Abstract
A novel 4H-benzo[h]chromene derivative was efficiently synthesized and structurally characterized as a β-enaminonitrile. Single-crystal X-ray diffraction confirmed its molecular structure, revealing a stable crystal lattice stabilized by intermolecular N–H···N hydrogen bonds and π–π stacking. The compound was evaluated for its inhibitory activity against both wild-type EGFR (EGFRWT) and the resistant T790M mutant (EGFRT790M). It exhibited moderate activity against EGFRWT (IC50 = 3.27 ± 0.72 μM) but demonstrated significantly enhanced potency against EGFRT790M (IC50 = 1.92 ± 0.05 μM), showing a low resistance factor compared to the reference drugs Erlotinib and Gefitinib. Comparative molecular docking studies against both wild-type and T790M mutant structures suggested that the compound maintains a stable binding mode involving key interactions with the hinge region residue Met769, rationalizing its ability to circumvent the T790M resistance mechanism. These findings identify the 4H-benzo[h]chromene scaffold as a promising lead for developing novel inhibitors to overcome EGFRT790M-mediated resistance.
Keywords:
4H-benzo[h]chromene; microwave irradiation; X-Ray; EGFR inhibitors; molecular docking; SAR 1. Introduction
A powerful technique essential for conducting in-depth structural analysis of materials across various fields, including materials science, chemistry, and biology, is single-crystal X-ray diffraction (SCXRD). Whether you’re researching new materials or the molecular structures of organic molecules, SCXRD offers precise information [1]. SCXRD research has uncovered the solid-state structures and conformation preferences of chemicals produced from pyrans. For example, 4-(2-ClC6H4) substituent of 6-Cl of 4H-chromene-benzene hybrid [2], and 4-(4-FC6H4), 4-(4-BrC6H4), 4-(4-NO2C6H4), 4-(2,3-Cl2C6H3) derivatives of 6-OMe of 4H-chromene-benzene hybrids [3,4,5,6]. Furthermore, N’-[3-CN-4-FC6H4)-6-OMe-4H-benzo[h]chromen-2-yl]-N,N-dimethylmethanimidamide [7], 4-(4-FC6H4) and 4-(4-BrC6H4) substituent of ethyl 6-OMe of 4H-chromene-benzene hybrid [8,9].
Additionally, 1-C6H5/1-(4-FC6H4)/1-(4-BrC6H4)/1-(4-OMeC6H4)/1-(2,6-F2C6H3)/1-(2,4-Cl2C6H3)/(2,5-Cl2C6H3)/1-(3,5-Br2-2-OMeC6H2) derivative of 8-OMe of 1H-chromene-benzene hybrids [10,11,12,13,14,15,16,17], 1-(4-OMeC6H4) of 1H-chromene-benzene hybrid [18], 1-C6H5/1-(4-FC6H4)/1-(4-ClC6H4)/1-(4-BrC6H4)/1-(4-OMeC6H4)/1-(4-MeC6H4) derivative of 9-OMe of 1H-chromene-benzene hybrids [19,20,21,22,23,24], and 1-(2,5-Cl2C6H3)/1-[3,4-(OMe)2C6H3] derivative of 8-Br of 1H-chromene-benzene hybrids [25,26].
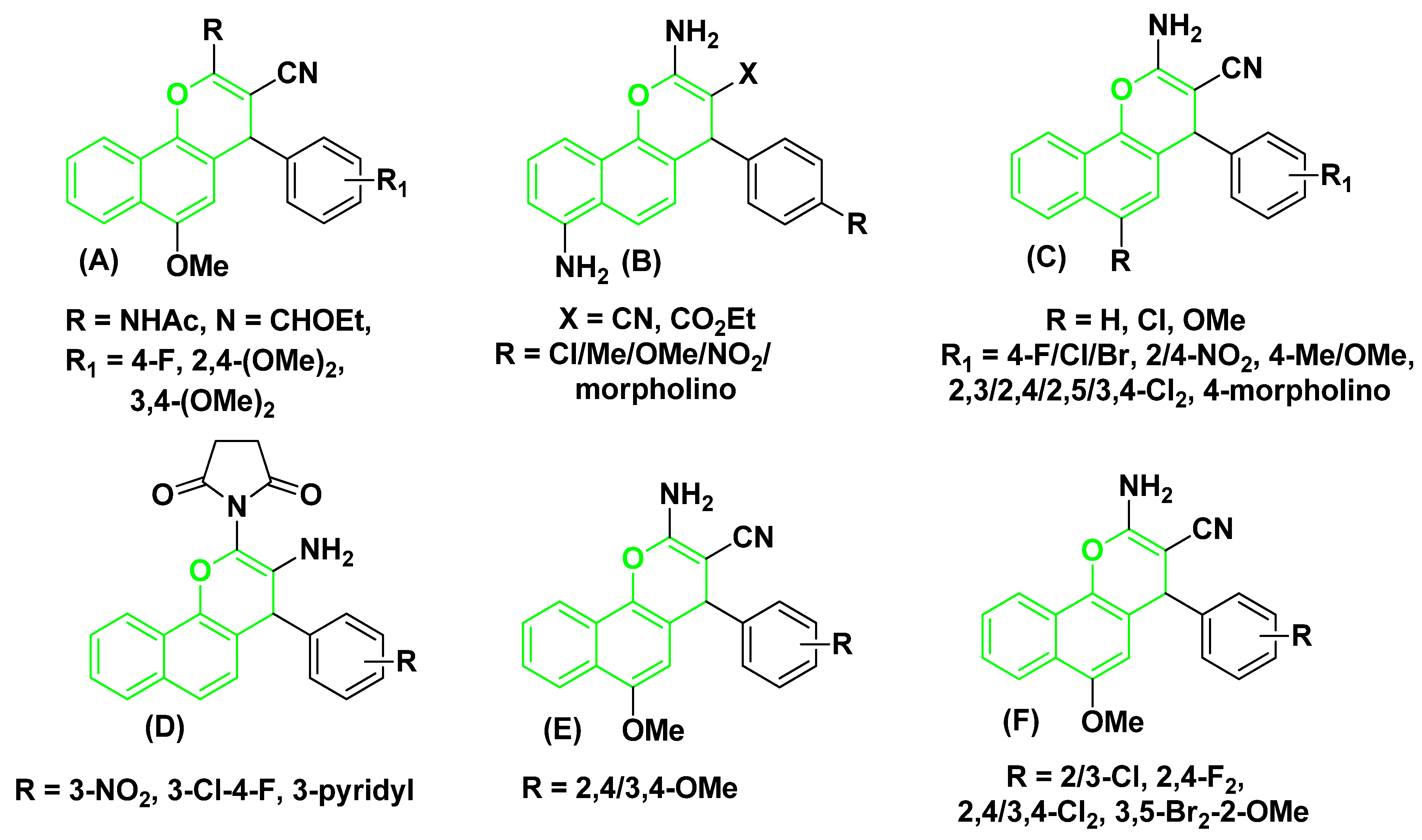
There are many uses for chromene-benzene hybrids in the production of common resources, pigments, modern medications, and agricultural products. According to several recent studies, most chemotherapy drugs promote apoptosis. For instance, the 4-aryl substituent of 2-NHCOCH3/N=CHOEt of 4H-chromene-benzene hybrids (A) stops the cell cycle. It causes cell death in human cancer cells by blocking topoisomerases and tubulin [27,28,29], and the 3-CN/CO2Et substituent of 7-NH2 of 4H-chromene-benzene hybrids (B), 4H-chromene-benzene hybrids and their 6-Cl/OMe (C) cause’s cell death in human cancer cells [30,31,32,33]. Additionally, as shown in Figure 1, the 2-N-succinimido derivatives (D) exhibit an anti-rheumatic effect [34]. At the same time, the [2,4/3,4-(OMe)2C6H3] substituent of 6-OMe of 4H-chromene-benzene hybrids (E) demonstrated antimicrobial effects [35], and halogen derivatives of 4H-chromene-benzene hybrids (F) produced strong anticancer analogs that targeted the c-Src Kinase enzyme [36].
Figure 1.
Structures of some 4H-chromene-benzene hybrids (A–F) (green-highlighted) with biological effects.
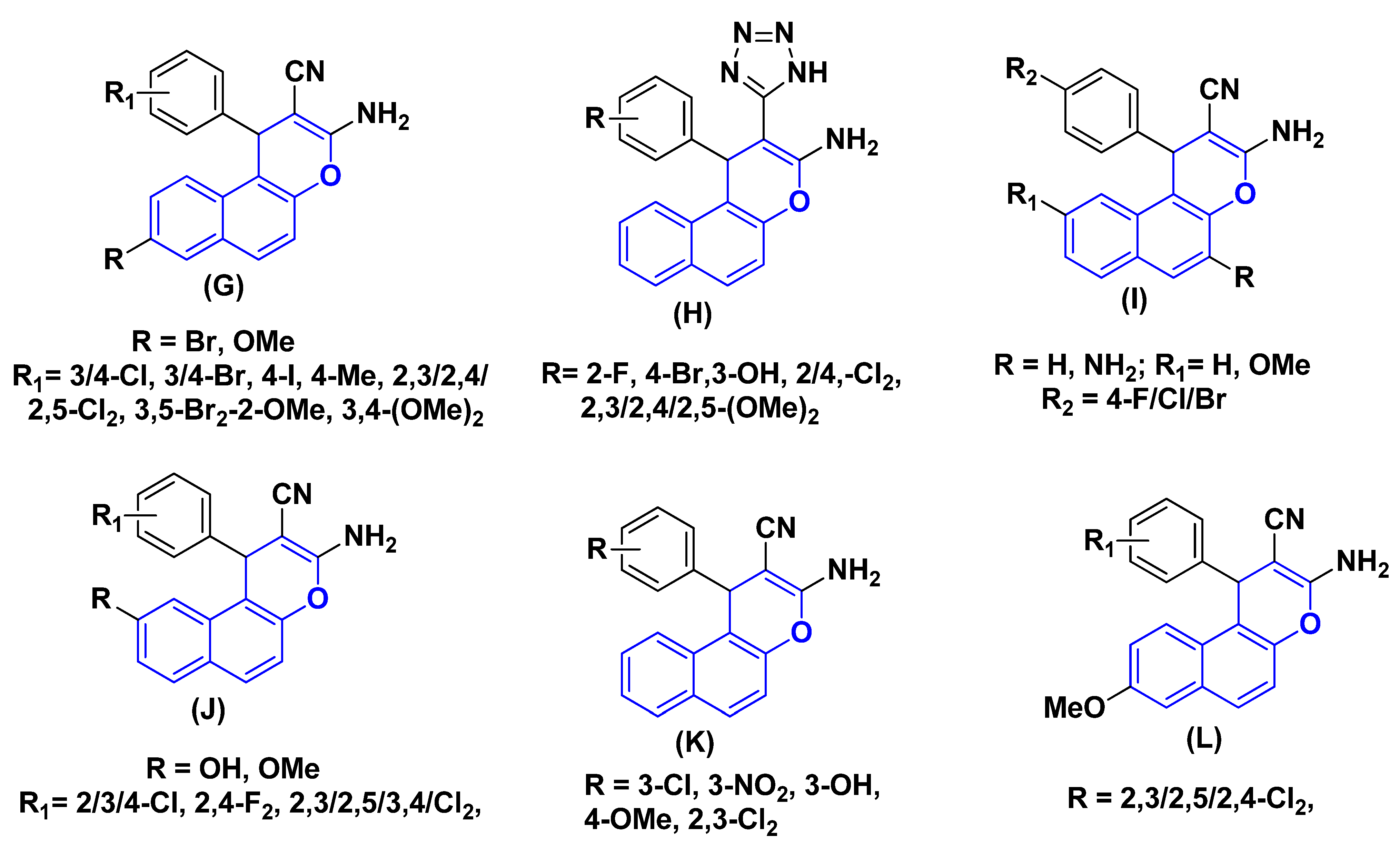
One of the most practical heterocyclic systems, 1H-chromene-benzene hybrids exhibits a range of pharmacological activities. For instant, the 8-Br/OMe substituent (G) causes cell death, stops the cell cycle in the G2/M, S, and S-G2/M phases, increases caspase production, and aids human cancer cells in their struggle against c-Src kinase by simultaneously blocking topoisomerase I and II [37,38,39,40,41]. Additionally, a series of 1-aryl-substituted 2-(1H-tetrazol-5-yl) of 1H-chromene-benzene hybrids (H), some derivatives of 3,5-diamino and 3-amino of 1H-chromene-benzene hybrids (I) have been reported to exhibit cytotoxic and apoptotic effects against a variety of human cancer cell lines [31,42,43,44], 9-OH/OMe substituents of 1H-chromene-benzene hybrids (J) have a strong effect on MCF7/ADR, have a strong effect on MCF7/ADR, block P-glycoprotein [24,45,46], and the β-enaminonitrile compounds (K) exhibit antiproliferative and c-Src kinase inhibitory activities [47]. Lastly, as seen in Figure 2, the dichloroaryl substituents of 1H-chromene-benzene hybrids (L) had strong antibacterial and anticancer properties [48].
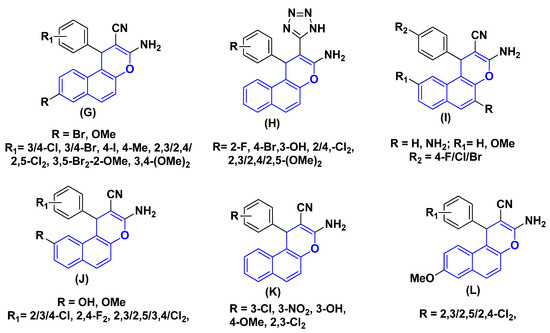
Figure 2.
Structure of some 1H-chromene-benzene hybrids (G–L) (blue highlighted) with biological effects.
The epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase that plays a critical role in regulating cell proliferation, survival, and differentiation. Its aberrant activation, due to overexpression or mutational activation, is a well-established driver in various cancers, particularly non-small cell lung cancer (NSCLC) [49,50]. This has made EGFR a prime target for cancer therapy, leading to the development of several generations of small-molecule tyrosine kinase inhibitors (TKIs). First-generation TKIs (e.g., Gefitinib, Erlotinib) are effective in patients with activating EGFR mutations (such as L858R); however, acquired resistance almost invariably develops, limiting their long-term efficacy [51,52,53].
