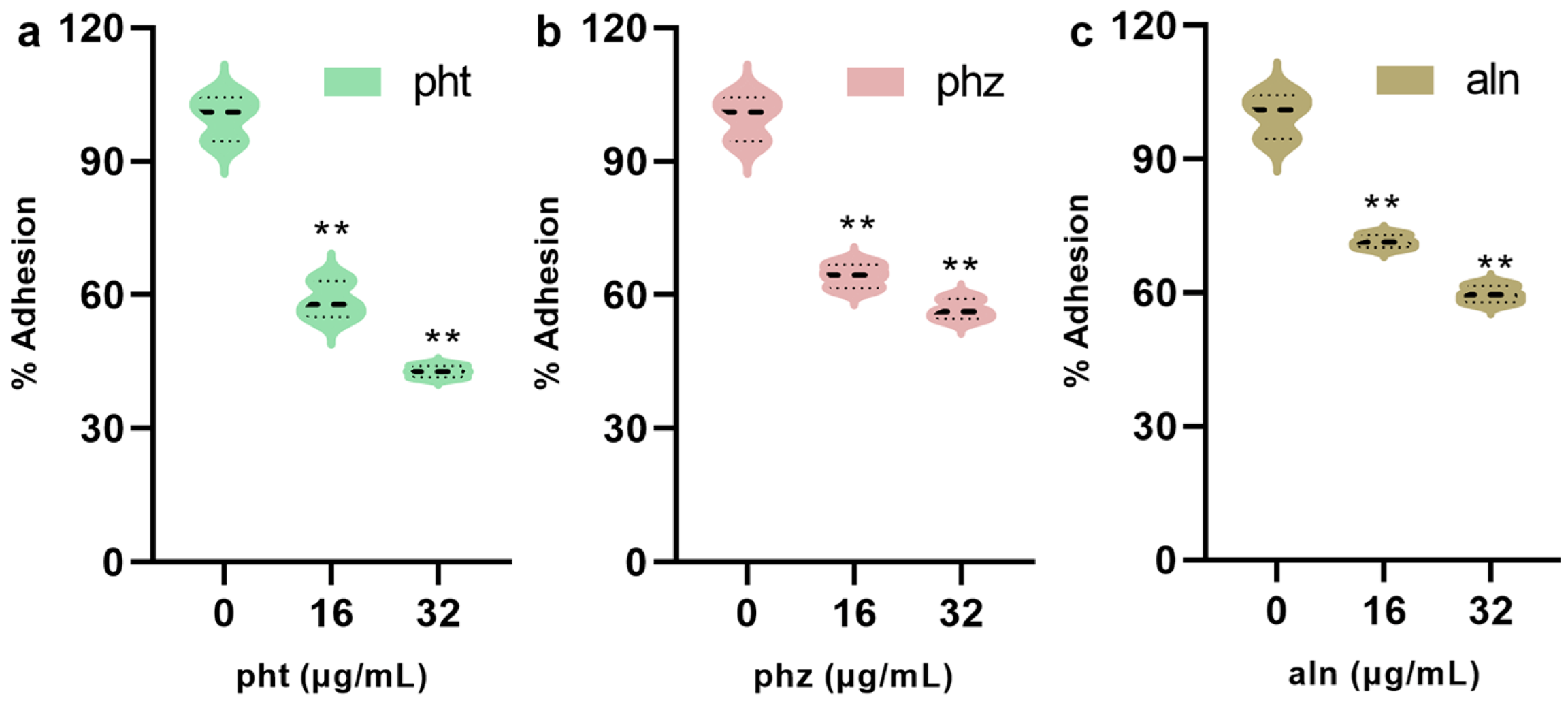
| Concentrations (µg/mL) | pht | phz | aln |
|---|---|---|---|
| 0 | 99.98 ± 4.25 | 100.00 ± 2.59 | 100.02 ± 3.34 |
| 16 | 50.95 ± 1.91 ** | 55.60 ± 2.76 ** | 68.46 ± 3.85 ** |
| 32 | 37.70 ± 3.95 ** | 47.14 ± 2.29 ** | 59.99 ± 2.89 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Lu, J.; Wen, J.; Duan, Y.; Zhou, H.; Peng, X.; Li, Z. Identification of Streptococcus pneumoniae Sortase A Inhibitors and the Interactive Mechanism. Crystals 2025, 15, 594. https://doi.org/10.3390/cryst15070594
Wang G, Lu J, Wen J, Duan Y, Zhou H, Peng X, Li Z. Identification of Streptococcus pneumoniae Sortase A Inhibitors and the Interactive Mechanism. Crystals. 2025; 15(7):594. https://doi.org/10.3390/cryst15070594
Chicago/Turabian StyleWang, Guizhen, Jiahui Lu, Jingyao Wen, Yifan Duan, Hanbing Zhou, Xinli Peng, and Zhandong Li. 2025. "Identification of Streptococcus pneumoniae Sortase A Inhibitors and the Interactive Mechanism" Crystals 15, no. 7: 594. https://doi.org/10.3390/cryst15070594
APA StyleWang, G., Lu, J., Wen, J., Duan, Y., Zhou, H., Peng, X., & Li, Z. (2025). Identification of Streptococcus pneumoniae Sortase A Inhibitors and the Interactive Mechanism. Crystals, 15(7), 594. https://doi.org/10.3390/cryst15070594





