The Crystallography of Enzymes: A Retrospective and Beyond
Abstract
1. Introduction
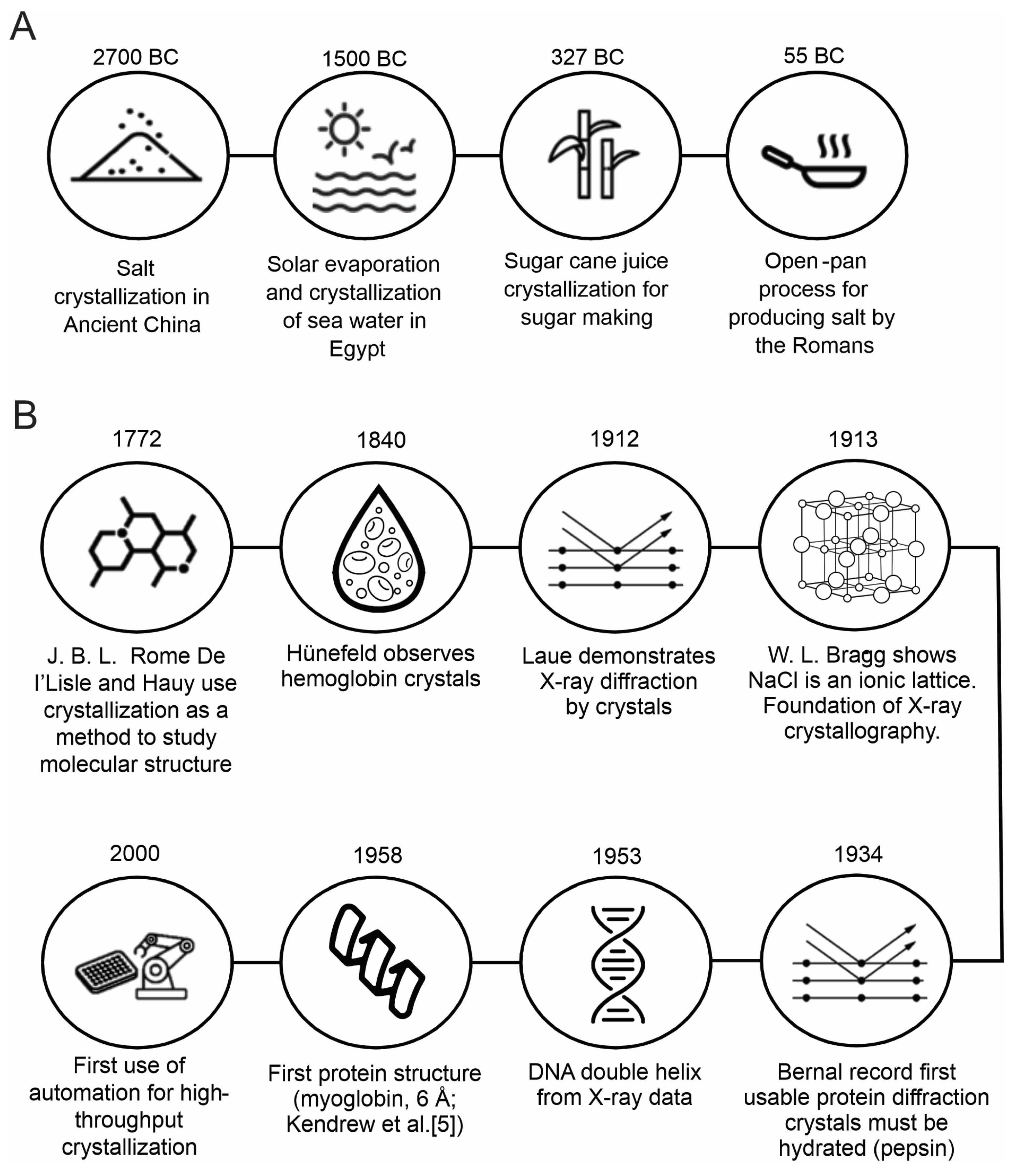
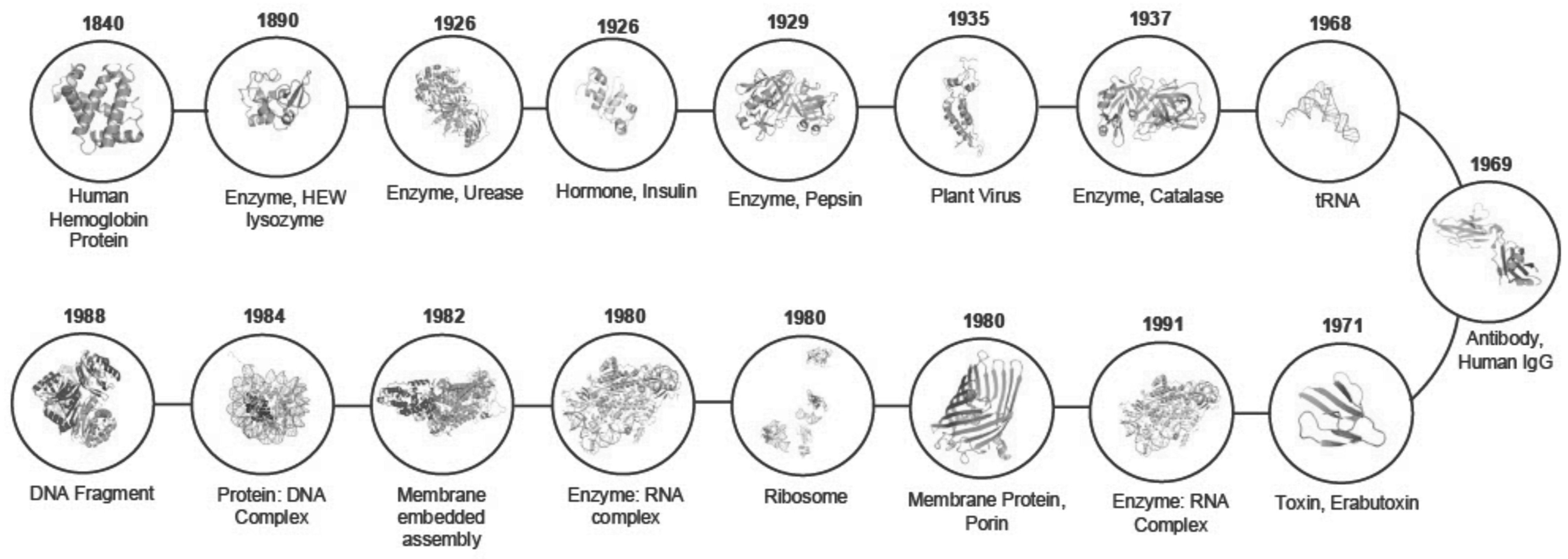
1.1. Historical Development of Crystallography
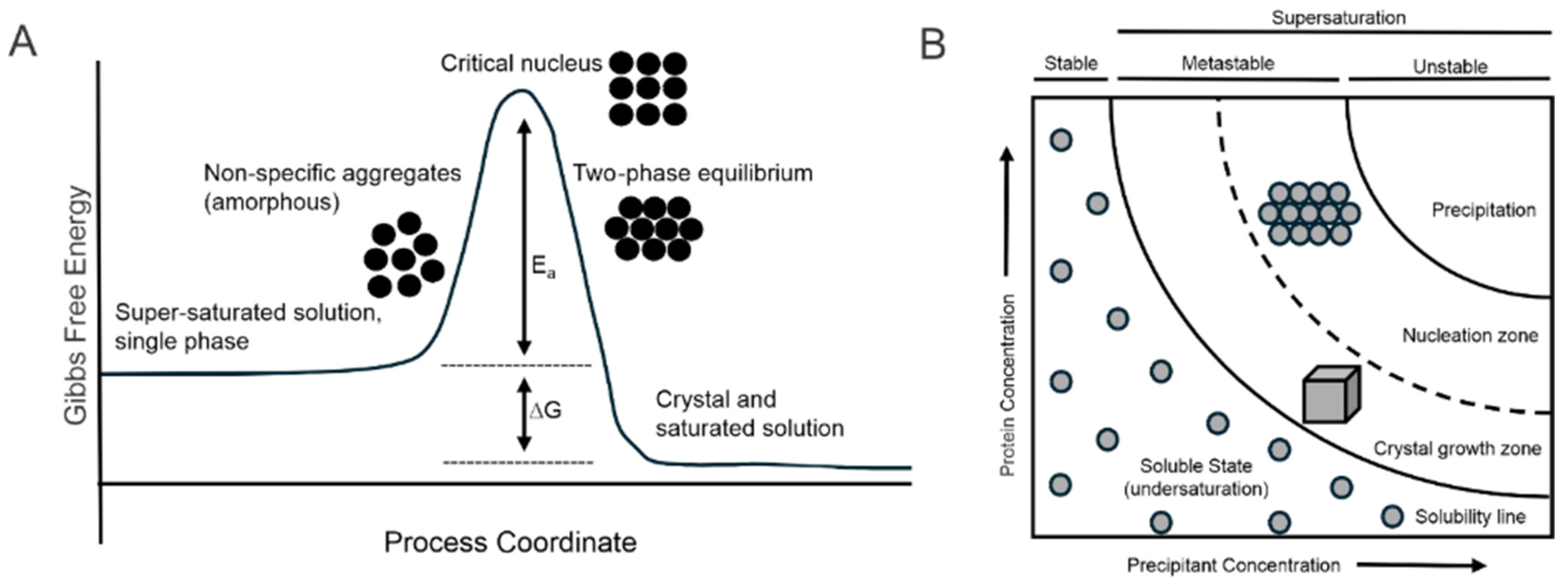
1.2. Principles of Crystallization
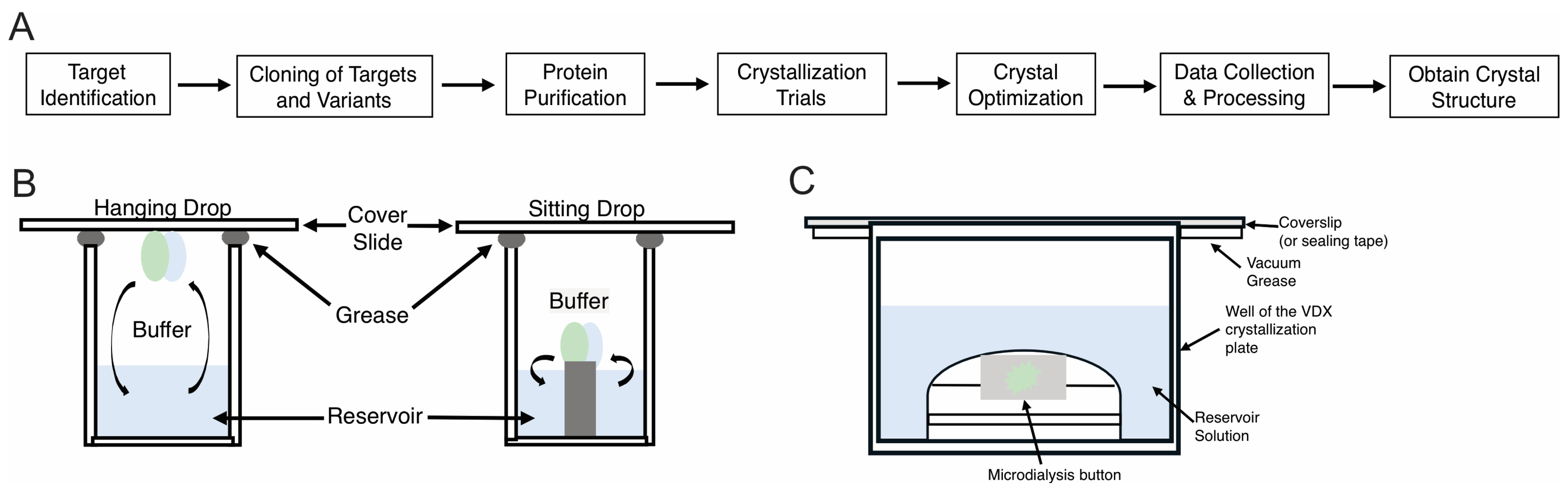
1.3. Practical Crystallization Methods
1.4. Insights into Enzymology
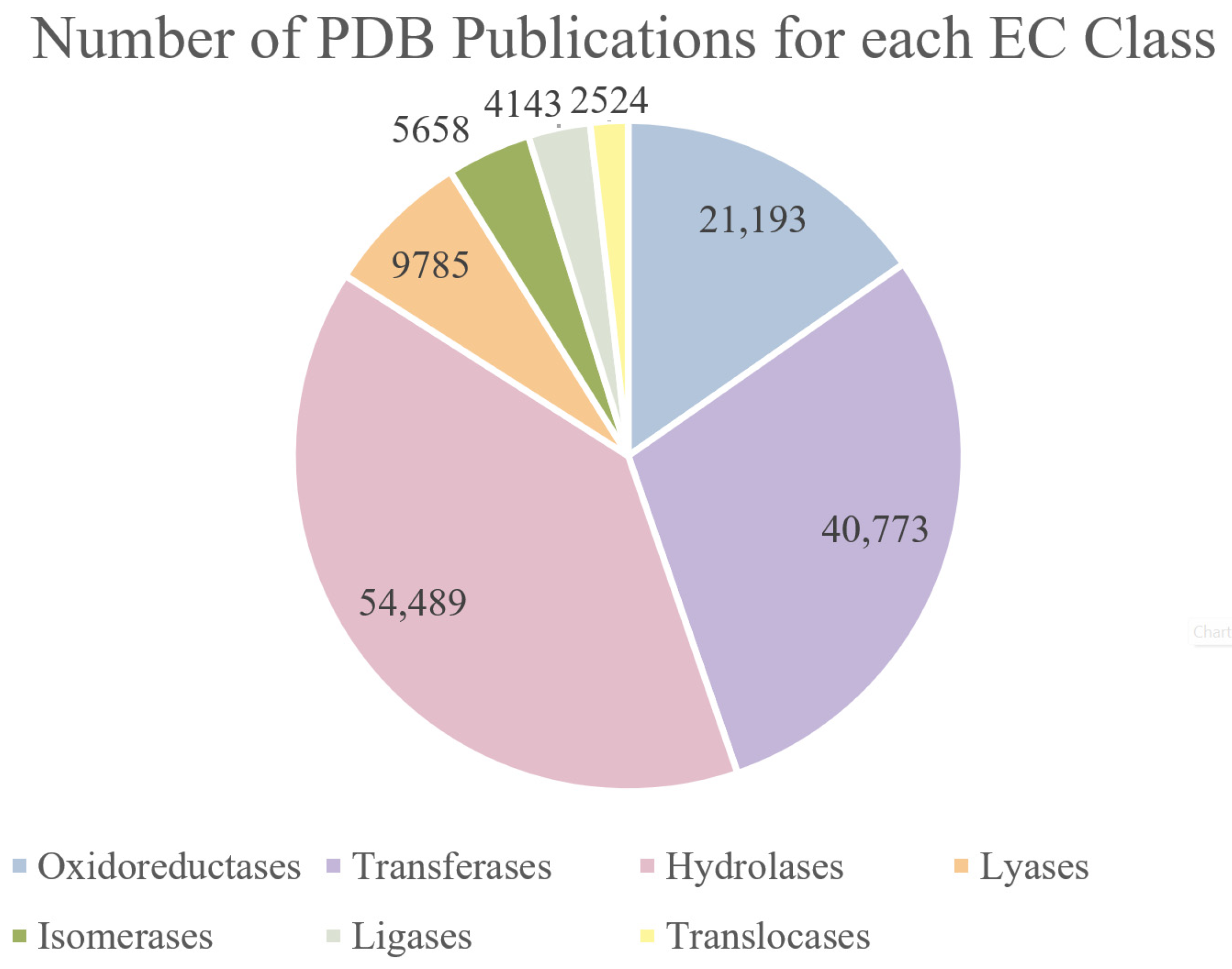
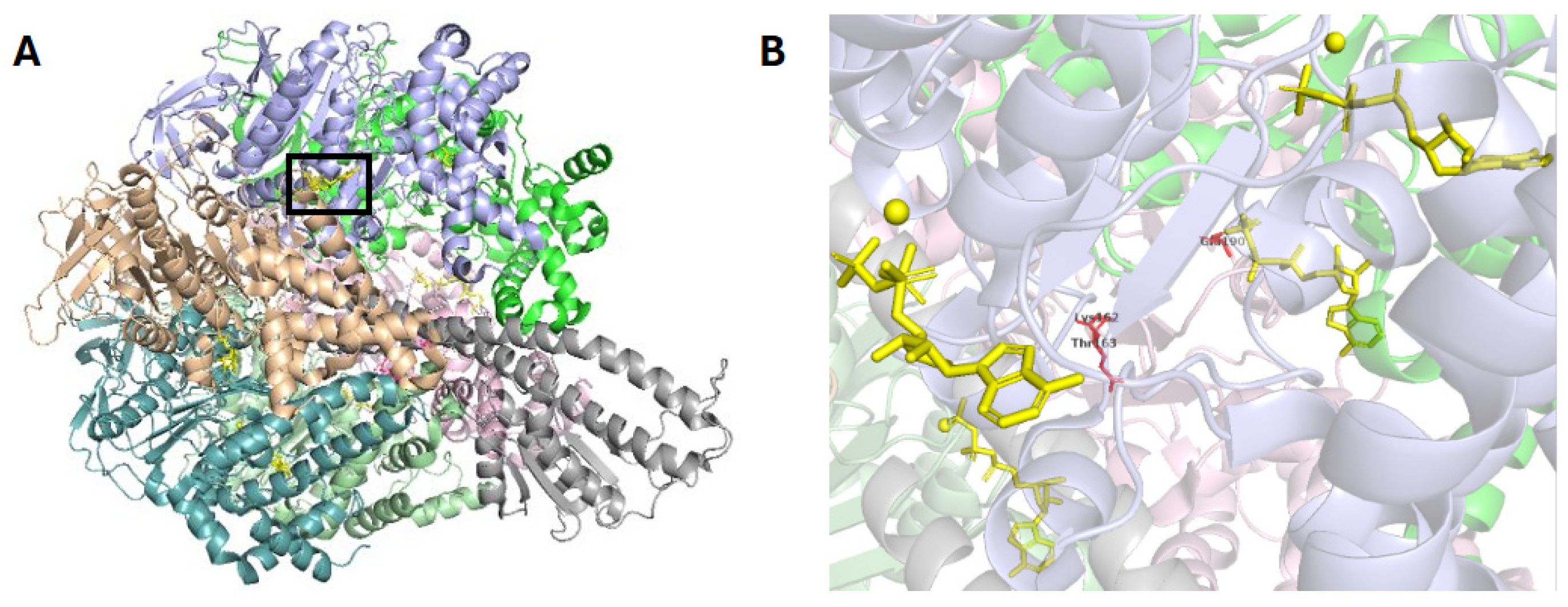
2. Crystallographic Insights into Enzyme Mechanisms
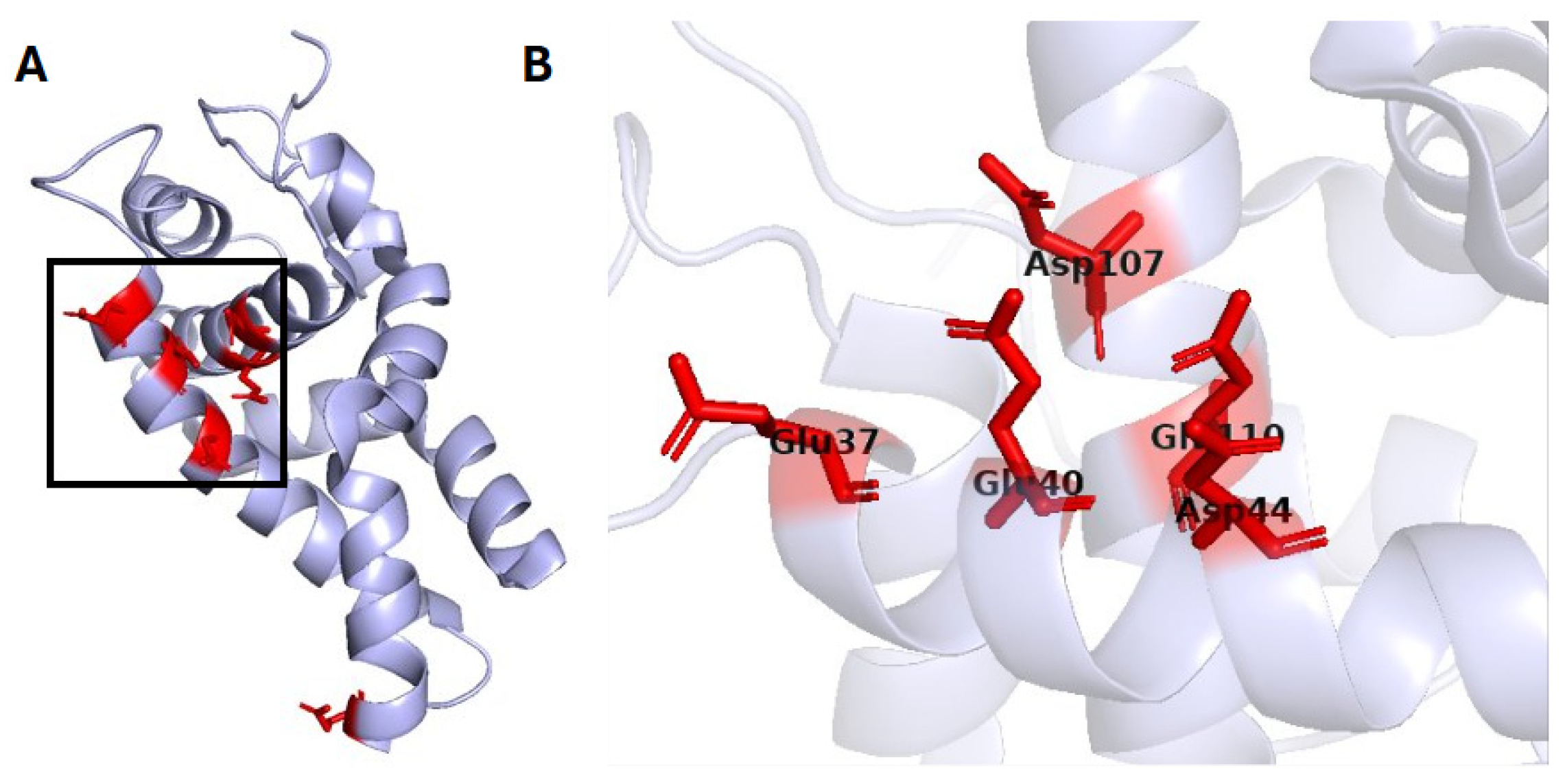
2.1. Oxidoreductases
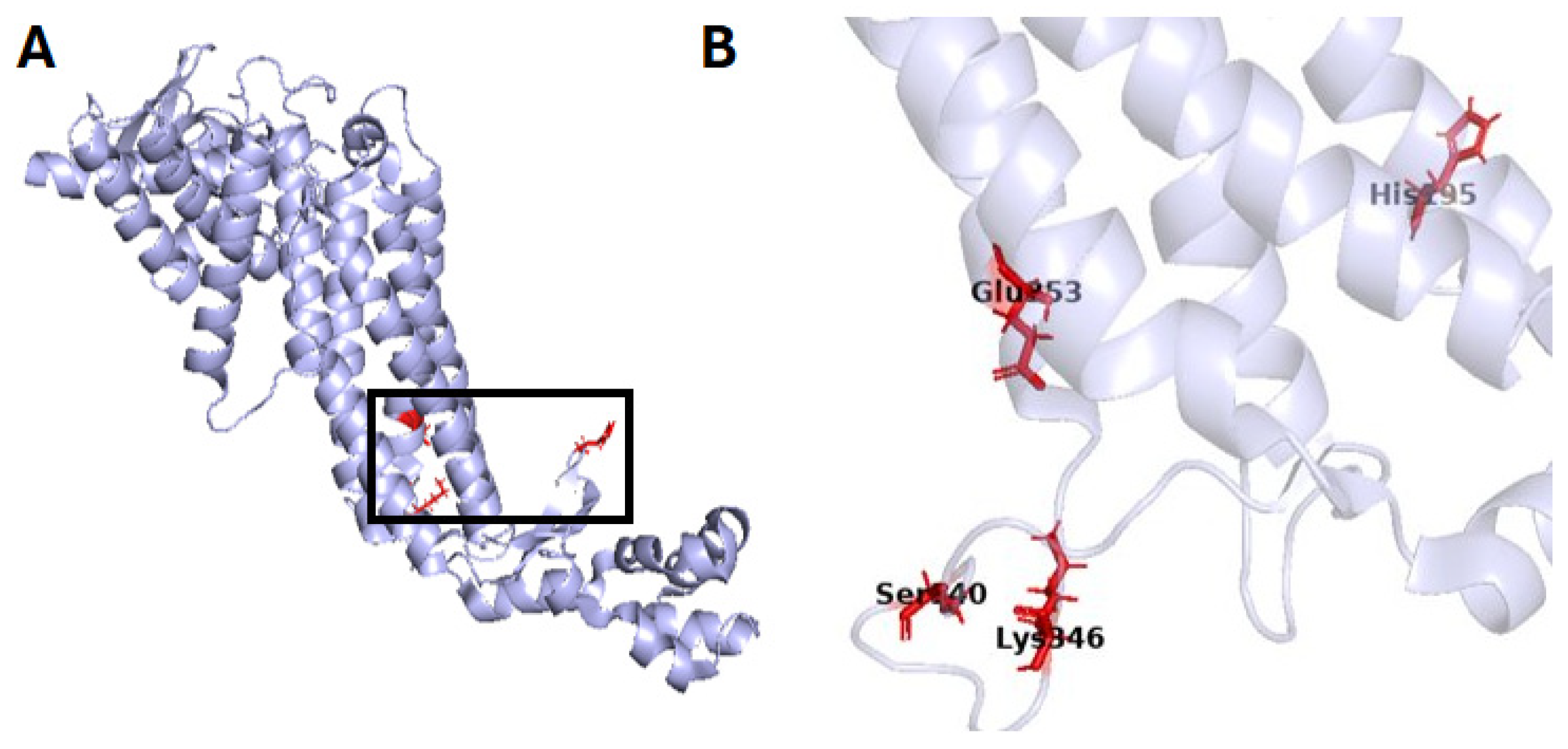
2.2. Transferases
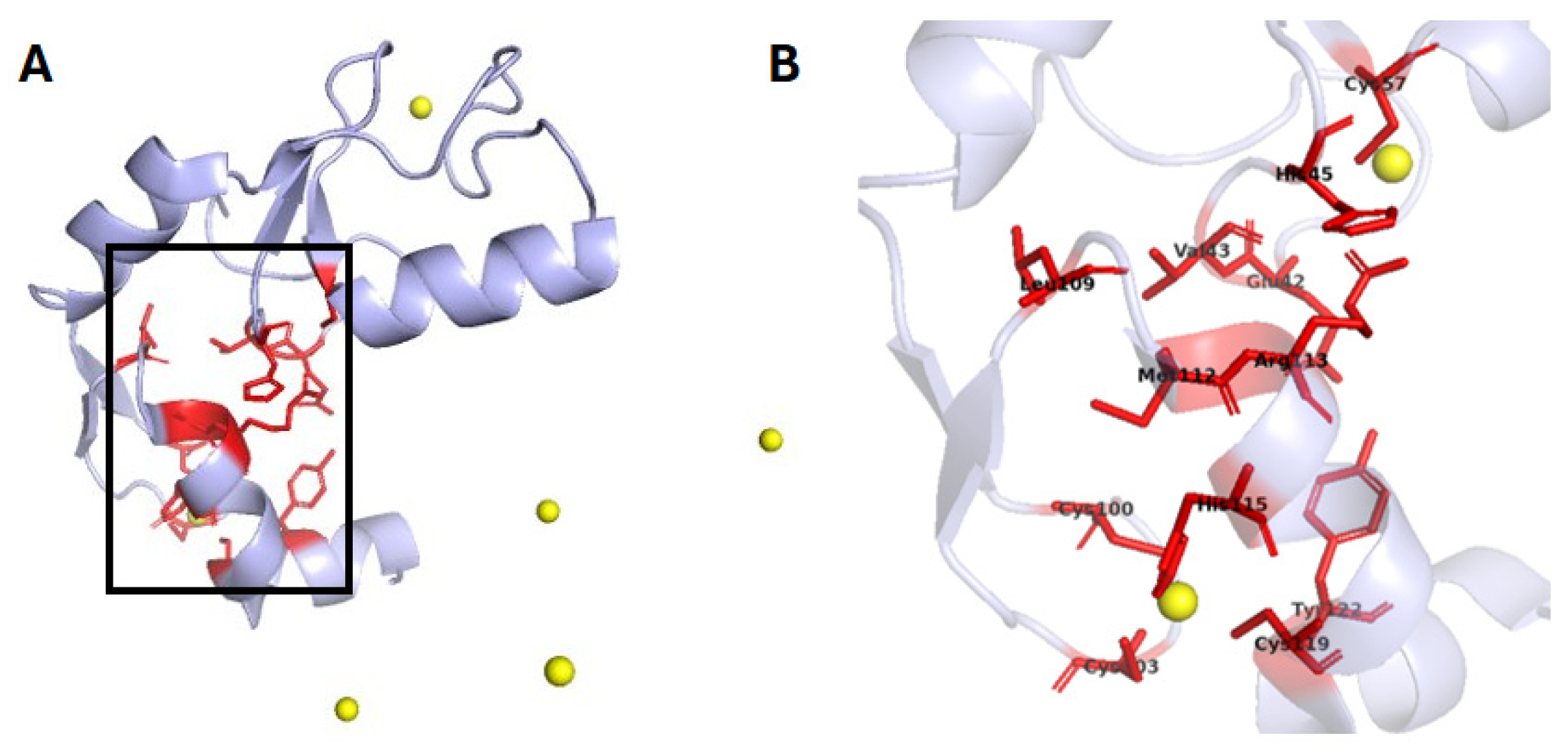
2.3. Hydrolases
2.4. Lyases
2.5. Isomerases
2.6. Ligases
2.7. Translocases
3. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PDB | Protein Data Bank |
| NAD+ | nicotinamide adenine dinucleotide (oxidized form) |
| NADH | nicotinamide adenine dinucleotide with hydrogen (reduced form) |
| OYE | old yellow enzyme |
| M.W. | molecular weight |
| PEP | phosphoenolpyruvate |
| PTS | phosphotransferase system |
| HPr | histidine phosphocarrier protein |
| RNase III | Ribonuclease 3 |
| FH | fumarate hydratase |
| HLRCC | hereditary leiomyomatosis and renal cell cancer |
| HIFs | hypoxia-inducible factors |
| U55 | Uridine 55 |
| TΨC | thymidine-pseudouridine-cytidine loop |
| snRNA | small nuclear RNA |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| E1 | ubiquitin-activating enzyme |
| E2 | ubiquitin-conjugating enzyme |
| E3 | ubiquitin ligase enzyme |
| PROTACs | proteolysis-targeting chimeras |
| ATP | adenosine triphosphate |
| ADP | adenosine diphosphate |
| Pi | inorganic phosphate |
| NMR | nuclear magnetic resonance |
| Cryo-EM | cryo-electron microscopy |