The most prevalent mechanism of this acquired resistance, accounting for approximately 50–60% of cases, is the T790M ‘gatekeeper’ mutation in exon 20 [54]. This mutation involves the substitution of a threonine residue with a bulkier methionine (T790M) within the ATP-binding pocket. This change confers resistance through two key mechanisms: it sterically hinders the binding of first-generation TKIs, and it increases the affinity of the pocket for ATP, thereby outcompeting the inhibitor [55]. Consequently, there is a critical and ongoing need for new inhibitors capable of effectively targeting EGFRT790M to overcome this clinical challenge. The development of such agents, including the third-generation TKI Osimertinib, represents a major advance in the field. Our work aims to contribute to this effort by evaluating a novel chromene-based scaffold for its potential to inhibit this therapeutically paramount resistant mutant.
The present study is an extension of our ongoing efforts and those of other groups towards developing potent pyran-based biological agents [56,57,58,59]. The numerous chromene-benzene hybrids reported in the literature, including those from our work and others, underscore the significance of this scaffold in medicinal chemistry. These prior studies demonstrate that substitutions at the 4-position of the 4H-chromene core and the 1-position of the 1H-chromene core are critical for modulating biological activity, and have yielded compounds with diverse and potent effects, including antitumor and kinase inhibitory properties. Building on this established structure-activity relationship (SAR), the current work independently focuses on the design and synthesis of a new 4H-chromene-benzene hybrid, 2-amino-6-methoxy-4-(2-bromophenyl)-4H-benzo-4H-chromene-3-carbonitrile (4), featuring a less common ortho-bromophenyl substituent. We then evaluate its inhibitory effects against both wild-type EGFR (EGFRWT) and the resistant T790M mutant (EGFRT790M), to explore its potential in overcoming a major mechanism of drug resistance.
2. Experimental Section
2.1. Materials and Equipment
Sigma-Aldrich Chemical Co., Ltd. (Sigma-Aldrich Corp., St. Louis, MO, USA) supplied all the chemicals and solvents. The progress of the reaction was monitored by analytical thin-layer chromatography (TLC, n-hexane/ethyl acetate 1:3) on pre-coated silica gel 60 F254 plates (Merck, Darmstadt, Germany). Visualization was achieved using a UV lamp at 254 nm. The uncorrected melting point was determined using equipment from Stuart Scientific Co., Ltd. (Companies House – GOV, UK) Fourier-Transform Infrared (FT-IR) Spectroscopy (JASCO International Co., Ltd., Tokyo, Japan) was performed on a Jasco FT/IR 460 plus spectrophotometer. Spectrum was recorded in the range of 4000–400 cm−1 using a potassium bromide (KBr) disk method. Nuclear Magnetic Resonance (NMR) Spectroscopy: 1H and 13C NMR spectra were recorded at 400 MHz and 100 MHz, respectively, on a Varian 400/100 MHz NMR spectrometer (Varian, Palo Alto, CA, USA). The NMR chemical shifts in ppm, which are the same as the remaining solvent peak (1H/13C-NMR) at δ 2.50 and 39.52 for DMSO-d6. Mass Spectrometry (MS): Electron-Impact (EI) mass spectrum was obtained using a Shimadzu GC/MS-QP5050A spectrometer with a direct inlet system. (Shimadzu Corporation, a Japanese company with headquarters in Kyoto, Japan). The ionization energy was 70 eV.
Single crystal X-ray diffraction data (SCXRD) was collected using a Rigaku Oxford Diffraction XtaLAB Synergy-S (Tokyo, Japan), equipped with two microfocus PhotonJet-S sources, Cu Kα radiation (λ = 1.54184 Å), Mo Kα radiation (λ = 0.71073 Å). Elemental Analysis (CHN) was performed on a PerkinElmer 2400 Series II CHNS/O Analyzer (Waltham, MA, USA). The results for carbon, hydrogen, and nitrogen are within ±0.06% of the theoretical values.
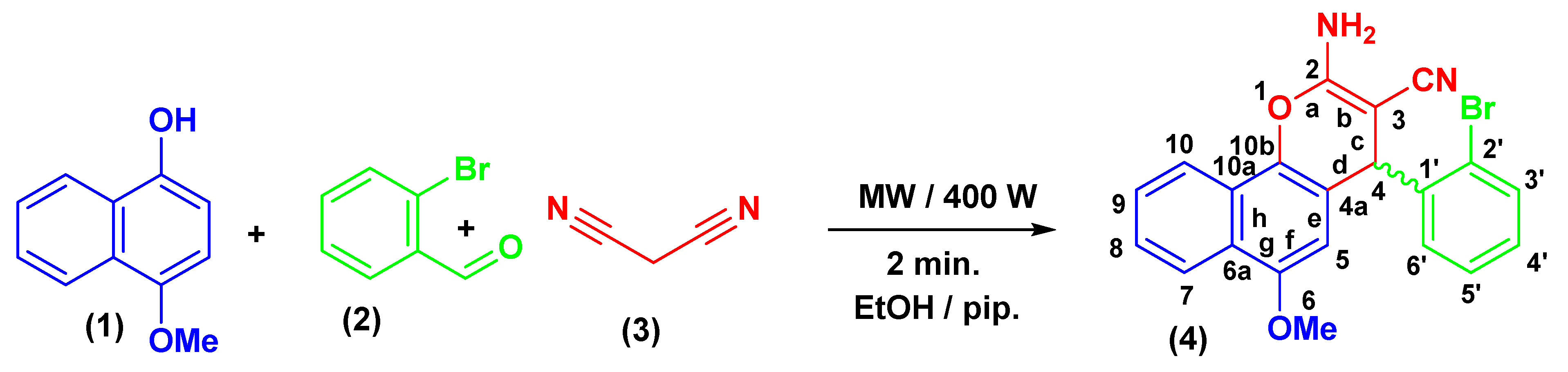
2.2. Synthesis of 2-amino-6-methoxy-4-(2-bromophenyl)-4H-benzo[h]chromene-3-carbonitrile (4)
4-Methoxynaphthalen-1-ol (1) (0.01 mol), 2-bromobenzaldehyde (2) (0.01 mol), malononitrile (3) (0.01 mol), and piperidine (0.5 mL) were combined with 30 mL of 100% ethanol using a 400 W Microwave irradiation probe set at 140 °C for two minutes. The reaction mixture was allowed to cool to room temperature, the solids that formed was filtered out, and then it was cleaned with methanol before being recrystallized using ethanol to give 4 as colorless crystals; yield 93%; m.p. 257–258 °C; IR (FTIR/KBr) υ (cm− 1): 3413, 3321, 3196 (NH2), 2201 (CN); 1H-NMR δ: 8.17–6.35 (m, 11H, Ar-H and NH2), 5.33 (s, 1H, 4H-pyran), 3.72 (s, 3H, OMe); 13C NMR δ: 161.01 (C-2), 151.70 (C-6), 144.29 (C-1′), 137.39 (C-10b), 133.40 (C-3’), 131.55 (C-6’), 129.58 (C-4’), 129.08 (C-5’), 127.84 (C-9), 126.85 (C-8), 124.97 (C-10a), 124.04 (C-6a), 122.76 (C-2’), 122.08 (C-7), 121.12 (C-10), 120.65 (C-4a), 117.01 (CN), 102.68 (C-5), 55.99 (Me), 55.35 (C-3), 41.18 (C-4); MS m/z (%): 408 (M+ +2, 3.11), 406 (M+, 3.23) with a base peak at 43 (100); Anal. Calcd for C21H15BrN2O2 (407.25): C, 61.93; H, 3.71; N, 6.88. Found: C, 61.99; H, 3.76; N, 6.94%. The high purity of the compound, as confirmed by elemental analysis and the absence of extraneous signals in the NMR spectra, ensured reliable biological evaluation.
2.3. X-Ray Crystallography Analysis
Single crystal X-ray diffraction data (SCXRD) was collected using a Rigaku Oxford Diffraction XtaLAB Synergy-S, equipped with two microfocus PhotonJet-S sources, Cu Kα radiation (λ = 1.54184 Å), Mo Kα radiation (λ = 0.71073 Å), and a HyPix-6000HE hybrid photon counting detector. Data processing and refinement were carried out using CrysA-lisPro and Olex2 software (Version: 38.46) (https://rigaku.com/products/crystallography/x-ray-diffraction/crysalispro and https://www.olexsys.org/olex2/, both accessed on 18 August 2024), respectively. Compound 4 was obtained as single crystals by slow evaporation from an ethanol solution of the pure compound at room temperature with CCDC 2424373. All crystallographic data of the compound 4 can be obtained free of charge from the Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif (accessed on 16 February 2025, 11:58 PM) and the CIF file was cited in Supplementary Materials.
2.4. Biological Screening
In Vitro EGFR Inhibition Assay
With Erlotinib and Gefitinib as positive control, the 4H-chromene-benzene hybrid (4) was tested for their activities against both EGFRWT and EGFRT790M, using Erlotinib and Gefitinib as positive controls. Homogeneous time resolved fluorescence (HTRF) assay was applied in this test [60] with EGFRWT and EGFRT790M (Sigma). The assay was performed using recombinant human EGFR kinase domains (EGFRWT and EGFRT790M) purchased from Sigma-Aldrich (USA). Firstly, EGFRWT and/or EGFRT790M and their substrates were incubated with the tested compound in enzymatic buffer for 5 min. ATP (1.65 μM) was added into the reaction mixture to allow the enzymatic reaction to start. The assay was conducted for 30 min at room temperature. The reaction was stopped by the addition of detection reagents which contain EDTA. The detection step continued for 1 h, and then the IC50 values were determined using GraphPad Prism 5.0. Three independent experiments were performed for each concentration.
2.5. Molecular Docking
Molecular docking simulations were performed to predict the binding mode, affinity, and key interactions of the synthesized compound 4 and the reference drug Erlotinib within the active site of the EGFR kinase domain (PDB ID: 4HJO). All computational studies were conducted using the AutoDock 4.2.6 software suite. The three-dimensional crystal structure of the EGFR tyrosine kinase domain in complex with a quinazoline inhibitor was retrieved from the Protein Data Bank (http://www.rcsb.org, PDB ID: 4HJO, accessed on 15 August 2025). The protein structure was prepared for docking by removing all water molecules, heteroatoms, and the native co-crystallized ligand. Polar hydrogen atoms were added, and Kollman united atom charges were assigned using AutoDock Tools (ADT) version 1.5.6. The protein file was then saved in the required PDBQT format. The three-dimensional structure of compound 4 was optimized using density functional theory (DFT) at the B3LYP/6-31G(d,p) level of theory to obtain its minimum energy conformation. The geometry-optimized structure was subsequently used for docking. The 3D structure of Erlotinib was obtained from the PubChem database (CID: 176870). Both ligand structures were prepared by adding Gasteiger charges and defining rotatable bonds. The final structures were converted to the PDBQT format using AutoDock Tools. A grid box was defined to encompass the active site of the EGFR protein, centered on the original co-crystallized ligand to ensure comprehensive sampling of all potential binding poses. The grid dimensions were set to 60 × 60 × 60 points with a grid spacing of 0.375 Å. The center coordinates of the grid box were set to x = 13.862, y = 5.907, z = 53.413, which adequately surrounded key catalytic and hinge region residues, including Met769, Gln767, Thr830, and Leu768. The resulting docking poses were clustered based on a 2.0 Å RMSD tolerance and ranked according to their calculated binding free energy (ΔG, kcal/mol). The pose with the most favorable binding energy within the most populous cluster was selected for detailed analysis of protein–ligand interactions. Hydrogen bonds, hydrophobic interactions, and binding distances were analyzed and visualized using PyMOL Molecular Graphics System. (Version: 3.1.0.).
3. Results and Discussion
3.1. Chemistry
The creation of 4-aryl-substituted derivative of the 4H-chromene-benzene hybrid (4) is shown in Scheme 1. A 400 W Microwave irradiation probe set at 140 °C for two minutes easily produced a substituted phenyl at the 4-position of 4H-chromene-benzene hybrid (4) in a high yield using a one-pot, three-component heterocyclocondensation process of 4-methoxynaphthalen-1-ol (1) with 2-bromobenzaldehydes (2) and malononitrile (3) in ethanolic piperidine solution, as shown in Scheme 1. Given that compound 4 is chiral, the precise rotation of the 4H-chromene-benzene moiety at position 4 was measured using the Carl Zeiss polarimeter. The purpose of this was to determine the racemic stereochemistry of that location. According to our research, chemical 4 does not spin as expected [61,62].
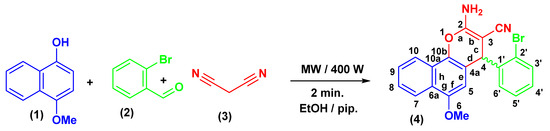
Scheme 1.
Synthesis of 4H-chromene-benzene hybrid (4).
3.2. Spectroscopic Data
Compound 4 IR spectra at u 3413, 3321, and 3196 cm−1 helped find the NH2 group. The CN group was found at u 2201 cm−1. The CH and MeO protons were found at δ 5.33 ppm and δ 3.72 ppm in the 1H-NMR data of 4, while the CH and MeO carbons at δ 41.18 ppm and δ 55.99 ppm were identified as the resonating signals in the 13 C-NMR data of 4. Furthermore, SCXRD and mass spectra of compounds 4 (see Supplementary Data Figures S1 and S2) provided a conclusive verification for the desired molecule.
