Abstract
Fragment screening directly on protein crystals has been applied using AnalytiCon’s collection of intermediates that have been utilized to generate libraries of larger synthetic natural product-like molecules. The fragments with well-balanced physicochemical properties show an impressively high hit rate for a screen using the aspartic protease endothiapepsin. The subsequent validation and expansion of the discovered fragment hits benefits from AnalytiCon’s comprehensive library design. Since the screened fragments are intermediates that share a common core with larger and closely related analogs with modulated substitution patterns, they allow for the retrieval of off-the-shelf follow-up compounds, which enable the development of design strategies for fragment optimization. A promising bicyclic core scaffold found in several fragment hits could be validated by selecting a set of enlarged follow-up compounds. Due to unexpected changes in binding mode and no significant improvement in ligand efficiency, this series was quickly deemed unsuitable and therefore discontinued. The structures of follow-up compounds of two other fragments helped to evaluate a putative fusion of two overlapping fragment hits. A design concept on how to fuse the two fragments could be proposed and helps to plan a suitable substitution pattern and promising central bridging element.
1. Introduction
Fragment-based lead discovery (FBLD) was first proposed more than 20 years ago as an efficient approach to discover novel ligands that modulate pharmacological function for macromolecular targets [1,2,3,4,5,6,7,8,9]. The results of recent fragment screening campaigns, technical advances at synchrotron sources, and improvements in crystal growth, data collection, and evaluation increasingly argue for placing significantly more emphasis on crystallography as the primary screening method to discover putative fragment binders [10,11,12,13,14,15,16]. In particular, modern synchrotron beamlines allow for the collection of several hundred data sets within an 8 h beamtime shift. These developments are increasingly transforming crystallographic fragment screening from a method previously used to validate promising hits at a later stage to an incipient method of exhaustively mapping protein binding sites using multiple small molecule probes [17,18,19]. This approach not only provides structurally validated starting points for subsequent optimization but also informs the medicinal chemist of promising molecular recognition options for the target protein under investigation.
Many fragment libraries have been suggested over the years [20,21,22,23]. However, most of these libraries consist of scaffolds typically used as starting materials in synthetic organic chemistry, usually containing well-decorated aromatic moieties. The design of such libraries adheres quite strictly to the “rule of 3”, allowing only some minor extensions [24,25]. In the current study, we focus on a less common selection and move into the realm of natural compound chemistry. Numerous studies have highlighted the superior properties of natural products as drug candidates [26,27,28,29,30,31]. However, natural compounds have received little attention as a pool for fragment-based screening due to their synthetic intractability and the challenging chemistry required for structural expansion and optimization. Nevertheless, fragments derived from natural products are believed to occupy parts of the chemical space that are not exhaustively covered by typical intermediates of classical organic synthesis [26]. Thus, a direct crystallographic screening campaign based on a collection of natural product-like fragments is expected to provide novel insights into protein binding, especially when compared to alternative campaigns based on conventional libraries following the rules of “classical synthetic organic chemistry”. It is expected that collections derived from natural compounds will suffer less from properties falling under the pan-assay interference compounds (PAINS) [32] criteria, show better solubility behavior, and have a higher “three-dimensionality” in terms of a pharmacophoric pattern.
AnalytiCon has built an impressive collection of natural product-like compounds over the years (NATx libraries). They have taken ideas from nature and transformed them into libraries of synthetic molecules based on natural product motifs [33,34]. The compilation of these libraries comprises a subset of synthetic intermediates that largely conform to the “rule of 3”. They are summarized in the so-called FRGx library [35], which in many cases, contains sets of closely related analogs with modulated substitution patterns at common core structures. In fact, a major advantage of this collection is the amount of synthetic validation that has gone into these intermediates, and the multiple follow-up options that are available off-the-shelf for possible synthetic transformations. This subset of fragment-sized molecules is available for testing, and once individual candidates are identified as promising hits in a screening campaign, they can be grown into larger analogs simply by screening the AnalytiCon NATx catalog.