| Cryo-ET | cryo-electron tomography |
| AF2 | AlphaFold2 |
| RMSD | root-mean-square deviation |
| RFAA | RoseTTAFold All-Atom |
| PET | polyethylene terephthalate |
| FEP | free energy perturbation |
| RCSB | Research Collaboratory for Structural Bioinformatics |
| HEW | hen egg-white (lysozyme) |
Appendix A
References
- Russo Krauss, I.; Merlino, A.; Vergara, A.; Sica, F. An overview of biological macromolecule crystallization. Int. J. Mol. Sci. 2013, 14, 11643–11691. [Google Scholar] [CrossRef]
- Schoen, H.M.; Grove, C.S.; Palermo, J.A. The early history of crystallization. J. Chem. Educ. 1956, 33, 373. [Google Scholar] [CrossRef]
- Bragg, W.L. The structure of some crystals as indicated by their diffraction of X-rays. Proc. R. Soc. A Math. Phys. Character 1997, 89, 248–277. [Google Scholar] [CrossRef]
- Smith, T. Early crystals. Nat. Struct. Biol. 1999, 6, 411. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Bodo, G.; Dintzis, H.M.; Parrish, R.G.; Wyckoff, H.; Phillips, D.C. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 1958, 181, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- McPherson, A.; Gavira, J.A. Introduction to protein crystallization. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 2–20. [Google Scholar] [CrossRef]
- Bijelic, A.; Rompel, A. Polyoxometalates: More than a phasing tool in protein crystallography. ChemTexts 2018, 4, 10. [Google Scholar] [CrossRef]
- Makita, H.; Zhang, M.; Yano, J.; Kern, J. Room temperature crystallography and X-ray spectroscopy of metalloenzymes. Methods Enzymol. 2023, 688, 307–348. [Google Scholar] [CrossRef]