3.3. Single-Crystal X-Ray Diffraction Analysis and Structural Elucidation of Compound 4
Single-crystal X-ray diffraction (SCXRD) is the definitive technique for unambiguously determining the 3-D molecular and supramolecular structure of crystalline materials. For novel synthetic 4H-chromene-benzene moiety (4), SCXRD provides critical information on molecular geometry, conformation, stereochemistry, and intermolecular interactions, all of which are essential for understanding its physicochemical properties and biological activity, particularly its role as an EGFR inhibitor as described in the accompanying manuscript. In the context of this study, the SCXRD analysis of compound 4 serves several key purposes: it unambiguously confirms the molecular structure synthesized; it reveals the solid-state conformation, which is used as the validated input for molecular docking studies; and it elucidates the supramolecular packing, which directly influences the compound’s physicochemical properties (e.g., melting point, solubility) and stability. This work presents a comprehensive analysis of the crystal structure of compound 4 (CCDC 2424373), elucidating its molecular geometry, key bond parameters, conformational features, and the crystal packing forces that stabilize its solid-state arrangement.
3.3.1. Crystal System and Symmetry
The compound 4 crystallizes in the monoclinic crystal system with the space group P21/c (No. 14), a common and centrosymmetric space group for organic molecules. The inversion center at (0, 0, 0) implies that every atom has a symmetric counterpart related by the operation (−x,−y,−z). The presence of inversion symmetry means the crystal is a racemic mixture; the asymmetric unit contains one molecule, and its enantiomer is generated by symmetry operations within the lattice. This is consistent with the polarimetry results, which indicated the bulk synthesized sample was racemic. This symmetry significantly influences the crystal packing and the distribution of molecular dipoles, potentially minimizing electrostatic repulsions and stabilizing the lattice through balanced intermolecular forces. The unit cell parameters are a = 13.915(2) Å, b = 13.1400(1) Å, c = 9.7627(1) Å, and β = 102.418(1)°. The asymmetric unit contains one discrete molecule of C21H15BrN2O2, confirming the molecular formula established by spectroscopic and elemental analysis. The crystal was measured at room temperature (298.0 K), and the refinement statistics indicate excellent data quality (R1 = 0.0388, wR2= 0.0782, GOF = 1.038), suggesting reliable atomic positions and displacement parameters (Table 1).

Table 1.
Experimental (SCXRD) crystal data and structure refinement for compound 4.
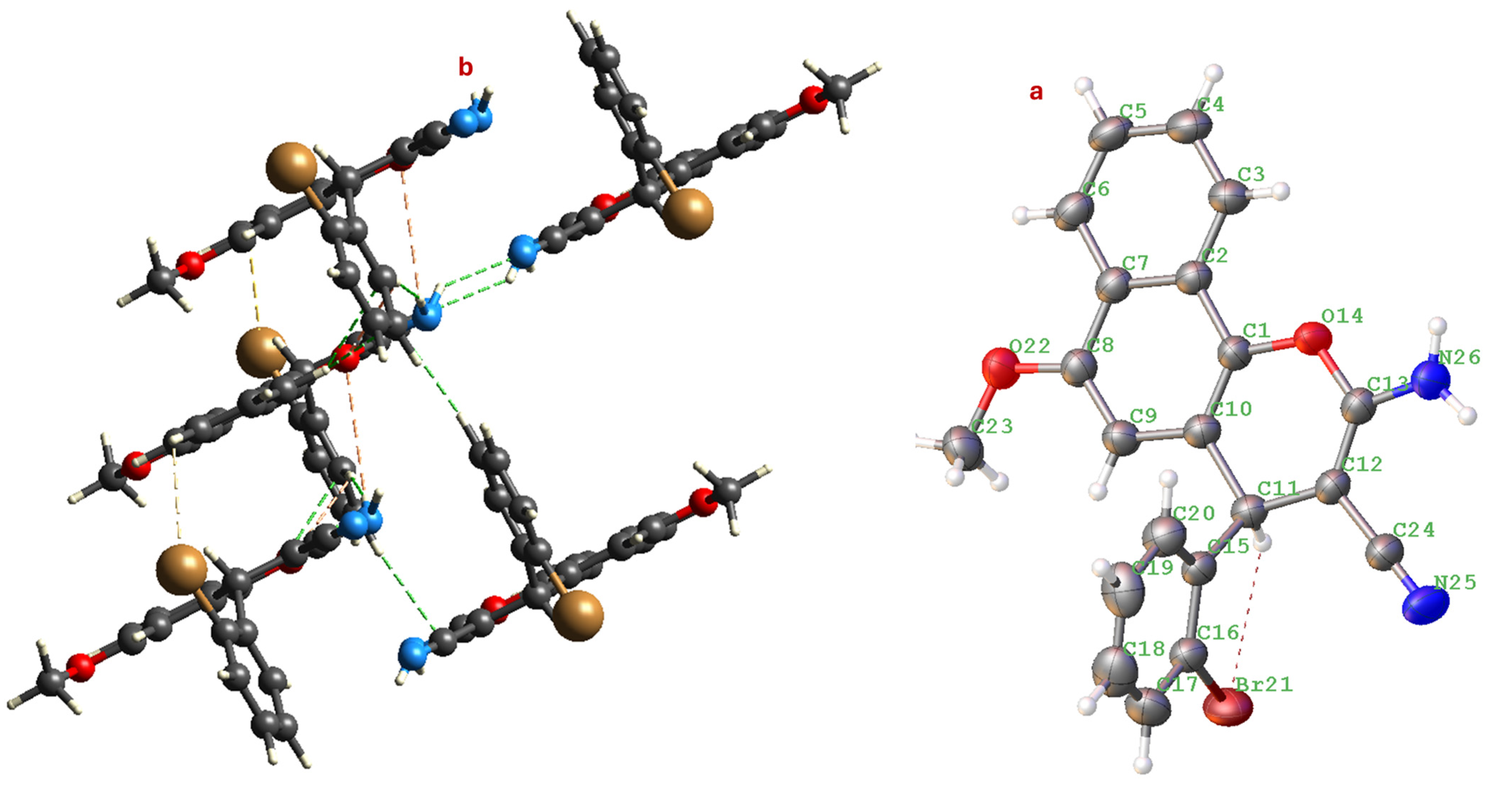
The molecular structure reveals that the central pyran ring adopts a slightly distorted half-chair conformation (Figure 3), a typical feature for 4H-chromene-benzene moiety (4). The mean plane of the naphthalene system is nearly planar, as expected. The 2-bromophenyl substituent at the 4-position of the pyran ring is oriented approximately perpendicular to the mean plane of the naphthalene moiety, a conformation likely stabilized to minimize steric clashes.
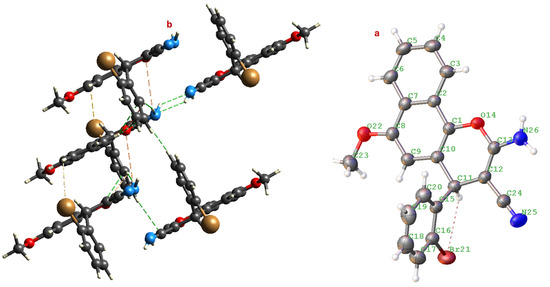
Figure 3.
(a) Molecular structure of compound 4 showing the atom numbering scheme. (b) View of the one-dimensional chain formed by N-H···N hydrogen bonds (shown as dashed blue lines) running along the c-axis. Displacement ellipsoids are drawn at the 50% probability level.
3.3.2. Key Geometric Parameters
Bond Lengths
Analysis of the bond lengths provides insight into the electronic structure and conjugation within the molecule. The C-Br bond length in C15-C16 is measured at 1.907(3) Å, which falls within the expected range for an aromatic C(sp2)-Br bond and is consistent with reported values for similar bromophenyl systems. The C-N bond length of the exocyclic amino group is 1.349(3) Å. This value is significantly shorter than a standard C-N single bond (~1.47 Å) and indicates substantial double-bond character due to resonance with the adjacent cyano group and the ring system (C12-C13-N26). This is a hallmark of an β-enaminonitrile system. The nitrile bond N25-C24 (C≡N) length is 1.144(6) Å, which is characteristic of a carbon-nitrogen triple bond. The bond C12-C24 (C-C≡N) connecting the cyano group to the chromene core is 1.415(4) Å. This length is intermediate between a single and double bond, consistent with the conjugation extending from the naphthalene system, through the pyran ring, into the β-enaminonitrile moiety (C12-C13-N26 and C12-C24≡N25). This extended conjugation is crucial for the electronic properties and potentially for the biological activity of the molecule. The C-O bond lengths in the pyran ring are 1.396(3) Å (C1-O14) and 1.352(3) Å (C13-O14). The difference is consistent with C13 being part of the β-enaminonitrile system, influencing its electron density.
Bond Angles
The bond angles around key atoms further define the molecular geometry. The endocyclic bond angle at the sp3-hybridized C4 atom (C3-C4-C5, C11-C4-C3, etc.) is approximately 109.5°, confirming its tetrahedral geometry. The bond angle at the ether oxygen O14 (C1-O14-C13) is 118.39(18)°, which is typical for an aromatic C-O-C linkage. The angles within the naphthalene system are all close to 120°, confirming the sp2 hybridization and planarity of these carbon atoms.
Torsion Angles and Molecular Conformation
Torsion angles define the conformation of the molecule. The most significant torsion angle involves the orientation of the 2-bromophenyl ring relative to the 4H-chromene-benzene system. The torsion angle C11-C15-C16-Br21 is −6.70°, indicating that the bromophenyl ring is nearly coplanar with the bond connecting it to the tetrahedral C4 atom. However, due to the ortho-bromo substituent, the entire phenyl ring is twisted out of the plane of the chromene system to avoid steric hindrance. This is observed in the torsion angle C20-C15-C16-Br21 (176.90°) and others, which show the ring is rotated.
The torsion angle C1-O14-C13-N26 is −177.64°, demonstrating that the amino group and the pyran oxygen are in an anti-periplanar conformation. This alignment is ideal for the resonance that stabilizes the β-enaminonitrile system.
3.3.3. Intermolecular Interactions and Crystal Packing
Intermolecular interactions and 2D- and 3D-crystal packing in 2-amino-6-methoxy-4-(2-bromophenyl)-4H-benzo[h]chromene-3-carbonitrile (4). The single-crystal X-ray diffraction study of compound 4 reveals a complex and well-organized crystal structure governed by a delicate balance of intermolecular interactions. These interactions—hydrogen bonding, π–π stacking, halogen bonding, and Van der Waals forces, dictate the molecular packing, stability, and physical properties of the solid-state assembly. This analysis integrates data (Table 2) to provide a comprehensive understanding of the intermolecular interactions and their manifestation in both two-dimensional (2D) layers and three-dimensional (3D) networks.

Table 2.
Selected non-covalent geometric parameters (Å, °) in compound 4.
Primary Supramolecular Synthon: The Robust N-H···N Hydrogen Bond
The most significant and directional intermolecular interaction in the crystal structure of 4 is the classical hydrogen bond between the exocyclic amino group (N26-H26A/B) and the nitrile nitrogen (N25) of a symmetry-related molecule. The N···N distance is significantly shorter than the sum of their van der Waals radii (3.10 Å), confirming a strong, attractive interaction. The angle, while not ideal (180° is ideal), is well within the acceptable range for a strong hydrogen bond. This interaction is generated by the symmetry operation (−x, −y, 1 − z), which translates the molecule and creates an inversion-related dimer linked by this bond. This N-H···N hydrogen bond acts as the primary supramolecular synthon. It links molecules head-to-tail into infinite one-dimensional (1D) chains propagating along the [001] direction (the c-axis). This is a common and highly effective strategy for creating linear, stable architectures in crystals of molecules featuring both hydrogen bond donors and acceptors.
Secondary and Weak Intermolecular Interactions: Halogen-Mediated Interactions (Type I and II)
The crystal structure is further consolidated by a network of weaker, but cumulatively very important, interactions that define the higher-dimensional packing. The bromine atom, being a large, polarizable halogen, plays an active role in the packing. A notable interaction occurs between the bromine atom (Br21) and the methoxy oxygen (O22) of a neighboring molecule (symmetry op: 1 − x, −1/2 + y, 1/2 − z). The Br···O distance is 3.187 Å, which is just slightly less than the sum of their Aan der Waals radii (3.37 Å). This qualifies it as a halogen-centric interaction. The geometry (C-Br···O angle 150°) suggests this is not a highly directional halogen bond (which prefers a 180° angle) but rather a Type I halogen contact, which is still electrostatically favorable and contributes to layer formation.
Several C-H groups (C-H···π Interactions) act as weak donors to oxygen acceptors. For instance, atom H11 (on C11) donates to the methoxy oxygen O22 (symmetry op: x, 1/2 − y, −1/2 + z) with an H···O distance of 2.87 Å. These interactions, while weak individually, form a pervasive network that “stitches together” the structure in all directions. The electron-rich aromatic systems (naphthalene and phenyl rings) attract weak hydrogen bonds from aliphatic and aromatic C-H groups. For example, the contact between H19 and the centroid of a nearby naphthalene ring (not explicitly listed but observable in the full structure) would be a typical C-H···π interaction, adding to the cohesion energy.
The molecules are nearly planar, facilitating face-to-face π-π stacking interactions between the extended benzo[h]chromene systems of adjacent molecules. The stacking is not perfectly eclipsed but is offset or slipped. This offset stacking maximizes the attractive dispersion forces between the π-electron clouds while minimizing repulsive interactions between the atoms. The interplanar distance between the mean planes of stacked naphthalene systems is approximately 3.4–3.6 Å, which is ideal for strong π-π interactions. This stacking is crucial for forming two-dimensional sheets in the ab-plane and provides significant stability to the crystal lattice, contributing to the compound’s high melting point (257–258 °C).
Construction of the 3D Architecture: From 1D Chains to a 3D Network
The crystal packing is an elegant hierarchical assembly driven by the interplay of the interactions described above. Step 1: Formation of 1D Chains: The strong N-H···N link molecules into robust linear chains running parallel to the c-axis ([001] direction). This is the first level of organization. Step 2: formation of 2D sheets via halogen contacts and π-stacking. These 1D chains do not exist in isolation. They are connected sideways (in the ab-plane) through two main mechanisms: The Br···O halogen contacts and various C-H···O interactions link adjacent chains together. Simultaneously, the offset π-π stacking between the large, flat chromene systems of molecules in neighboring chains efficiently bundles the chains into corrugated 2D layers lying approximately parallel to the (100) plane. Step 3: Interlayer stacking into a 3d framework, finally, these 2D layers stack along the a-axis. The layers are held together primarily by Van der Waals forces between the hydrophobic regions of the molecules. the interdigitation of the 2-bromophenyl substituents from one layer into the “pockets” of the adjacent layer. Additional, weaker C-H···π and C-H···Br interactions that provide stability to the stack. This results in a highly stable, densely packed, and complex 3-D supramolecular architecture.