As a feasibility study for later follow-up studies, a selection of the AnalytiCon fragment library was screened against endothiapepsin (EP) as a model protein for pepsin-like proteases [36]. This class of enzymes includes several important drug targets, such as renin, HIV protease, or BACE [37,38,39]. Furthermore, we chose this enzyme because we discovered many fragment complexes based on exhaustive screening campaigns using libraries composed of typical “synthetic organic chemistry fragments” [40,41,42]. Based on the initial hits from the FRGx screen, we selected a series of follow-up compounds from AnalytiCon’s large NATx database. They all contained a promising bicyclic parent scaffold found in three of the initially screened fragment structures. In addition, two discovered hits suggested a promising merging strategy to fuse two fragments into a larger candidate. To validate the binding mode of the fragments to be fused, some analogs were extracted from the NATx library. Considering these additional follow-up compounds, a design proposal of a putative fusion product is presented. Our study demonstrates the suitability of fragments based on natural product motifs for initial screening and subsequent validation by retrieving already available follow-up ligands from the NATx collection, thus avoiding the need for time-consuming synthesis of possible validation or optimization candidates.
2. Results and Discussion
2.1. Fragment Test Set
An initial set of 73 fragments was selected from AnalytiCon’s FRGx library [33,34,35]. The selection was based on the availability and diversity of the fragments for our crystallographic experiments. Fragment complex structures were obtained through individual soaking experiments of the selected compounds in preformed monoclinic EP crystals. A subsequent structural analysis revealed that 15 fragments were bound to EP and provided well-diffracting crystals (see Figure 1 and Table S1: 56, 62, 75, 80, 81, 134, 140, 164, 166, 175, 203, 245, 270, 274 and 283, hereafter referred to by their reference numbers only). For the full identification in AnalytiCon’s FRGx library, refer to Table S5.
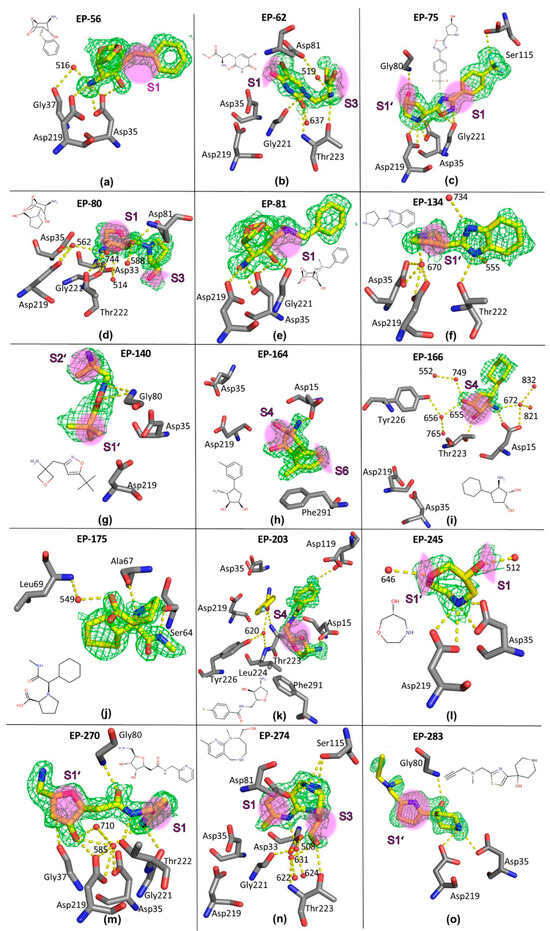
Figure 1.
(a–o). Crystallographically determined binding modes of the 15 fragments bound in complex with EP. The fragments are shown as yellow sticks, with the fragment ID in bold and the chemical formulas, contoured by the unbiased (Fo−Fc)-difference electron density, shown as green meshes (contour level at 2 σ). EP residues of the final refined EP complex structure are shown as gray sticks. The atoms are labeled with atom type colors (PyMOL default). The hydrogen bonds up to a donor-acceptor distance of 3.2 Å are shown as yellow dashed lines and involve interstitial water molecules. Waters are depicted as red spheres and labeled with numbers according to the corresponding PDB files (see Table S1). The spatial position of the binding sites, S1–S6, S1’, and S2’, are approximately marked as magenta areas. Fragment 175 is found at the protein surface distal from any of the substrate binding sites.