- Chayen, N.E.; Saridakis, E. Protein crystallization: From purified protein to diffraction-quality crystal. Nat. Methods 2008, 5, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Junius, N.; Oksanen, E.; Terrien, M.; Berzin, C.; Ferrer, J.L.; Budayova-Spano, M. A crystallization apparatus for temperature-controlled flow-cell dialysis with real-time visualization. J. Appl. Crystallogr. 2016, 49, 806–813. [Google Scholar] [CrossRef]
- Weinberg, C.E.; Weinberg, Z.; Hammann, C. Novel ribozymes: Discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019, 47, 9480–9494. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Martinez Cuesta, S.; Rahman, S.A.; Furnham, N.; Thornton, J.M. The Classification and Evolution of Enzyme Function. Biophys. J. 2015, 109, 1082–1086. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.G.; Tipton, K.F. Enzyme nomenclature and classification: The state of the art. FEBS J. 2023, 290, 2214–2231. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Hardas, S.S.; Lange, M.L. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease: Many pathways to neurodegeneration. J. Alzheimers Dis. 2010, 20, 369–393. [Google Scholar] [CrossRef]
- Shampo, M.A.; Kyle, R.A. Hugo Theorell--Nobel Prize for study of enzymes. Mayo Clin. Proc. 1998, 73, 147. [Google Scholar] [CrossRef]
- Akeson, A.; Theorell, H. Molecular weight and FMN content of crystallin old yellow enzyme. Arch. Biochem. Biophys. 1956, 65, 439–448. [Google Scholar] [CrossRef]
- Fox, K.M.; Karplus, P.A. Old yellow enzyme at 2 Å resolution: Overall structure, ligand binding, and comparison with related flavoproteins. Structure 1994, 2, 1089–1105. [Google Scholar] [CrossRef]
- Fox, K.M.; Karplus, P.A. Crystallization of Old Yellow Enzyme illustrates an effective strategy for increasing protein crystal size. J. Mol. Biol. 1993, 234, 502–507. [Google Scholar] [CrossRef]