This intricate packing explains the excellent quality of the crystals obtained and their suitability for high-resolution SCXRD analysis. From a functional perspective, the confirmed planar conformation of the chromene core is a critical feature for its biological activity, as it mimics the flat heterocyclic systems of known ATP-competitive kinase inhibitors like Erlotinib. The specific orientation of the 2-bromophenyl group, locked in place by both intramolecular sterics and intermolecular Br···O contacts, could be a key determinant of its selectivity profile against the EGFRT790M mutant, potentially facilitating unique interactions within the hydrophobic pocket of the mutated enzyme.
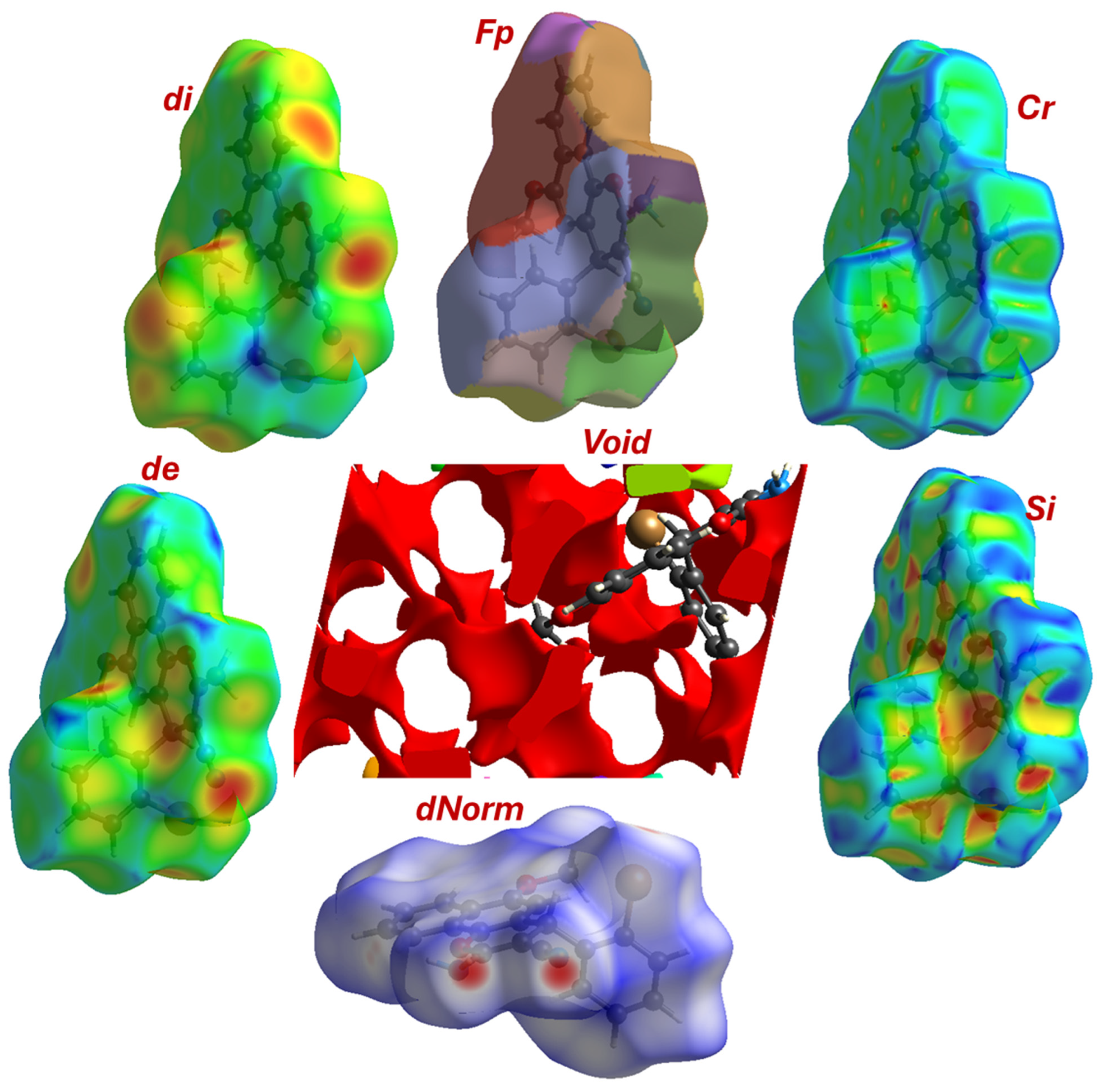
3.4. Molecular Packing for Compound 4 Based on Hirschfeld (HF) Profile
Hirshfeld surface (HS) analysis is a powerful computational technique that provides a visual and quantitative representation of intermolecular interactions in a crystal lattice by partitioning space into regions where the electron distribution of a sum of spherical atoms for the molecule of interest dominates over that of the surrounding molecules. This method offers an intuitive way to decode the complex network of interactions that define crystal packing, complementing traditional X-ray diffraction analysis. Surfaces are typically mapped with properties like dnorm into two-dimensional fingerprint plots that quantify the contribution of each atom pair interaction to the total surface. This analysis is presented for the title compound 4 to quantitatively deconstruct its supramolecular architecture.
3.4.1. General Surface Information and Molecular Shape
The HF was generated with a standard high resolution and an isosurface value of 0.5 e·Å3. The calculated surface has a volume of 428.03 Å3 and a surface area of 377.16 Å2.
The Globularity (0.728) parameter measures how spherical the molecule is, with a value of 1.0 representing a perfect sphere. The value of 0.728 indicates a significant deviation from sphericity. This is entirely expected given the complex, multi-ring, and substituted structure of 4, which features a flat, planar 4H-benzo[h]chromene core with protruding substituents (the bromophenyl and amino-nitrile groups), resulting in an elongated and anisotropic molecular shape.
The asphericity (0.109) value quantifies the deviation of the molecule’s shape from that of a perfect sphere. A value of zero indicates a spherical shape. The positive value of 0.109 further confirms the non-spherical, anisotropic nature of the molecule, consistent with its extended π-conjugated system and the steric bulk of the bromine atom.
3.4.2. Topological Features
The HF was mapped with key properties to visualize intermolecular contacts, dᵢ and dₑ: These represent the distance from the surface to the nearest atom interior (dᵢ) and exterior (dₑ) to the surface. The similar mean values (~1.76 Å) for both parameters suggest a relatively balanced and efficient packing of molecules around the surface of the central molecule (Figure 4). dₙₒᵣₘ is the most informative mapping. It is defined by the equation dnorm = (dᵢ − rᵢʷᵈᵂ)/rᵢʷᵈᵂ + (dₑ − rₑʷᵈᵂ)/rₑʷᵈᵂ, where rʷᵈᵂ is the Van der Waals radius. Regions where dₙₒᵣₘ is negative indicate contacts shorter than the sum of Van der Waals radii (strong interactions), while values around zero represent Van der Waals contacts, and positive values indicate longer, weaker contacts.
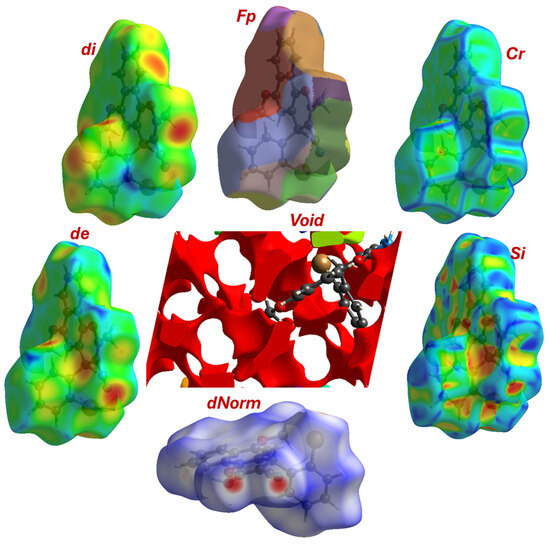
Figure 4.
Topological maps parameters including, Cr; Si, dnorm, de, di, and FP.
The mean dnorm value of 0.482 indicates that the majority of the surface is involved in contacts around the sum of Van der Waals radii. The minimum value of −0.337 and the maximum of 1.333 confirm the presence of a range of interactions, from significantly shorter-than-VdW (strong, attractive) to longer-than-VdW (weak) contacts. The red spots on the dnorm surface (corresponding to the most negative values) would pinpoint the locations of the strongest interactions, such as the N-H···N hydrogen bond and the Br···O contact identified in the crystallographic analysis (Figure 4). The Shape Index (SI) and Curvedness (Cr) parameters are sensitive to π-π stacking interactions. The shape index ranges from −1.0 to 1.0, with adjacent red and blue triangles indicating complementary “bumps” and “hollows” characteristic of stacked aromatic rings. The curvedness measures how flat or curved the surface is, with low, flat regions indicating planar stacking. The reported range of both values (SI: −0.998 to 0.998; Cr: −3.950 to 0.355) is strongly indicative of significant face-to-face π-π stacking between the large, flat 4H-benzo[h]chromene systems in the crystal lattice.
3.4.3. Analysis of Crystal Voids and Packing
The analysis of void spaces within a crystal structure is critical for understanding its packing efficiency and stability. Voids are the empty spaces between molecules in a crystal lattice. Their size, shape, volume, and distribution are direct consequences of the molecular shape and the nature of the intermolecular interactions that dictate the packing arrangement. Tight, efficient packing with minimal void space is often associated with high crystal density and stability. This analysis examines the voids in the crystal structure of (4). The void analysis was performed with a high-resolution isosurface value of 0.002 au, a standard value for mapping the outer limits of electron density in interstitial regions. Total Void Volume: 193.90 Å3, while total Void Surface Area: 624.81 Å2. The very low globularity (0.259) value indicates that the overall void space is highly non-spherical. It is not a single, round cavity but rather a complex, interconnected channel or a series of flattened, slit-like pores. This is a classic signature of a structure packed primarily through flat, planar 4H-benzo[h]chromene core with protruding groups, creating irregular gaps. While the void is non-spherical (globularity << 1) with 0.067, this relatively low asphericity value suggests that the void’s shape, while complex, does not deviate into extremely elongated or highly irregular forms. It indicates a certain degree of consistency in the packing motif throughout the crystal.
Breakdown of Void Domains
The analysis identifies 9 distinct void domains; their properties are summarized in Table 3. In standard visualization CrystalExplorer, these domains are typically represented by unique colors (Figure 4). To quantify the packing efficiency, we calculate the percentage of the unit cell volume that is occupied by void space. To quantify the packing efficiency, the void percentage is calculated relative to the total unit cell volume (V = 1743.34 Å3 from the crystallographic data). This means the crystal is 88.89% filled with molecule 4 (atoms), leaving only 11.12% empty space, which is typical for organic crystals with significant molecular bulk and planar structures.

Table 3.
Void domain analysis.
The red regions dominate the void surface, accounting for ~98% of the total surface area (606.13 Å2) and ~99% of the total volume (190.73 Å3). These areas represent large, open cavities with minimal obstruction from neighboring molecules. The high surface-to-volume ratio suggests that these voids are shallow and expansive, likely formed between adjacent layers of molecules rather than deep tunnels. The molecules do not create numerous isolated cages but instead assemble in a way that creates a connected network of channels or layers of empty space. This is consistent with the previously identified packing driven by 1D chains linked via N-H···N bonds and stabilized by π-π stacking. The void likely exists in the regions between these stacked layers of molecules. The shape of the major void with very low globularity (0.259) confirms this void is not a pore but a slit. This is the expected result for crystals of flat, plate-like molecules. The π-π stacking forces the molecules to pack in layers, and the void takes the form of the empty space between these layers. The irregular surface area and volume are caused by the protrusion of the 2-bromophenyl and methoxy groups into this interlayer space, sculpting the void’s complex shape.
Orange and yellow regions (8.1–8.49 Å2) indicate smaller, localized voids, possibly due to steric gaps between methyl groups or hydrogen atoms. Green, cyan, blue, purple, and pink contribute negligibly (<1% of total area/volume), suggesting they represent microscopic pores or edge effects at the boundary of the unit cell. The predominance of red indicates that the crystal structure contains significant unoccupied space, which may influence physical properties such as solubility, diffusion, and mechanical strength. The size and shape of the void are a testament to the strength and directionality of the intermolecular forces, The strong N-H···N hydrogen bonds pull molecules into specific, well-defined positions, creating a predictable and efficient framework. The extensive offset π-π stacking brings the large aromatic surfaces into very close contact, minimizing the void space in the stacking direction and creating the characteristic slit-like morphology. The Br···O halogen interaction and various C-H···O contacts act as “cross-links” that further tighten the packing, reducing the void volume to a minimum.
3.4.4. Analysis of Fragment Patch Information and Crystal Packing in Compound 4
To further deconstruct the contributions of different chemical moieties to the overall intermolecular interaction landscape, a fragment patch analysis was performed. This method partitions the HF into distinct regions, or “patches,” corresponding to predefined chemical fragments within the molecule. By quantifying the surface area of each patch, this analysis reveals which parts of the molecule are most exposed to the crystalline environment and are therefore most active in forming intermolecular contacts. Figure 4 visualizes the surface colored according to these fragment patches. Each color represents a distinct region of the molecule’s “interactive skin.” This visualization provides immediate qualitative insights. The largest continuous patches correspond to the bulkiest and most exposed parts of the molecule. A large orange patch (75.5 Å2) clearly covers the 2-bromophenyl substituent, while a large blue-toned patch (e.g., 40.8 Å2) is associated with the planar naphthalene core. The region encompassing the dihydropyran ring and its crucial substituents (the amino and nitrile groups) is represented by a combination of patches, predominantly colored in various shades of green. Smaller fragments, such as the methoxy group, are represented by smaller, distinct patches like the one colored red (14.7 Å2).