2.2. Crystal Structures of Fragment Complexes
Overall, the data resolution (1.12–1.76 Å) and refinement statistics indicate a rather high quality of the obtained structures. Consequently, this implies a fairly reliable characterization of fragment binding modes (see Section 4). Refinement of fragment occupancy resulted in occupancy values ranging from 50 to 99%. In some cases, small parts of the geometry and solvation pattern of the uncomplexed enzyme are still visible in the residual electron density. A short description of the individual binding modes of the discovered fragment hits is given below. Please note that for single atoms, the atomic nomenclature of the PDB file [43] is used to unambiguously describe the observed ligand–protein interactions (e.g., O, OG1, OD2, N, N1). Water molecules are labeled according to the PDB file. A consistent labeling scheme for water molecules across all structures is impossible to achieve, as individual water molecules can sometimes be strongly shifted, replaced by the bound ligand, or newly recruited. Below, the fragments are grouped according to similarities in chemical structure or binding orientation. In Figure 1, the fragments are listed according to their ascending reference number in the FRGx library.
EP-56 complex (Figure 1a, PDB-code: 5QB5). The fragment is the first of three related examples that share a bicyclic parent scaffold. It binds at the apex between the catalytic dyad with its likely protonated and charged primary amino nitrogen. This atom displaces the catalytic water molecule [11,42] and forms hydrogen bonds with both catalytic aspartate residues. While both carboxylate atoms of Asp35 are involved in hydrogen bonding (3.05 and 3.20 Å), OD2 of Asp219 forms an H-bond with a length of 3.03 Å, and OD1 is with 3.29 Å at the upper limit of a hydrogen bonding distance. Since the fragment occupancy is refined to 65%, some residual density suggests the presence of the catalytic water molecule in some parts of the studied crystal, reflecting the geometry of the uncomplexed enzyme. In addition, water 516 mediates a hydrogen bonding interaction between the amino group of the fragment and the carbonyl oxygen of Gly37. Furthermore, the hydroxyl group of the fragment forms an H-bond with the carboxylate atom OD2 of Asp35 (2.76 Å). The linker between the two ring moieties of the fragment is located in the S1 pocket.
EP-80 complex (Figure 1d, PDB-code: 5QB8). This fragment has the same bicyclic scaffold as fragment 56. However, it adopts a different binding position. It binds with its most likely charged primary amino group via the catalytic water molecule 562 (2.69 Å) to the two aspartic residues, Asp35-OD1/OD2 (2.74 and 3.22 Å) and Asp219-OD1/OD2 (2.98 and 2.84 Å). The basic nitrogen atom is also H-bonded to Gly221-O (2.78 Å) and water 744, which mediates a hydrogen bond to Thr222-OG1. The pyrrolidine nitrogen of the fragment is likely protonated and interacts with Asp81-OD2 (2.65 Å), while the hydroxyl oxygen O2 on the bicyclic ring forms hydrogen bonds to Asp81-OD1 (3.01 Å) and water 588 (2.72 Å). The ether oxygen in the short ring bridge interacts with Gly221-O (3.07 Å) and water 514 (3.10 Å). The latter mediates a hydrogen bond with Asp33-OD2 (2.53 Å). The larger methylene–oxygen bridge is oriented toward the S1 pocket, while the methylene–hydroxy substituent on the pyrrolidine ring is oriented toward the S3 pocket.
EP-81 complex (Figure 1e, PDB-code: 5QB9). This fragment is a third hit that shares the bicyclic scaffold. It adopts a similar orientation as 56, but its extended benzyl substituent is placed in a slightly different orientation. It also uses its primary, probably charged, amino group to displace the catalytic water molecule and forms hydrogen bonds with the carboxyl oxygen atoms of both catalytic aspartates at distances of 2.91 to 3.20 Å; the OD1 of Asp219 is at a 3.30 Å distance. Some residual density suggests the presence of the catalytic water molecule in a small amount of the uncomplexed enzyme. The hydroxyl oxygen O2 on the bicyclic backbone interacts with Asp35-OD2 (2.90 Å) and Gly221-O. The attached benzyl substituent is oriented toward the S1 pocket.