- Kitzing, K.; Fitzpatrick, T.B.; Wilken, C.; Sawa, J.; Bourenkov, G.P.; Macheroux, P.; Clausen, T. The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of old yellow enzymes. J. Biol. Chem. 2005, 280, 27904–27913. [Google Scholar] [CrossRef]
- Saier, M.H.; Hvorup, R.N.; Barabote, R.D. Evolution of the bacterial phosphotransferase system: From carriers and enzymes to group translocators. Biochem. Soc. Trans. 2005, 33, 220–224. [Google Scholar] [CrossRef]
- Roth, P.; Jeckelmann, J.-M.; Fender, I.; Ucurum, Z.; Lemmin, T.; Fotiadis, D. Structure and mechanism of a phosphotransferase system glucose transporter. Nat. Commun. 2024, 15, 7992. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.J.; de Meis, L. Role of water in the energy of hydrolysis of phosphoanhydride and phosphoester bonds. J. Biol. Chem. 1989, 264, 7869–7873. [Google Scholar] [CrossRef]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. FAT SIGNALS-lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Court, D.L.; Gan, J.; Liang, Y.H.; Shaw, G.X.; Tropea, J.E.; Costantino, N.; Waugh, D.S.; Ji, X. RNase III: Genetics and function; structure and mechanism. Annu. Rev. Genet. 2013, 47, 405–431. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999, 23, 371–390. [Google Scholar] [CrossRef]
- Blaszczyk, J.; Tropea, J.E.; Bubunenko, M.; Routzahn, K.M.; Waugh, D.S.; Court, D.L.; Ji, X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 2001, 9, 1225–1236. [Google Scholar] [CrossRef]
- Nicholson, A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 2014, 5, 31–48. [Google Scholar] [CrossRef]