The accompanying table provides a quantitative breakdown of the surface area for each of the 16 calculated fragment patches (Table 4). By correlating these areas with the visualization, we can assess the quantitative contribution of each major chemical fragment to the molecule’s total surface area and, by extension, to the crystal packing. The analysis immediately highlights the profound steric influence of the 2-bromophenyl group. Represented by the expansive orange patch, this moiety commands the single largest contiguous surface area of 75.5 Å2. This significant exposure is a direct consequence of its steric bulk and its pronounced torsional twist away from the molecular backbone. Far from being an inert bystander, this vast interactive surface acts as a crucial locus for a multitude of weaker, yet cumulatively significant, interactions, including the C-H···Br, C-H···π, and C-H···N contacts that meticulously stitch the crystal lattice together in three dimensions. In a testament to its functional importance, the dihydropyran ring and its crucial β-enaminonitrile substituent collectively present the largest total surface area, a combined 123.9 Å2 (visualized predominantly in shades of green and purple). The dominant green patch, with an area of 70.8 Å2, specifically envelops the amino and nitrile groups. This extensive and highly accessible surface underscores their central role as the primary architects of the crystal packing, providing the exposed donor and acceptor sites essential for forming the robust, structure-directing N-H···N hydrogen-bonded dimers. The rigid, planar naphthalene core contributes a substantial 99.7 Å2 (blue-toned patches), providing a broad platform for stabilizing Van der Waals forces and weak C-H···π interactions, which are vital for cohesion between the stacked molecular layers. Finally, the smaller but functionally specific methoxy group is represented by a distinct red patch of 14.7 Å2. While modest in size, this exposed surface is the precise location of the C-H···O interactions that serve as critical linkers between the primary dimer units, demonstrating that even minor fragments can play targeted and indispensable roles in the supramolecular assembly.

Table 4.
FP analysis and contribution in crystal package of compound 4.
Ultimately, the fragment patch analysis provides a compelling, quantitative narrative of molecular self-assembly. It reveals a sophisticated synergy between different molecular regions. The crystal architecture is not dictated by a single dominant force but is rather the emergent property of a cooperative interplay: the strong, directional hydrogen bonds of the β-enaminonitrile system provide the primary structural motif, while the vast, exposed surfaces of the sterically demanding bromophenyl group and the planar naphthalene core maximize a network of weaker interactions to achieve overall thermodynamic stability. This detailed partitioning of the molecular surface offers a nuanced and powerful confirmation of the forces driving the solid-state structure of 4.
3.4.5. Deconvolution of Intermolecular Contacts via 2D Fingerprint Plots
For a granular, quantitative deconvolution of the intermolecular interactions governing the crystal packing of compound 4, a comprehensive analysis of the 2D Hirshfeld fingerprint plots was performed. This powerful tool serves as a unique two-dimensional histogram of all close contacts, plotting the distance from the surface to the nearest atom nucleus inside the surface (di) against the distance to the nearest nucleus outside the surface (de). By filtering these plots by the specific atom pairs involved, we can precisely quantify the contribution of each interaction type to the overall crystal stability and identify their characteristic geometric features.
The interactions can be categorized into a clear hierarchy, from the dominant, structure-directing forces to the weaker, supporting contacts and the general Van der Waals background. the H···H (37.2%) is characteristic for hydrogen-rich organic structures, the non-specific H···H contacts represent the largest single contribution to the HF surface. The fingerprint plot shows a large, diffuse cloud of points, indicating a multitude of contacts distributed over a wide range of distances (Figure 5). This represents the general Van der Waals background, which is crucial for maximizing packing efficiency and overall cohesion. The second most abundant interaction C···H/H···C (23.7%), these contacts also largely represent non-directional Van der Waals forces. The plot features two prominent “wings,” characteristic of interactions involving the hydrogen atoms on aromatic rings making contact with the faces and edges of neighboring aromatic systems (weak C-H···π interactions). N···H/H···N (15.3%) plot is the most diagnostically significant feature of the supramolecular assembly. It is dominated by a pair of prominent, sharp, and symmetrical spikes. This is the unequivocal hallmark of a strong, specific, and highly directional hydrogen bond. The tip of these spikes corresponds to the shortest di and de values, signifying the N-H donor and N acceptor of the N-H···N hydrogen bond. Their symmetry reflects the reciprocal nature of this interaction in forming the R22(8) supramolecular synthon that defines the primary centrosymmetric dimer. The sharpness of the spikes indicates a very well-defined contact distance, reinforcing its critical role as the primary architect of the crystal packing. Br···H/H···Br (11.0%) accounting for a substantial 11% of the surface, this plot confirms the importance of halogen bonding. The plot shows two distinct, wing-like features corresponding to close contact between the bromine atom and hydrogen atoms on adjacent molecules (Figure S3). These represent the weak C-H···Br hydrogen bond previously identified. Their significant contribution quantitatively validates their role as a major stabilizing force, responsible for linking the primary N-H···N bonded dimers into the final three-dimensional network. Although a minor contributor for O···H/H···O (2.4%) interaction is structurally pertinent. The plot shows a scattered region of points at relatively short contact distances, confirming the presence of weak C-H···O hydrogen bond. These interactions serve a supporting role, further reinforcing the cohesion between the supramolecular synthons. The C···C (4.3%) fingerprint is crucial for what it lacks. It does not show significant features at short di/de values. Instead, the points are concentrated at distances corresponding to the sum of the Van der Waals radii or greater. The characteristic “dagger” or “wing” shape at the top right of the plot is indicative of contacts between offset aromatic rings. This conspicuously confirms the absence of strong, face-to-face π-π stacking interactions, which aligns perfectly with the geometric analysis showing a twisted molecular conformation. Trace contacts (Br···O, N···N, N···C, Br···C): The remaining interactions contribute minimally to the overall surface (1.9% down to 0.3%).
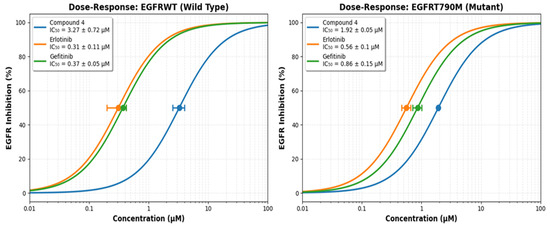
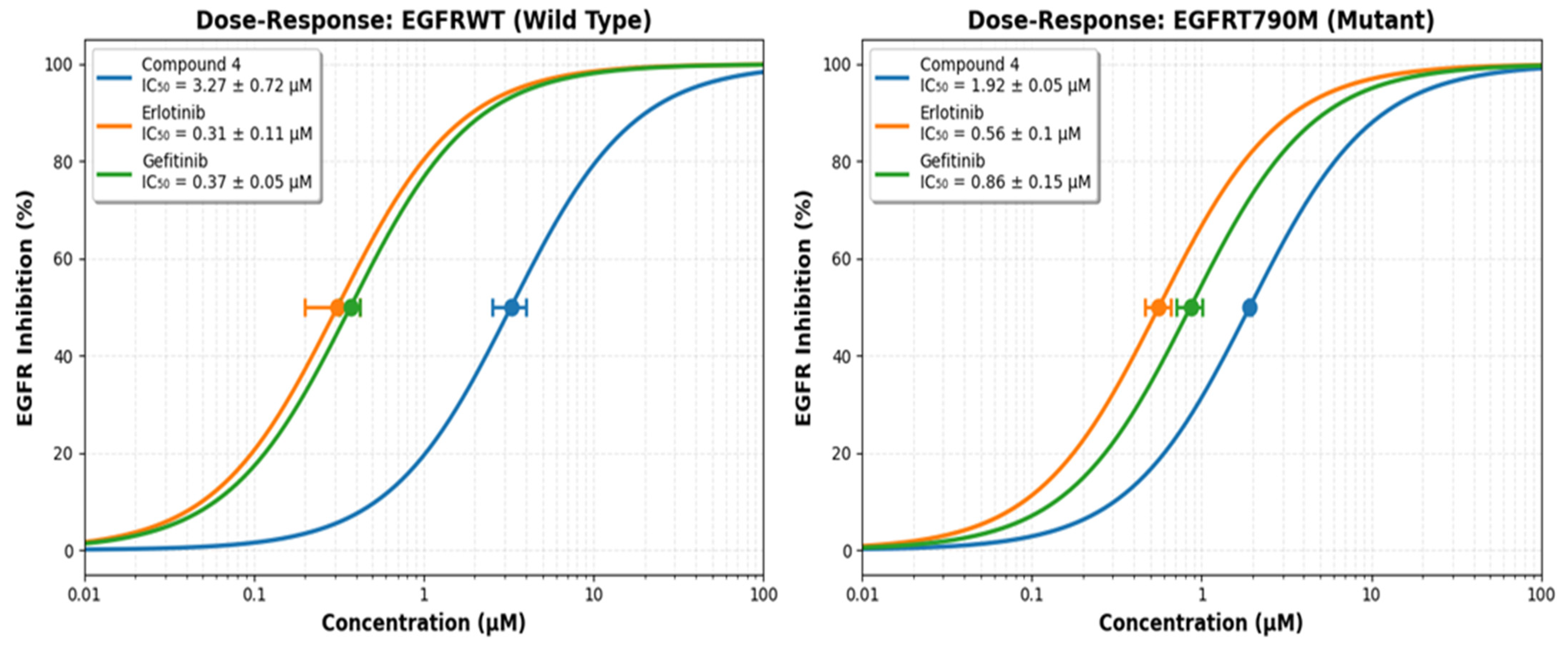
Figure 5.
The dose–response compound 4, Erlotinib and Gefitinib ability to inhibit EGFRWT and EGFRT790M.
3.4.6. Quantitative Analysis of the Crystal’s Energetic Architecture
Crystal packing is stabilized by a complex interplay of non-covalent interactions, including electrostatic (Coulomb), dispersion (Van der Waals), hydrogen bonding, and halogen bonding. While traditional crystallography reveals atomic positions, energy decomposition methods such as those used here allow for the quantification of these forces. The diagrams presented illustrate the total interaction energy between molecules in the unit cell, decomposed into Coulomb and dispersion contributions. This approach is essential for rationalizing crystal structure formation and predicting properties such as solubility, mechanical strength, and thermal stability.
The “Total energy” diagram shows the net stabilization energy (in kJ/mol) between adjacent molecules in the crystal lattice. Negative values indicate attractive interactions, while positive values would suggest repulsion (Figure 6). The strongest N–H···N H-interactions are observed between the amino group (N26–H) and the cyano group (C≡N), with energies ranging from –62.6 to –62.0 kJ/mol. These values are consistent with moderate-strength hydrogen bonds, comparable to those found in N–H···O systems. The C–H···Br Halogen Interactions involving the bromine atom (Br21) and C–H bonds exhibit energies of –37.2 to –38.3 kJ/mol, indicating significant stabilizing contributions from halogen bonding. The face-to-face stacking of aromatic rings contributes energies of –19.8 to –10.0 kJ/mol, reflecting the cumulative effect of dispersion forces and dipole–dipole interactions. The Weaker C–H···O interactions between methoxy oxygen atoms (O14, O22) and aromatic C–H bonds contribute –7.0 to –10.0 kJ/mol. The dominance of hydrogen and halogen bonding suggests that directional interactions play a critical role in determining the crystal’s topology, while dispersion forces provide the bulk of cohesive energy (Figure S4).
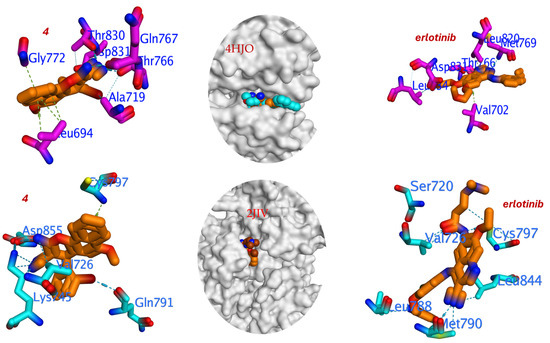
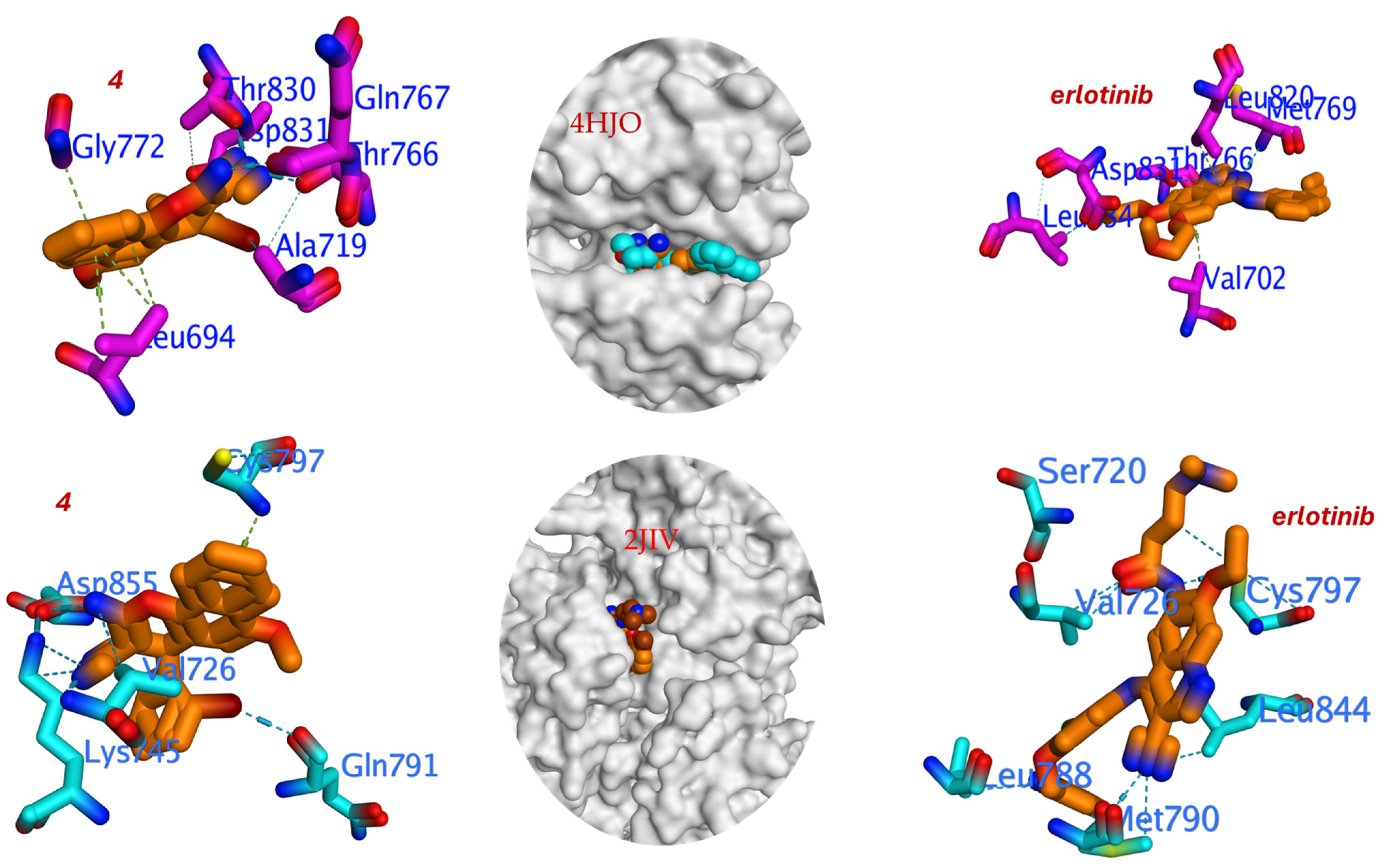
Figure 6.