EP-75 complex (Figure 1c, PDB-code: 5QB7). The secondary amino group of the terminal pyrrolidine ring of fragment 75 is located at the apex between the two catalytic aspartates and forms hydrogen bonds with the four carboxylate oxygen atoms of Asp35 and Asp219 at distances between 2.87 and 2.99 Å. The exocyclic hydroxyl oxygen on the pyrrolidine ring forms only one hydrogen bond to Gly80-N (3.04 Å), while the oxygen atom of the central oxadiazole moiety is at a hydrogen bond distance (2.84 Å) to Asp35-OD2, which makes an uncharged state of Asp35 residue likely in this case. Furthermore, the oxadiazole oxygen and the adjacent ring nitrogen form electrostatic interactions with the carbonyl oxygen of Gly221 (2.95, 3.19 Å). Finally, the fluorine atom F1 of the fragment interacts with Ser115-OG. The central part of the fragment occupies the S1 pocket, and the pyrrolidine ring points toward the S1’ pocket, suggesting a putative vector for the synthetic expansion of the fragment in this direction.
EP-245 complex (Figure 1l, PDB-code: 5QBG). This seven-membered cyclic fragment binds with its secondary amino group at the apex between the two catalytic aspartates. It displaces the catalytic water molecule and involves all four carboxylate oxygens in H-bonds at distances between 2.87 and 3.15 Å. The exocyclic hydroxyl oxygen contacts water 512 (2.49 Å), and the endocyclic ether oxygen recruits the water molecule 646 (2.94 Å). This fragment is located between pockets S1 and S1’.
EP-283 complex (Figure 1o, PDB-code: 8PXI). Fragment 283 also binds close to the catalytic dyad, placing its likely protonated basic nitrogen between the catalytic aspartates. It displaces the catalytic water molecule and forms one hydrogen bond to Asp219-OD2 (2.94 Å), as well as Asp35-OD1 (3.23 Å). Another hydrogen bond connects the hydroxyl oxygen atom of the fragment to the amide nitrogen of Gly80 (2.79 Å). The central part of the fragment bridges the S1’ pocket with its five-membered thiazole ring.
EP-62 complex (Figure 1b, PDB-code: 5QB6). This fragment does not directly bind to the catalytic dyad but interacts with its primary amino group (N2) via a hydrogen bond to Thr223-OG1 (3.13 Å). A second H-bond is formed via water 519 (2.58 Å) to Asp81-OD2 (2.86 Å). The urea-type oxygen (O3) of the fragment is hydrogen-bonded to Thr223-N (3.12 Å) and water 637 (2.81 Å). While the bromine atom faces the S1 pocket, the side chain of the fragment occupies the S3 pocket. A close polar contact is formed between the tertiary amino nitrogen and the carbonyl oxygen of Gly221 (3.06 Å).
EP-134 complex (Figure 1f, PDB-code: 5QBA). The secondary amino group (N2) of the pyrrolidine ring of the fragment interacts indirectly with the catalytic aspartates via the catalytic water molecule 670 (2.80 Å). The benzimidazole moiety is used by one of its nitrogens to form a hydrogen bond with water 734 (2.92 Å), while the second nitrogen (N1) interacts with water 555 (2.67 Å) and Thr222-OG1 (2.75 Å). A part of the terminal pyrrolidine ring occupies the S1’ pocket, while the benzimidazole faces the S2 pocket.
EP-140 complex (Figure 1g, PDB-code: 5QBB). The fragment binds remotely from the dyad and forms two hydrogen bonds through its isoxazole moiety to the adjacent amide nitrogen of Gly80, both at the extended distance of 3.08 Å. The attached tertiary butyl group partially occupies the S1’ pocket, while the four-membered ring at the opposite end of the fragment is oriented toward the S2’ pocket.
EP-164 complex (Figure 1h, PDB-code: 5QBC). Fragment 164 also binds remotely from the catalytic center and forms a π-stacking interaction with the phenyl ring of Phe291 (3.95–4.44 Å). In addition, its primary amino group forms a hydrogen bond at a distance of 3.12 Å to one of the carboxylate oxygens of Asp15. Part of the five-membered ring of the fragment, including one of the attached hydroxyl groups, occupies the S4 pocket. The methyl-substituted phenyl ring is located adjacent to the S6 pocket.