- Akey, D.L.; Berger, J.M. Structure of the nuclease domain of ribonuclease III from M. tuberculosis at 2.1 A. Protein Sci. 2005, 14, 2744–2750. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Northrop, J.H.; Technical Assistance of Marie, K. The Effect of Trypsin, Chymotrypsin, Ribonuclease, and Desoxyribonuclease on Active, Inactive, and Reversibly Inactivated Megatherium Phage. J. Gen. Physiol. 1955, 39, 251–258. [Google Scholar] [CrossRef]
- Brovetto, M.; Gamenara, D.; Mendez, P.S.; Seoane, G.A. C-C bond-forming lyases in organic synthesis. Chem. Rev. 2011, 111, 4346–4403. [Google Scholar] [CrossRef] [PubMed]
- Yogev, O.; Yogev, O.; Singer, E.; Shaulian, E.; Goldberg, M.; Fox, T.D.; Pines, O. Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010, 8, e1000328. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Goncalves, E.; Johnson, T.I.; Zecchini, V.R.; da Costa, A.S.; Gaude, E.; Drubbel, A.V.; Theobald, S.J.; Abbo, S.R.; Tran, M.G.; et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016, 537, 544–547. [Google Scholar] [CrossRef]
- Isaacs, J.S.; Jung, Y.J.; Mole, D.R.; Lee, S.; Torres-Cabala, C.; Chung, Y.L.; Merino, M.; Trepel, J.; Zbar, B.; Toro, J.; et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 2005, 8, 143–153. [Google Scholar] [CrossRef]
- Yang, M.; Soga, T.; Pollard, P.J.; Adam, J. The emerging role of fumarate as an oncometabolite. Front. Oncol. 2012, 2, 85. [Google Scholar] [CrossRef]
- Mizobata, T.; Fujioka, T.; Yamasaki, F.; Hidaka, M.; Nagai, J.; Kawata, Y. Purification and characterization of a thermostable class II fumarase from Thermus thermophilus. Arch. Biochem. Biophys. 1998, 355, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Ajalla Aleixo, M.A.; Rangel, V.L.; Rustiguel, J.K.; de Padua, R.A.P.; Nonato, M.C. Structural, biochemical and biophysical characterization of recombinant human fumarate hydratase. FEBS J. 2019, 286, 1925–1940. [Google Scholar] [CrossRef]