Three-dimensional binding interaction for compound 4 and Erlotinib against T790M mutant and wild-type EGFR.
Dispersion energy arises from instantaneous dipole-induced dipole interactions and is the primary contributor to the stability of organic crystals. The green tubes in the “Dispersion energy” diagram highlight the regions where dispersion forces are most significant. The largest dispersion contributions occur between the chromene core and the bromophenyl ring, with energies exceeding −50 kJ/mol. These interactions are responsible for the efficient packing of planar molecules and the formation of π–π stacked layers. The methyl group (C23–H) interacts with neighboring aromatic rings via dispersion, contributing ~−10 kJ/mol. Despite being polarized, the bromine atom contributes modestly to dispersion due to its localization at the periphery. The extensive network of dispersion forces ensures high density and low porosity, which is reflected in the calculated crystal density of 1.545 g/cm3.
Coulomb energy reflects the electrostatic attraction or repulsion between charged or polarized atoms. The red tubes in the “Coulomb energy” diagram show the direction and magnitude of these interactions. The strong negative Coulomb energy (−62.6 kJ/mol) indicates a significant electrostatic component in the N–H···N interaction, driven by the partial positive charge on the hydrogen and the partial negative charge on the nitrogen. The −37.2 kJ/mol contribution confirms the presence of a dipole–dipole interaction for C–H···Br Halogen Bonding, where the electron-deficient C–H bond is attracted to the electron-rich lone pairs of bromine. The overall dipole moment of the molecule, arising from the polar functional groups (amino, cyano, methoxy), leads to favorable alignment of molecules in the lattice. The balance between Coulomb and dispersion energies determines the directionality and strength of the interactions. In this case, the dominance of Coulomb energy in hydrogen and halogen bonding suggests that these interactions are not only energetically significant but also highly directional. The energy decomposition results are fully consistent with the structural data from the X-ray analysis, The N–H···N hydrogen bond (IC50 = 1.92 μM against EGFRT790M) is supported by both the short distance (2.98 Å) and high energy (−62.6 kJ/mol). The C–H···Br interaction (distance = 3.20 Å) correlates with a moderate energy (−37.2 kJ/mol), confirming its role in stabilizing the crystal. The layered packing observed in the Hirshfeld surface analysis is mirrored in the energy diagram, where interactions are strongest within planes and weaker between them. This multi-level stabilization ensures a highly stable crystal lattice with minimal void volume and optimal molecular connectivity. The insights gained from this analysis can guide the design of new materials with tailored properties, such as improved bioavailability or enhanced optoelectronic performance.
3.5. Biological Evaluation
Compound 4 was assessed for their in-vitro EGFR inhibitory activity (EGFRWT and EGFRT790M) using Erlotinib and Gefitinib as positive control Figure 5 and the values of IC50 of compound 4 against EGFR inhibitory activity (EGFRWT and EGFRT790M) was cited in (Supplementary Data Table S1).
The enzymatic inhibitory potential of the newly synthesized 4H-benzo[h]chromene derivative (4), was rigorously evaluated against both the wild-type epidermal growth factor receptor (EGFRWT) and its clinically recalcitrant T790M mutant variant (EGFRT790M). This mutation, characterized by a methionine-for-threonine substitution at the “gatekeeper” residue, sterically hinders the binding of first-generation TKIs and is a predominant mechanism of acquired resistance in non-small cell lung cancer (NSCLC) [51,54]. The assay was benchmarked against the first-generation inhibitors Erlotinib and Gefitinib, which served as positive controls to contextualize the potency of the novel compound.
The results indicate that compound 4 possesses a definitive and promising biochemical efficacy profile. Against EGFRWT, the compound exhibited moderate inhibitory activity with an IC50 value of 3.27 ± 0.72 μM. Although this potency is approximately an order of magnitude lower than that of the established drugs Erlotinib (IC50 = 0.31 ± 0.11 μM) and Gefitinib (IC50 = 0.37 ± 0.05 μM), it unequivocally demonstrates that the molecular scaffold of 4 possesses an inherent affinity for the ATP-binding pocket of the kinase domain. This activity is non-trivial and suggests that the core structure serves as a viable pharmacophore.
The more significant finding emerges from the evaluation against the resistant EGFRT790M mutant. Compound 4 demonstrated enhanced inhibitory activity with an IC50 of 1.92 ± 0.05 μM. The paramount metric for assessing efficacy against resistant mutants is not merely the absolute IC50 value, but the selectivity ratio (IC50 EGFRT790M/IC50 EGFRWT), which quantifies the loss in potency incurred by the mutation [63,64]. The critical metric for assessing a compound’s ability to overcome the T790M resistance mechanism is the Resistance Factor (RF), defined as RF = IC50(EGFRT790M)/IC50 (EGFRWT). This ratio quantifies the loss in potency caused specifically by the mutation. For compound 4, the RF is 0.59. An RF < 1 indicates that the compound is slightly more potent against the mutant than the wild-type. More importantly, this low RF demonstrates that the T790M mutation does not confer resistance to compound 4. This profile stands in stark contrast to the first-generation TKIs, Erlotinib and Gefitinib, which suffer a significant loss of potency against the mutant, evidenced by high Resistance Factors of 1.81 and 2.32, respectively. Therefore, while compound 4 is active against both enzyme forms, its promising attribute is not mutant selectivity, but its low resistance factor. This suggests that its binding mode is less compromised by the structural changes in the T790M mutant pocket compared to first-generation inhibitors, positioning it as a potential lead for overcoming T790M-mediated resistance.
This differential activity profile suggests that compound 4 may interact with the kinase domain in a manner distinct from that of the first-generation TKIs. The T790M mutation is known to increase the affinity of the pocket for ATP and sterically clash with the aniline quinazoline core of Gefitinib and Erlotinib [65]. The fact that the potency of 4 is less adversely affected implies that its binding mode might be less dependent on interactions with the gatekeeper residue or that its specific orientation mitigates the steric clash. The structural features of 4—including the bulky 2-bromophenyl group at the 4-position and the hydrogen-bond-donating β-enaminonitrile moiety—could facilitate alternative interactions with conserved residues in the binding pocket, such as those in the hinge region or the front cleft, thereby partially circumventing the mechanism of resistance [66].
In conclusion, while compound 4 exhibits moderate potency against EGFRWT, its superior profile against the EGFRT790M mutant is highly noteworthy. The low selectivity ratio indicates a promising lack of cross-resistance to a major mechanism of TKI failure. This structure-activity relationship (SAR) insight positions the 4H-benzo[h]chromene scaffold, particularly with specific substitutions at the 4-position, as a valuable template for the future development of inhibitors targeting EGFR with T790M-mediated resistance. Further investigations, including cellular assays, kinome-wide selectivity profiling, and co-crystallization studies to elucidate the exact binding mode, are strongly warranted to fully validate its therapeutic potential.
3.6. Molecular Docking Analysis of Compound 4 Against EGFR
Molecular docking serves as a pivotal computational tool for elucidating the potential binding modes and interaction mechanisms of ligands within the active site of target proteins. Herein, we discuss the structure-activity relationship (SAR) and docking analysis of compound 4 against both type of epidermal growth factor receptor in the wild-type (PDB: 2JIV [67]) and the T790M mutant (PDB: 4HJO [68,69], in comparison with the reference inhibitor Erlotinib. The primary objective is to rationalize the experimental inhibitory activity through a detailed examination of binding affinities, interaction energies, and key residue engagements. It is important to note that while this analysis provides a plausible binding mode consistent with the inhibitory activity, a detailed comparative docking study with a T790M mutant structure to elucidate the structural basis for the low resistance factor will be an invaluable focus of future computational work. Compound 4 exhibits a favorable binding free energy (ΔG = −5.861 kcal/mol), marginally lower than that of erlotinib (ΔG = −5.847 kcal/mol). This suggests comparable thermodynamic stability within the EGFR binding pocket. The low RMSD values (~1.5 Å) for both ligands indicate stable docking poses with minimal deviation from the crystallographic reference, affirming the reliability of the predicted binding modes (Table 5).

Table 5.
EGFR Inhibitory activity and binding parameters for compound 4 and Erlotinib against T790M mutant and wild-type EGFR.
The pose for compound 4 with the most favorable binding energy belonged to a cluster with a 45% population, indicating a well-converged and reproducible binding mode. The docking analysis reveals that compound 4 engages with critical residues in the ATP-binding site of EGFR, including the following. Met769 forms a crucial hydrogen bond via the β-enaminonitrile moiety (–NH2 group), contributing to anchor the ligand in the hinge region. Thr830 participates in hydrogen bonding with the methoxy group, enhancing binding specificity. Gln767 and Thr766, Stabilize the ligand through hydrophobic and Van der Waals interactions. Val702, Ala719, and Leu820, engage in hydrophobic contacts with the bromophenyl substituent, facilitating enhanced binding affinity (Figure 6). Notably, the hydrogen bond energy (H.B = −138.137 kcal/mol) for compound 4 is significantly more favorable than that of Erlotinib (H.B = −81.251 kcal/mol), indicating stronger and more numerous hydrogen bonding interactions. This is likely attributable to the presence of the β-enaminonitrile core (–NH2 and –CN) and the methoxy substituent, which act as hydrogen bond donors and acceptors, respectively.
The 2-bromophenyl group at the 4-position of the chromene scaffold plays a pivotal role in enhancing binding affinity and mutant selectivity. The bulky bromine atom engages in hydrophobic interactions with residues Leu820 and Val702, filling a hydrophobic subpocket adjacent to the gatekeeper residue Thr790. May induce slight conformational adjustments in the mutant T790M receptor, mitigating steric clashes and preserving binding potency, as evidenced by the improved IC50 against EGFRT790M (1.92 μM) compared to EGFRWT (3.27 μM).
The critical finding emerges from the docking into the wild-type protein (PDB: 2JIV). Here, the binding affinity of compound 4 remained largely consistent (ΔG = −5.166 kcal/mol). This minimal change in affinity between the wild-type and mutant forms computationally corroborates the low resistance factor observed experimentally. In stark contrast, Erlotinib suffered a significant loss of binding affinity in the wild-type docking (ΔG = −6.483 kcal/mol) compared to the mutant, which is consistent with its known high resistance factor. The binding of compound 4 within the wild-type EGFR active site is stabilized by a different set of interactions (Figure 6). Key hydrogen bonds are formed with the side chains of Asp855, Lys745, and Gln791. The bulky 2-bromophenyl group is accommodated in a hydrophobic pocket defined by residues including Val726. Notably, a crucial interaction is observed with the gatekeeper residue Cys797. However, the overall hydrogen bond energy is significantly weaker (H.B = −29.644 kcal/mol) compared to its interaction with the T790M mutant, which accounts for the less favorable binding energy.
While Erlotinib demonstrates superior potency against EGFRWT (IC50 = 0.31 μM), its efficacy diminishes against EGFRT790M (IC50 = 0.56 μM). In contrast, compound 4 shows reduced wild-type potency but enhanced mutant selectivity (IC50 ratio T790M/WT = 0.59 vs. 1.81 for Erlotinib). This suggests that the chromene scaffold, adorned with the 2-bromophenyl group, may adopt a binding mode less susceptible to steric hindrance imposed by the methionine substitution at residue 790.
This dual-docking analysis strongly suggests that the structural framework of compound 4, particularly the β-enaminonitrile core and the 2-bromophenyl substituent, allows it to adopt a binding conformation that is less susceptible to the steric hindrance from the T790M mutation and can form more stable interactions within the mutant active site. These computational insights provide a robust structural foundation for the observed biological activity and position the 4H-benzo[h]chromene scaffold as a valuable template for developing next-generation inhibitors targeting resistant EGFR mutations.
3.7. Structure-Activity Relationship (SAR) Insights
β-Enaminonitrile motif serves as a hydrogen bond donor/acceptor system, mimicking the quinazoline core of erlotinib, and is critical for hinge region binding (e.g., with Met769). Methoxy group enhances solubility and forms additional hydrogen bonds with Thr830, improving binding specificity. 2-Bromophenyl Substituent contributes to hydrophobic complementarity and may promote mutant selectivity by optimizing van der Waals contacts with hydrophobic residues near the gatekeeper region. Chromene Core provides a planar, rigid structure that facilitates π–π stacking with Phe832 and other aromatic residues, akin to the quinazoline ring in Erlotinib. The molecular docking analysis elucidates that compound 4 binds stably within the EGFR active site through a network of hydrogen bonds and hydrophobic interactions. The presence of the 2-bromophenyl group and the β-enaminonitrile functionality underpins its improved selectivity against the T790M mutant. Although less potent than erlotinib against EGFRWT, the distinct binding mode of compound 4 offers a promising strategy for circumventing T790M-mediated resistance. Future efforts should focus on optimizing the chromene scaffold through strategic substitutions to enhance potency while maintaining mutant selectivity.