EP-166 complex (Figure 1i, PDB-code: 5QBD). Fragment 166 is similarly positioned as 164, with its cyclohexyl ring occupying almost the same space as the methyl-substituted phenyl ring of 164. Although identically substituted as 164, its three polar groups of 166 participate in a rather different hydrogen bonding network. Its primary amino nitrogen forms hydrogen bonds to Asp15-OD1 (3.14 Å) and water 672 (2.83 Å), which mediates a second hydrogen bond to Asp15-OD2 together with two other water molecules 821 and 832. One hydroxyl oxygen atom on the five-membered ring interacts with Thr223-OG1 (3.02 Å) and water 655 (2.80 Å), while the second oxygen atom forms a hydrogen bond to water 749 (3.02 Å), which further bonds to water 552. Another H-bond is observed in water 656 (3.13 Å), which mediates contact with Tyr226-OH and water 765. Similar to 164, the cyclopentyl ring of 166 is located in the S4 pocket, and the six-membered ring is adjacent to the S6 pocket.
EP-270 complex (Figure 1m, PDB-code: 5QBH). This fragment binds close to the catalytic center and extends from the active site in both directions, occupying either the S1 pocket with its terminal pyridine or the S1’ pocket with its tetrahydrofuran moiety. It forms a distinct hydrogen bonding network with the catalytic dyad of EP. The pyridine nitrogen binds to the carboxyl oxygens of the catalytic aspartates via the catalytic water molecule 585 (2.85 Å). One of the exocyclic hydroxyl oxygens (O3) on the tetrahydrofuran ring also interacts with the dyad via the catalytic water molecule 585 (2.92 Å). It also binds directly to Asp219-OD2 and to water 710, which is linked to the catalytic water 585 and Asp35-OD2. The second hydroxyl oxygen (O2) forms a hydrogen bond to the backbone carbonyl oxygen of Gly37 (2.79 Å). The central amide linker of the fragment involves the amide NH of Gly80 (2.70 Å) via its carbonyl oxygen and the adjacent amide NH group Thr222-OG1 in hydrogen bonds.
EP-203 complex (Figure 1k, PDB-code: 5QBF). This fragment is observed about 9 Å remote from the catalytic center and almost 5 Å away from Phe291. It binds with its exocyclic primary amino group on the tetrahydrofuran ring to the acid function of Asp15 (2.82 Å). This part of the fragment binds to the S4 pocket. The central amide nitrogen interacts with Thr223-OG1 (3.03 Å). The fluoro-substituted phenyl ring of 203, located in the S3 pocket, forms a π-stacking interaction with Asp15 at about 4.7 Å, and the terminal fluorine atom binds to Asp119-OD2 (alternative conformation B) at a distance of 3.18 Å. A second copy of 203 is found in the crystal structure adjacent to the S2 pocket. In the density, the fluoro-substituted phenyl ring and the attached amide bond of the second fragment copy are visible in the electron density. They bind adjacent to the first, fully defined fragment molecule. The visible part of the second fragment copy confirms an interaction via its amide carbonyl oxygen O with Thr223-N (3.15 Å) and water 620 (2.79 Å), which further binds to Leu224-N and Tyr226-OH.
EP-274 complex (Figure 1n, PDB-code: 5QBI). Located at least 4 Å away from the catalytic site, 274 interacts via its pyridine-type nitrogen with water 508 (3.18 Å) that connects to water 622, Asp33-OD2, and Gly221-O. Furthermore, the hydroxy–methylene–oxygen atom of the -CH2OH substituent forms hydrogen bonds to Thr223-OG1 (2.92 Å) and water 631 (2.82 Å). The latter connects to water 624 and Thr223-N. The eight-membered ring of the fragment appears in two mutually superimposed conformations. In the first conformer, the secondary ring amino group (N2) interacts with Ser115-OG (2.98 Å), whereas in the second conformation, this nitrogen interacts with Ser115-OG (3.04 Å) and Asp81-OD2 (2.52 Å). The fused pyridine moiety with its attached methyl group occupies the S1 pocket, while the methylene–hydroxy substituent at the eight-membered ring stretches out into the S3 pocket.