- Martinez Cuesta, S.; Rahman, S.A.; Thornton, J.M. Exploring the chemistry and evolution of the isomerases. Proc. Natl. Acad. Sci. USA 2016, 113, 1796–1801. [Google Scholar] [CrossRef] [PubMed]
- Garus, A.; Autexier, C. Dyskerin: An essential pseudouridine synthase with multifaceted roles in ribosome biogenesis, splicing, and telomere maintenance. RNA 2021, 27, 1441–1458. [Google Scholar] [CrossRef]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [CrossRef]
- Wu, G.; Radwan, M.K.; Xiao, M.; Adachi, H.; Fan, J.; Yu, Y.T. The TOR signaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA. RNA 2016, 22, 1146–1152. [Google Scholar] [CrossRef]
- Hoang, C.; Hamilton, C.S.; Mueller, E.G.; Ferre-D’Amare, A.R. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci. 2005, 14, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Agarwalla, S.; Moustakas, D.T.; Finer-Moore, J.; Stroud, R.M. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. USA 2003, 100, 12648–12653. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Gray, M.W. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000, 28, 2342–2352. [Google Scholar] [CrossRef][Green Version]
- Kirwan, M.; Dokal, I. Dyskeratosis congenita, stem cells and telomeres. Biochim. Biophys. Acta 2009, 1792, 371–379. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.T. The Critical Contribution of Pseudouridine to mRNA COVID-19 Vaccines. Front. Cell Dev. Biol. 2021, 9, 789427. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Bijlmakers, M.J.; Teixeira, J.M.; Boer, R.; Mayzel, M.; Puig-Sarries, P.; Karlsson, G.; Coll, M.; Pons, M.; Crosas, B. A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125. Sci. Rep. 2016, 6, 29232. [Google Scholar] [CrossRef]