4. Conclusions
In conclusion, the present study successfully demonstrates the design, synthesis, and comprehensive characterization of a novel β-enaminonitrile (4). The efficient microwave-assisted synthetic pathway afforded the target compound in high yield, while its structural elucidation was unequivocally confirmed through single-crystal X-ray diffraction analysis, revealing a highly stable and efficiently packed crystalline arrangement stabilized by a robust network of N–H···N hydrogen bonds and π–π stacking interactions. The biological evaluation revealed a compelling inhibitory profile for compound 4. While it exhibited moderate activity against EGFRWT, its significantly enhanced potency and reduced selectivity loss against the challenging EGFRT790M mutant variant underscores its potential as a promising scaffold for overcoming TKI resistance. This advantageous activity was rationalized through molecular docking studies, which identified a favorable binding mode within the ATP-binding pocket, characterized by critical hydrogen bonding with Met769 and extensive hydrophobic interactions facilitated by the strategic 2-bromophenyl substituent.
The synergistic integration of synthetic chemistry detailed structural analysis, biological screening, and computational modeling provides a solid foundation for future work. The 4H-benzo[h]chromene core, particularly with specific aromatic substitutions at the 4-position, emerges as a highly viable and versatile pharmacophore for the development of a new class of EGFR inhibitors. Consequently, this work not only presents a potent lead compound but also delivers valuable insights for the ongoing pursuit of effective therapies against resistant forms of non-small cell lung cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst15110935/s1, Figure S1. 1H-NMR spectrum of compound 4; Figure S2. 13C-NMR spectrum of compound 4; Figure S3. Percentage contribution of specific intermolecular contacts to the total HF surface area for compound 4; Figure S4. Energy frameworks: Visualizing the interaction topology for compound 4; Table S1. Compound 4 ability to inhibit EGFRWT and EGFRT790M.
Funding
This research received no external funding.
Data Availability Statement
Data is available in this article and Supplementary Materials.
Acknowledgments
The author gratefully acknowledges Ziad Moussa and Ahmed A. Elhenawy for their drawings and registering the crystal of 4.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Bragg, W.H. The structure of organic crystals. Proc. Phys. Soc. Lond. 1921, 34, 33–50. [Google Scholar] [CrossRef]
- Okasha, R.M.; El-Agrody, A.M.; Amr, A.-E.G.E.; Al-Omar, M.A.; Ghabbour, H.A. Synthesis and X-ray Single Crystals Char-acterizations of 2-Amino-4-(2-chlorophenyl)-6-Chloro-4H-Benzo[h]Chromene-3-Carbonitrile. J. Comput. Theor. Nanosci. 2017, 14, 5286–5291. [Google Scholar] [CrossRef]
- Al-Dies, A.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Chia, T.S.; Fun, H.-K. 2-Amino-4-(4-fluorophenyl)-6-methoxy-4H-benzo[h]chromene-3-carbonitrile. Acta Crystallogr. 2012, E68, o1934–o1935. [Google Scholar] [CrossRef]
- Al-Dies, A.M.; El-Agrody, A.M.; Al-Omar, M.A.; Amr, A.-E.G.E.; Ng, S.W.; Tiekink, E.R.T. 2-Amino-4-(4-bromophenyl)-6-methoxy-4H-benzo[h]chromene-3-carbonitrile. Acta Crystallogr. 2013, E69, o480–o481. [Google Scholar] [CrossRef] [PubMed]
- Al-Dies, A.M.; Abd El-Wahab, A.H.F.; Alamri, A.A.; Borik, R.M.A.; Mohamed, H.M.; Assirey, E.A.; Alsehli, M.H.; Moussa, Z.; Alzamly, A.; Mehany, A.B.M.; et al. Synthesis, crystal structure, DFT studies, molecular docking, of 2-amino-6-methoxy-4-(4-nitrophenyl)-4H-benzo[h]chromene-3-carbonitrile as tyrosinase inhibitor. J. Mol. Struct. 2025, 1322, 140289. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Fouda, A.M.; Mohamed, H.M.; Alshahrani, M.Y.; Ghabbour, H.A.; Amr, A.-E.G.E.; Okasha, R.M.; Naglah, A.M.; Almehizia, A.A.; Elhenawy, A.A. The Crystal structure of 2-amino-4-(2,3-dichlorophenyl)-6-methoxy-4H-benzo[h]chromene-3-carbonitrile: Antitumor and tyrosine kinase receptor inhibition mechanism study. Crystals 2022, 12, 737. [Google Scholar] [CrossRef]
- Al-Dies, A.M.; Al-Omar, M.A.; Amr, A.-E.G.E.; El-Agrody, A.M.; Ng, S.W.; Tiekink, E.R.T. N′-[3-Cyano-4-(4-fluorophenyl)-6-methoxy-4H-benzo[h]chromen-2-yl]-N,N-dimethylmethanimidamide. Acta Crystallogr. 2013, E69, o482–o483. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Al-Omar, M.A.; Amr, A.-G.E.; Chia, T.S.; Fun, H.-K. Ethyl 2-amino-4-(4-fluorophenyl)-6-methoxy-4H-benzo[h]chromene-3-carboxylate. Acta Crystallogr. 2012, E68, o1803–o1804. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Fouda, A.M.; Al-Omar, M.A.; Ng, S.W.; Tiekink, E.R.T. Ethyl 2-amino-4-bromophenyl)-6-methoxy-4H-benzo[h]chromene-3-carboxylate. Acta Crystallogr. 2013, E69, o435–o436. [Google Scholar] [CrossRef]
- Okasha, R.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A. Crystal structure of 3-amino-8-methoxy-1-phenyl-1H-benzo[f]chromene-2-carbonitrile, C21H16N2O. Z. Krist.-New Cryst. Struct. 2017, 232, 497–499. [Google Scholar]
- El-Agrody, A.M.; Al-Omar, M.A.; Amr, A.-E.G.E.; Ng, S.W.; Tiekink, E.R.T. 3-Amino-1-(4-fluorophenyl)-8-methoxy-1H-benzo[f]chromene-2-carbonitrile. Acta Crystallogr. 2013, E69, o476–o477. [Google Scholar] [CrossRef]
- Alsehli, M.H.; Al-Harbi, L.M.; Okasha, R.M.; Fouda, A.M.; Ghabbour, H.A.; Amr, A.-E.G.E.; Elhenawy, A.A.; El-Agrody, A.M. Synthesis, cytotoxic activity, crystal structure, DFT, molecular docking study of β-enaminonitrile incorporating 1H-benzo[f]chromene moiety. Crystals 2023, 13, 24. [Google Scholar] [CrossRef]
- Fouda, A.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A. Crystal structure of 3-amino-8-methoxy-1-(4-methoxyphenyl)-1H-benzo[f]chromene-2-carbonitrile, C22H18N2O3. Z. Krist.-New Cryst. Struct. 2017, 232, 567–569. [Google Scholar]
- Halawa, A.H.; El-Agrody, A.M.; Amr, A.-E.G.E.; Al-Omar, M.A.; Ghabbour, H.A. X-ray Characterizations of new synthesized 3-amino-1-(2,6-difluorophenyl)-8-methoxy-1H-benzo[f]chromene-2-carbonitrile. J. Comput Theor. Nanosci. 2017, 14, 3994–3999. [Google Scholar] [CrossRef]
- Okasha, R.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A. Synthesis, X-ray characterization and antimicrobial activity of 3-amino-1-(2,4-dichlorophenyl)-8-methoxy-1H-benzo[f]chromene-2-carbonitrile. J. Comput. Theor. Nanosci. 2017, 14, 5717–5721. [Google Scholar] [CrossRef]
- ElGaafary, M.; Syrovets, T.; Mohamed, H.M.; Elhenawy, A.A.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A.; Almehizia, A.A. Synthesis, cytotoxic activity, crystal structure, DFT studies and molecular docking of 3-amino-1-(2,5-dichlorophenyl)-8-methoxy-1H-benzo[f]chromene-2-carbonitrile. Crystals. 2021, 11, 184. [Google Scholar]
- Amr, A.-E.G.E.; Abd El-Mawgoud, H.K.; El-Agrody, A.M.; Al-Omar, M.A.; Alsultan, M.S. X-ray, microwave assisted synthesis and spectral data of 3-amino-1-(3,5-dibromo-2-methoxyphenyl)-8-methoxy-1H- benzo[f]chromene-2-carbonitrile. J. Comput. Theor. Nanosci. 2017, 14, 3930–3935. [Google Scholar] [CrossRef]
- S Mohamed, K.P.; Horton, N.; Akkurt, M.; Younese, S.H.H.; Albayatif, M.R. Crystal structure of 3-amino-1-(4-methoxyphenyl)-1H-benzo[f]chromene-2-carbonitrile. Acta Crystallogr. 2015, E71, o468–o469. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Amr, A.-E.G.E.; Sabry, N.M.; Al-Omar, M.A.; Ghabbour, H.A. Crystal structure of 3-amino-9-methoxy-1-phenyl-1H-benzo[f]chromene-2-carbonitrile, C21H16N2O2. Z. Krist.-New Cryst. Struct. 2016, 231, 1193–1195. [Google Scholar] [CrossRef][Green Version]
- Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ng, S.W.; Tiekink, E.R. 3-Amino-1-(4-fluorophenyl)-9-methoxy-1H-benzo[f]chromene-2-carbonitrile. Acta Crystallogr. 2013, 69, o478–o479. [Google Scholar] [CrossRef]
- Okasha, R.M.; Fouda, A.M.; Bajaber, M.A.; Ghabbour, H.A.; Amr, A.-E.G.E.; El-Agrody, A.M.; Naglah, A.A.; Almehizia, A.A.; Elhenawy, A.M.; El-Agrody, A.M. The Crystal structure of 3-amino-1-(4-chlorophenyl)-9-methoxy-1H-benzo[f]chromene-2-carbonitrile: Antimicrobial activity and docking studies. Crystals 2022, 12, 982. [Google Scholar] [CrossRef]
- Mohamed, H.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A. Crystal structure of 3-amino- 1-(4-bromophenyl)-9-methoxy-1H-benzo[f]chromene-2-carbonitrile, C21H15BrN2O2. Z. Krist.-New Cryst. Struct. 2017, 232, 561–563. [Google Scholar] [CrossRef][Green Version]
- Okasha, R.M.; Alblewi, F.F.; Assiri, M.A.; Amr, A.-E.G.E.; Ghabbour, H.A.; Afifi, T.H.; El-Agrody, A.M. Crystal structure and spectral studies of 3-amino-9-methoxy-1-(4-methoxyphenyl)-1H-benzo[f]chromene-2-carbonitrile. J. Comput. Theor. Nanosci. 2018, 15, 1835–1838. [Google Scholar] [CrossRef]
- Al-Harbi, L.M.; Al-Harbi, E.A.; Okasha, R.M.; El-Eisawy, R.A.; El-Nassag, M.A.A.; Mohamed, H.M.; Fouda, A.M.; Elhenawy, A.A.; Mora, A.; El-Agrody, A.M.; et al. Discovery of benzochromene derivatives first example with dual cytotoxic activity against the resistant cancer cell MCF-7/ADR and inhibitory effect of the P-glycoprotein expression levels. J. Enzyme Inhib. Med. Chem. 2023, 38, 2155814. [Google Scholar] [CrossRef] [PubMed]
- Fouda, A.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Al-Omar, M.A.; Ghabbour, H.A. Spectroscopic data, single X-ray and antimicrobial activity of microwave synthesized 3-amino-8-bromo-1-(2,5-dichlorophenyl)-1H-benzo[f]chromene-2-carbonitrile. J. Comput. Theor. Nanosci. 2017, 14, 3831–3836. [Google Scholar] [CrossRef]
- Afifi, T.H.; El-Agrody, A.M.; Amr, A.-E.G.E.; El-Agrody, A.M.; Ghabbour, H.A. X-ray characterization and antimicrobial activity of synthesized new 3-amino-8-bromo-1-(3,4-dimethoxyphenyl)-1H-benzo[f]chromene-2-carbonitrile. J. Comput. Theor. Nanosci. 2017, 14, 3924–3929. [Google Scholar] [CrossRef]
- Halawa, A.H.; Fouda, A.M.; Al-Dies, A.M.; El-Agrody, A.M. Synthesis, biological evaluation and molecular docking studies of 4H-benzo[h]chromenes, 7H-benzo[h]chromeno [2,3-d]Pyrimidines as antitumor agents. Lett. Drug. Des. Discov. 2016, 1, 77–88. [Google Scholar]
- Alblewi, F.F.; Okasha, R.M.; Eskandrani, A.A.; Afifi, T.H.; Mohamed, H.M.; Halawa, A.H.; Fouda, A.M.; Al-Dies, A.M.; Mora, A.; El-Agrody, A.M. Design and synthesis of novel heterocyclic-based 4H-benzo[h]chromene moieties: Targeting antitumor caspase 3/7 activities and cell cycle analysis. Molecules 2019, 24, 1060. [Google Scholar] [CrossRef] [PubMed]
- Alblewi, F.F.; Okasha, R.M.; Hritani, Z.M.; Mohamed, H.M.; El-Nassag, M.A.A.; Halawa, A.H.; Mora, A.; Fouda, A.M.; Assiri, M.A.; Al-Dies, A.M.; et al. antiproliferative effect, cell cycle arrest and apoptosis generation of novel synthesized anticancer heterocyclic derivatives based 4H-benzo[h]chromene. Bioorg. Chem. 2019, 87, 560–571. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Abd-El-Mawgoud, H.K.; Fouda, A.M.; Khattab, E.S.A.E.H. Synthesis, in-vitro cytotoxicity of 4H-benzo[h]chromene derivatives and structure–activity relationships of 4-aryl group and 3-, 7-positions. Chem. Pap. 2016, 70, 1279–1292. [Google Scholar] [CrossRef]