EP-175 complex (Figure 1j, PDB-code: 5QBE). Fragment 175 is found to bind to the surface of EP remote from the substrate-binding cleft. However, the hydrogen bonding interactions do not extend to symmetry-related molecules in the crystal packing. They involve the terminal amide nitrogen in an H-bond with the backbone carbonyl oxygen of Ser64 (2.73 Å), while the likely protonated tertiary nitrogen of the pyrrolidine ring interacts with the carbonyl oxygen of Ala67. The carboxylate group of the fragment is connected to the amide nitrogen of Leu69 via water 549 (2.69 Å).
2.3. Comparative Analysis
All of the discovered fragment hits contain a basic nitrogen functionality that most likely binds to EP in a protonated state. This reflects the properties of the EP binding site, which contains four aspartate residues (Asp15, Asp33, Asp81, Asp119) in addition to the two aspartates of the catalytic dyad (Asp35, Asp219). Most of the fragments are involved in binding to the catalytic dyad, with some replacing the catalytic water and occupying its position. Others leave the catalytic water in its position but establish water-mediated contact with the dyad residues. Three fragments (56, 80, 81) were found to share the same bicyclic parent scaffold. Two of them (56, 81) place their primary, probably charged, amino nitrogen at the position occupied by the catalytic water (Figure 2a–c). Surprisingly, 80 adopts a different orientation and interacts with the catalytic dyad via the catalytic water molecule. If 80 were to bind with a similar orientation as 56 and 81, the attached pyrrolidine ring would collide with Tyr79 and Phe116. This steric conflict is thought to explain the different orientation in the binding pocket. Furthermore, the formation of an H-bond to Asp81 further stabilizes the altered binding pose of 80 and underscores the preference for fragments to interact with one of the aspartates in the EP substrate binding site.
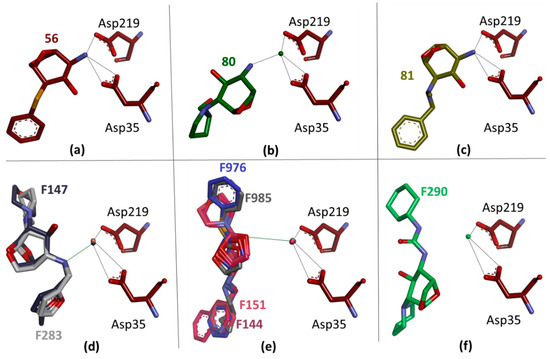
Figure 2.
The three fragments, 56 (a), 80 (b), and 81 (c), share a bicyclic parent scaffold but adopt different binding positions with the protein. While 56 and 81 have rather similar orientations and form a direct interaction with both aspartates of the catalytic dyad via their presumably protonated and charged amino group, 80 interacts with the dyad via the catalytic water molecule. The latter is displaced by the other two fragments. Comparing the binding modes of seven follow-up compounds with the same central parent scaffold, three different binding poses are observed. For F147 and F283 (d), the basic nitrogen of the ligands forms a water-mediated interaction with the aspartate residues in the catalytic center. F144, F151, F976, and F985 (e) adopt a second binding mode and bind slightly further away from the dyad. A weak polar interaction is formed by the bridging ether oxygen in the central bicycle of the ligands to the aspartates mediated through the catalytic water molecule. A third binding mode is found for F290 (f), which binds even further away (>3.8 Å) from the catalytic water and the dyad. This ligand does not have basic nitrogen that could serve as an H-bond donor to the catalytic center.
2.4. Crystal Structures and Validation Using Follow-Up Compounds in Complex with EP
To investigate whether the binding geometry of the bicyclic parent scaffold is retained with a higher substituted follow-up compound, we retrieved seven ligands, F144, F147, F151, F283, F290, F976, and F985, from the AnalytiCon library (hereafter “F” indicates a “follow-up” compound retrieved from the NATx library; the full IDs in the AnalytiCon library are listed in Table S5). In these follow-up compounds, an additional substituent has been added to the amino group of the fragment, changing it from a primary to a secondary group. This bi-substitution will also allow the original fragments to bridge the catalytic center and grow into two opposing binding pockets. With the exception of F290, we have chosen these compounds in such a way as to preserve the basic character of the fragment, which may be essential for the formation of charged interactions with EP. In F290, however, the basic amino group was replaced by an uncharged urea motif in order to study the importance of a charged nitrogen atom at this position. On the opposite side of the bicyclic ring system, we selected two ligands, F976 and F985, which carry the same thiophenyl substituent as found in fragment 56. The other compounds were chosen to carry basic nitrogen embedded in an aliphatic heterocycle (morpholino: F144, F147, F151 or piperidino: F283, F290).