- Dou, H.; Buetow, L.; Sibbet, G.J.; Cameron, K.; Huang, D.T. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012, 19, 876–883. [Google Scholar] [CrossRef]
- Petroski, M.D. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008, 9 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA, 2015. [Google Scholar]
- Morales-Rios, E.; Montgomery, M.G.; Leslie, A.G.; Walker, J.E. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc. Natl. Acad. Sci. USA 2015, 112, 13231–13236. [Google Scholar] [CrossRef]
- Abrahams, J.P.; Leslie, A.G.; Lutter, R.; Walker, J.E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 1994, 370, 621–628. [Google Scholar] [CrossRef]
- Sobti, M.; Ueno, H.; Noji, H.; Stewart, A.G. The six steps of the complete F(1)-ATPase rotary catalytic cycle. Nat. Commun. 2021, 12, 4690. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Rubinstein, J.L. Structure of ATP synthase under strain during catalysis. Nat. Commun. 2022, 13, 2232. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, J.; Neutze, R.; Sjogren, T.; Edman, K.; Szoke, A.; Wilmouth, R.C.; Wilmot, C.M. Analyzing protein functions in four dimensions. Nat. Struct. Biol. 2000, 7, 1006–1012. [Google Scholar] [CrossRef]
- Yamashita, A.; Endo, M.; Higashi, T.; Nakatsu, T.; Yamada, Y.; Oda, J.; Kato, H. Capturing enzyme structure prior to reaction initiation: Tropinone reductase-II-substrate complexes. Biochemistry 2003, 42, 5566–5573. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-Driven Deep Learning Techniques in Protein Structure Prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Krishna, R.; Wang, J.; Ahern, W.; Sturmfels, P.; Venkatesh, P.; Kalvet, I.; Lee, G.R.; Morey-Burrows, F.S.; Anishchenko, I.; Humphreys, I.R.; et al. Generalized biomolecular modeling and design with RoseTTAFold All-Atom. Science 2024, 384, eadl2528. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.; Vazzana, G.; Savojardo, C.; Martelli, P.L.; Casadio, R. AlphaFold2 and ESMFold: A large-scale pairwise model comparison of human enzymes upon Pfam functional annotation. Comput. Struct. Biotechnol. J. 2025, 27, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, Y.; Sun, J.; Zhu, T.; Pang, H.; Li, C.; Geng, W.C.; Wu, B. Computational redesign of a hydrolase for nearly complete PET depolymerization at industrially relevant high-solids loading. Nat. Commun. 2024, 15, 1417. [Google Scholar] [CrossRef] [PubMed]
- Passaro, S.; Corso, G.; Wohlwend, J.; Reveiz, M.; Thaler, S.; Somnath, V.R.; Getz, N.; Portnoi, T.; Roy, J.; Stark, H.; et al. Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction. bioRxiv 2025. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, T.; Khan, J.; Lakhani, S.; Li, A.; Vyas, A.; Hunt, J.; Espinosa Garcia, S.A.; Liang, B. The Crystallography of Enzymes: A Retrospective and Beyond. Crystals 2025, 15, 966. https://doi.org/10.3390/cryst15110966
Huang T, Khan J, Lakhani S, Li A, Vyas A, Hunt J, Espinosa Garcia SA, Liang B. The Crystallography of Enzymes: A Retrospective and Beyond. Crystals. 2025; 15(11):966. https://doi.org/10.3390/cryst15110966
Chicago/Turabian StyleHuang, Tianyi, Jannat Khan, Sheryar Lakhani, Albert Li, Aditya Vyas, Julia Hunt, Sara Andrea Espinosa Garcia, and Bo Liang. 2025. "The Crystallography of Enzymes: A Retrospective and Beyond" Crystals 15, no. 11: 966. https://doi.org/10.3390/cryst15110966
APA StyleHuang, T., Khan, J., Lakhani, S., Li, A., Vyas, A., Hunt, J., Espinosa Garcia, S. A., & Liang, B. (2025). The Crystallography of Enzymes: A Retrospective and Beyond. Crystals, 15(11), 966. https://doi.org/10.3390/cryst15110966