- Kheirollahi, A.; Pordeli, M.; Safavi, M.; Mashkouri, S.; Naimi-Jamal, M.R.; Ardestani, S.K. Cytotoxic and apoptotic effects of synthetic benzochromene derivatives on human cancer cell lines. NaunynSchmiedeberg’s Arch. Pharmacol. 2014, 387, 1199–1208. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Fouda, A.M.; Khattab, E.S.A.E.H. Halogenated 2-amino-4H-benzo[h]chromene derivatives as antitumor agents and the relationship between lipophilicity and antitumor activity. Med. Chem. Res. 2017, 26, 691–700. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Fouda, A.M.; Khattab, E.S.A.E.H. Synthesis, antitumor activity of 2-amino-4H-benzo[h]chromene derivatives, and structure–activity relationships of the 3- and 4-positions. Med. Chem. Res. 2013, 22, 6105–6120. [Google Scholar] [CrossRef]
- Smith, C.W.; Bailey, J.M.; Billingham, M.E.J.; Chandrasekhar, S.; Dell, C.P.; Harvey, A.K.; Hicks, C.A.; Kingston, A.E.; Wishart, G.N. The anti-rheumatic potential of a series of 2,4-disubstituted-4H-naphtho [1,2-b]pyran-3-carbonitriles. Bioorg. Med. Chem. Lett. 1995, 5, 2783–2788. [Google Scholar] [CrossRef]
- Fouda, A.M.; Hassan, A.H.; Eliwa, E.M.; Ahmed, H.E.A.; Al-Dies, A.M.; Omar, A.M.; Nassar, H.S.; Halawa, A.H.; Aljuhani, N.; El-Agrody, A.M. Targeted potent antimicrobial benzochromene-based analogues: Synthesis, computational studies, and inhibitory effect against 14α-demethylase and DNA gyrase. Bioorg. Chem. 2020, 105, 104387. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.E.A.; El-Nassag, M.A.A.; Hassan, A.H.; Mohamed, H.M.; Halawa, A.H.; Okasha, R.M.; Ihmaid, S.; Abd El-Gilil, S.M.; Khattab, E.S.A.E.H.; Fouda, A.M.; et al. Developing lipophilic aromatic halogenated fused systems with specific ring orientations, leading to potent anticancer analogs and targeting the c-Src Kinase enzyme. J. Mol. Struct. 2019, 1186, 212–223. [Google Scholar] [CrossRef]
- Ahmed, H.E.A.; El-Nassag, M.A.A.; Hassan, A.H.; Okasha, R.M.; Ihmaid, S.; Fouda, A.M.; Afifi, T.H.; Aljuhani, A.; El-Agrody, A.M. Introducing novel potent anticancer agents of 1H-benzo[f]chromene scaffolds, targeting c-Src kinase enzyme with MDA-MB-231 cell line anti-invasion effect. J. Enzym. Inhib. Med. Chem. 2018, 33, 1074–1088. [Google Scholar] [CrossRef]
- Fouda, A.M.; Okasha, R.M.; Alblewi, F.F.; Mora, A.; Afifi, T.H.; El-Agrody, A.M. A proficient microwave synthesis with structure elucidation and the exploitation of the biological behavior of the newly halogenated 3-amino-1H-benzo[f]chromene molecules, targeting dual inhibition of topoisomerase II and microtubules. Bioorg. Chem. 2020, 95, 103549. [Google Scholar] [CrossRef]
- Elgaafary, M.; Fouda, A.M.; Mohamed, H.M.; Hamed, A.; El-Mawgoud, H.K.A.; Jin, L.; Ulrich, J.; Simmet, T.; Syrovets, T.; El-Agrody, A.M. Synthesis of β-enaminonitriles linked 8-methoxy-1H-benzo[f]chromene moieties and analysis of their antitumor mechanisms. Front. Chem. 2021, 9, 759149. [Google Scholar] [CrossRef] [PubMed]
- Fouda, A.M.; Assiri, M.A.; Mora, A.; Ali, T.E.; Afifi, T.H.; El-Agrody, A.M. Microwave synthesis of novel halogenated β-enaminonitriles linked 9-bromo-1H-benzo[f]chromene moieties: Induces cell cycle arrest and apoptosis in human cancer cells via dual inhibition of topoisomerase I and II. Bioorg. Chem. 2019, 93, 103289. [Google Scholar] [CrossRef]
- Elgaafary, M.; Lehner, J.; Fouda, A.M.; Hamed, A.; El-Mawgoud, H.K.A.; Ulrich, J.; Simmet, T.; Syrovets, T.; El-Agrody, A.M. Synthesis and evaluation of antitumor activity of 9-methoxy-1H-benzo[f]chromene derivatives. Bioorg. Chem. 2021, 116, 105402. [Google Scholar]
- Gorle, S.; Maddila, S.; Maddila, S.; Naicker, K.; Singh, M.; Singh, P.; Jonnalagadda, S. Synthesis, molecular docking study and in vitro anticancer activity of tetrazole linked benzochromene derivatives. Anticancer Agents Med. Chem. 2017, 17, 464–470. [Google Scholar] [CrossRef]
- Afifi, T.H.; Okasha, R.M.; Ahmed, H.E.A.; Ilas, J.; Saleh, T.; Abd-El-Aziz, A.S. Structure-activity relationships and molecular docking studies of chromene and chromene based azo chromophores: A novel series of potent antimicrobial and anticancer agents. EXCLI J. 2017, 16, 868–902. [Google Scholar]
- Ahagh, M.H.; Dehghan, G.; Mehdipour, M.; Teimuri-Mofrad, R.; Payami, E.; Sheibani, N.; Ghaffari, M.; Asadi, M. Synthesis, characterization, anti-proliferative properties and DNA binding of benzochromene derivatives: Increased Bax/Bcl-2 ratio and caspase dependent apoptosis in colorectal cancer cell line. Bioorg. Chem. 2019, 93, 103329. [Google Scholar] [CrossRef]
- Abd El-Wahab, A.H.; Borik, R.M.; Al-Dies, A.A.M.; Fouda, A.M.; Mohamed, H.M.; El-Eisawy, R.A.; Mora, A.; El-Nassag, M.A.; Abd Elhady, A.M.; Elhenawy, A.A.; et al. Design, synthesis and bioactivity study on oxygen-heterocyclic-based pyran analogues as effective P-glycoprotein-mediated multidrug resistance in MCF-7/ADR cell. Sci. Rep. 2024, 14, 7589. [Google Scholar] [CrossRef]
- Albalawi, F.F.; El-Nassag, M.A.A.; El-Eisawy, R.A.; Mohamed, M.B.I.; Fouda, A.M.; Afifi, T.H.; Elhenawy, A.A.; Mora, A.; El-Agrody, A.M.; El-Mawgoud, H.K.A. Synthesis of 9-Hydroxy-1H-Benzo[f]chromene Derivatives with Effective Cytotoxic Activity on MCF7/ADR, P-Glycoprotein Inhibitors, Cell Cycle Arrest and Apoptosis Effects. Int. J. Mol. Sci. 2022, 24, 49. [Google Scholar] [CrossRef] [PubMed]
- Rafinejad, A.; Fallah-Tafti, A.; Tiwari, R.; Shirazi, A.N.; Mandal, D.; Shafiee, A.; Parang, K.; Foroumadi, A.; Akbarzadeh, T. 4-Aryl-4H-naphthopyrans derivatives: One-pot synthesis, evaluation of Src kinase inhibitory and anti-proliferative activities. Daru J. Pharm. Sci. 2012, 20, 100–107. [Google Scholar] [CrossRef]
- Abd El-Wahab, A.H.F.; Borik, R.M.A.; Al-Dies, A.M.; Fouda, A.M.; Mohamed, H.M.; El-Eisawy, R.A.; Sharaf, M.H.; Alzahrani, A.Y.A.; Elhenawy, A.A.; El-Agrody, A.M. Targeted potent antimicrobial and antitumor oxygen heterocyclic based pyran analogues: Synthesis and computational studies. Sci. Rep. 2024, 14, 9862. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar]
- Ahmed, S.A.; Kamel, M.S.; Aboelez, M.O.; Ma, X.; Al-Karmalawy, A.A.; Mousa, S.A.S.; Shokr, E.K.; Abdel-Ghany, H.; Belal, A.; El Hamd, M.A.; et al. Thieno[2, 3-b] thiophene derivatives as potential EGFRWT and EGFRT790M inhibitors with antioxidant activities: Microwave-assisted synthesis and quantitative in vitro and in silico studies. ACS Omega 2022, 7, 45535–45544. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005, 2, e73. [Google Scholar] [CrossRef] [PubMed]
- Yadav, T.; Moin, S.G.; Kumar, M.A. Review on Fused Pyrimidine Systems as EGFR Inhibitors and Their Structure-Activity Relationship. Front. Chem. 2022, 2, 861288. [Google Scholar] [CrossRef]
- Eno, M.S.; Brubaker, J.D.; Campbell, J.E.; De Savi, C.; Guzi, T.J.; Williams, B.D.; Wilson, D.; Wilson, K.; Brooijmans, N.; Kim, J.; et al. Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer. J. Med. Chem. 2022, 65, 9662–9677. [Google Scholar] [CrossRef]
- Yun, C.H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.K.; Meyerson, M.; Eck, M.J. The T790M Mutation in EGFR Kinase Causes Drug Resistance by Increasing the Affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Jänne, P.A.; Yang, J.C.H.; Kim, D.W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ahn, M.J.; Kim, S.W.; Su, W.C.; Horn, L.; et al. AZD9291 in EGFR inhibitor–resistant non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef]
- Helal, M.H.; Alsehli, M.H.; Al-Dies, A.-A.M.; Moussa, Z.; Elgammal, W.E.; Halawa, A.H.; Elhenawy, A.A.; El-Agrody, A.M. Unraveling the interaction of pyranocoumarins with P-glycoprotein: Implications for overcoming multidrug resistance in cancer therapy. Bioorg. Chem. 2025, 158, 108314. [Google Scholar] [CrossRef]
- Al-Dies, A.-A.M.; Alsehli, M.H.; Alsharif, H.; Ibrahim, I.; Rafrafi, S.; Moussa, Z.; Afifi, T.H.; Elgammal, W.E.; Halawa, A.H.; Elhenawy, A.A.; et al. Discovery of pyranocoumarin derivatives as dual cytotoxic activity against the resistant cancer cell MCF-7/ADR and inhibitory effect of the P-glycoprotein expression levels and function. J. Mol. Struct. 2025, 1337, 142007. [Google Scholar] [CrossRef]
- Al-Dies, A.M.; Alblewi, F.F.; Okasha, R.M.; Alsehli, M.H.; Borik, R.M.A.; Ihmaid, S.; Amr, A.E.G.E.; Ghabbour, H.A.; Elhenawy, A.A.; El-Agrody, A.M. Synthesis, crystal structure, hirshfeld study, DFT analysis, molecular docking study, antimicrobial activity of β-enaminonitrile bearing 1H-pyran. Discov. Appl. Sci. 2025, 7, 71. [Google Scholar] [CrossRef]
- Al-Dies, A.M.; Alsehli, M.H.; Assirey, E.A.; Okasha, R.M.; Rafrafi, S.; Ibrahim, I.; Moussa, Z.; Alzamly, A.; Elhenawy, A.A.; El-Agrody, A.M. Synthesis, EGFR and VEGFR-2 inhibitors, crystal structure, DFT analysis, molecular docking study of β-enaminonitrile incorporating 1H-benzo[f]-chromene-2-carbonitrile. J. Mol. Struct. 2025, 1336, 142030. [Google Scholar] [CrossRef]
- Jia, Y.; Quinn, C.M.; Gagnon, A.I.; Talanian, R. Homogeneous time-resolved fluorescence and its applications for kinase assays in drug discovery. Anal. Biochem. 2006, 356, 273–281. [Google Scholar] [CrossRef] [PubMed]
- El-Mawgoud, H.K.A.; Fouda, A.M.; El-Nassag, M.A.A.; Elhenawy, A.A.; Alshahrani, M.Y.; El-Agrody, A.M. Discovery of novel rigid analogs of 2-naphthol with potent anticancer activity through multi-target topoisomerase I & II and tyrosine kinase receptor EGFR & VEGFR-2 inhibition mechanism. Chem.-Biol. Interact. 2022, 355, 109838. [Google Scholar]
- Fouda, A.M.; El-Nassag, M.A.A.; Elhenawy, A.A.; Shati, A.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Alam, M.M.; El-Agrody, A.M. Synthesis of 1,4-dihydropyrano [2,3-c]pyrazole derivatives and exploring molecular and cytotoxic properties based on DFT and molecular docking studies. J. Mol. Struct. 2022, 1249, 131555. [Google Scholar] [CrossRef]
- Cross, D.A.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.; Ward, R.A.; Mellor, M.J.; et al. AZD9291, an Irreversible EGFR TKI, Overcomes T790M-Mediated Resistance to EGFR Inhibitors in Lung Cancer. Cancer Discov. 2014, 4, 1046–1061. [Google Scholar] [CrossRef]
- Remon, J.; Steuer, C.E.; Ramalingam, S.S.; Felip, E. Osimertinib and Other Third-Generation EGFR TKI in EGFR-Mutant NSCLC Patients. Ann. Oncol. 2018, 29 (Suppl. S1), i20–i27. [Google Scholar] [CrossRef]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR Mutation and Resistance of Non–Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
- Omar, A.M.M.; AboulWafa, O.M.; Amr, M.E.; El-Shoukrofy, M.S. Antiproliferative activity, enzymatic inhibition and apoptosispromoting effects of benzoxazole-based hybrids on human breast cancer cells. Bioorg. Chem. 2021, 109, 104752. [Google Scholar] [CrossRef] [PubMed]
- McTigue, M.B.; Murray, W.; Chen, J.H.; Deng, Y.L.; Solowiej, J.; Kania, R.S. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc. Natl. Acad. Sci. USA 2012, 109, 18281–18289. [Google Scholar] [CrossRef]
- Park, J.H.; Liu, Y.; Lemmon, M.A.; Radhakrishnan, R. Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem. J. 2012, 448 Pt 3, 417. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).