Prior to soaking the selected follow-up compounds into crystals of EP, a biochemical binding assay was performed (see Section 4). All selected ligands turned out to be EP binders, and their crystal structures could be determined (a detailed description of their binding geometry can be found in the Supplementary Materials, and their chemical structure and observed binding mode are shown in Figure 3).
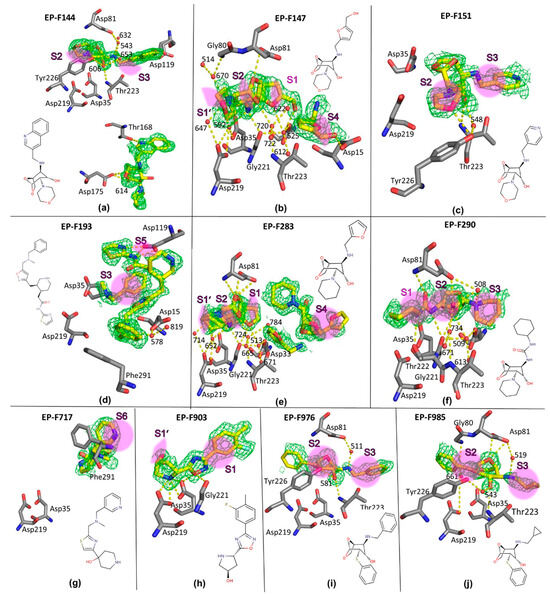
Figure 3.
(a–j). Crystallographically determined binding modes of ten follow-up compounds in EP. The follow-up compounds are shown as yellow sticks, with their IDs in bold (“F” stands for “follow-up”), contoured by the unbiased (Fo−Fc)-difference electron density, shown as green meshes (contour level at 2 σ). EP residues of the final refined EP complex structure are shown as gray sticks. The atoms are labeled with atom type colors (PyMOL default). The hydrogen bonds up to a donor-acceptor distance of 3.2 Å are shown as yellow dashed lines and involve interstitial water molecules. Waters are depicted as red spheres and labeled with numbers according to the corresponding PDB files (see Table S1). The spatial position of the binding sites S1–S6 and S1’, S2’ are approximately marked as magenta areas.
Disappointingly, none of the follow-up ligands comprising the bicyclic parent scaffold adopted a similar binding mode as the initial fragments. In total, three different binding orientations were found (Figure 2d–f). In the first one, adopted by F147 and F283, the ligands use, as planned, their polar secondary amino nitrogen to contact the catalytic dyad, however not directly, but rather through the catalytic water molecule (Figure 2d). Surprisingly, compared to the binding mode of 80, the orientation of the molecules is reversed (Figure 2b). For F147, two copies are bound to the enzyme; however, the second molecule is only partially visible in the electron density. The first copy occupies the S1 pocket, with part of its terminal furan ring and the attached methylene-hydroxy group facing the S3 pocket. The morpholino ring is located between the S1’ and S2 pockets (Figure 3b). Follow-up compound F283 is even present in three copies. One of these interacts with the catalytic dyad and adopts a binding mode virtually identical to that of the ligand F147.
In the second binding mode adopted by F144, F151, F976, and F985, the bicyclic parent scaffold binds close to the catalytic center. Again, the catalytic water remains in the structure (Figure 2e). However, it forms a long polar contact with the ether oxygen of the -OCH2- bridge of the bicyclic scaffold.
In the third binding mode found for F290, the ligand occupies approximately the same area as in the first binding mode (Figure 2f). However, probably due to the reduced basic character of the urea-type amino nitrogen, there is no direct polar contact with the catalytic water molecule (>3.8 Å) and the catalytic dyad.
Based on our experience in previous fragment studies on EP, direct contact with the catalytic dyad seems to be critical to achieving potent binding. Any indirect contact mediated by the catalytic water is more likely to be too weak to ensure a conserved binding pose, as found for 56 and 81 in the initial fragment screening when growing them into a larger follow-up compound. Based on the structural findings with the selected follow-up compounds, we decided not to proceed with the initially promising-looking bicyclic basic scaffold.
However, the pair of fragments, 75 and 283, caught our special interest. Both interact with the catalytic aspartates through charge-assisted hydrogen bonds formed by their basic amino nitrogen atoms located in either the terminal pyrrolidine or piperidine ring. Superimposing the corresponding crystal structures reveals an almost perfect match of the two heterocycles (Figure 4a). The remaining parts of the fragments are oriented toward either the S1 or S1’ pocket. This suggests the design hypothesis of fusing the two fragments by unifying the terminal carbocycles. Merging two independent fragments with adjacent binding poses into a single lead is a promising concept in fragment-based lead discovery [44,45,46]. This is expected to increase the binding affinity by at least about 3RT (six degrees of freedom), but values up to approximately 15–20 kJ/mol [47] have been suggested without adding new contacts to the protein beyond those already observed for the separated fragments. Successful examples have been described in the literature, e.g., by Nazare et al. [48] and Borsi et al. [49]. They studied the coupling of two non-overlapping fragments binding to factor Xa or matrix metalloproteinase MMP12 and found experimental values of 14–15 kJ/mol. The enhancement is due to the fact that the coupling of two fragments into one “supermolecule” restricts fewer rotational and translational degrees of freedom upon binding the ligands to the protein for entropic reasons. The entropic contributions involved are important in the thermodynamic inventory of binding and will improve the Gibbs free energy of binding. However, the success of this concept requires that the fragments to be bound remain in the positions observed for the individual fragments in their crystal structures and no unfavorable conformational strain is experienced in the designed “supermolecule”. To assess the chances of successful fusing, we tested the follow-up compounds F717 and F903 from the AnalytiCon library. Both ligands showed binding to EP, and the corresponding crystal structures were determined.
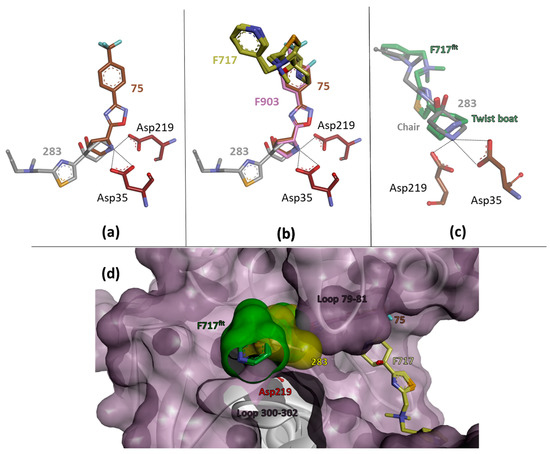
Figure 4.
(a) In a structural superposition, the two fragments, 75 (brown) and 283 (gray), formally overlap in the center by a pyrrolidine and a piperidine ring. Both rings use their polar nitrogen to interact directly with the aspartates of the catalytic dyad. (b) The follow-up ligand F903 (pink) confirms the binding mode of 75 (brown). Surprisingly, F717 (yellow), which is structurally related to 283 (gray), adopts a completely different binding mode at a distant position. It adopts two slightly different conformations at the remote site. (c) The follow-up compound F717 (green) shows the terminal piperidin-4-ol moiety in a twist boat conformation, whereas a chair conformation is observed for 283 (gray). Only in the chair conformation can a favorable H-bond be formed to the aspartates of the dyad. To demonstrate this difference, F717 has been fitted onto the binding mode of fragment 283. (d) The protein is shown with a purple surface. In 283 (yellow surface), there is a propargyl substituent that fits into a small crevice between loops 79–81 and loops 300–302. When F717fit (green) is artificially superimposed on the position of 283 (yellow), its terminal pyridine moiety (green surface) would sterically collide with the loop formed by residues 300–302 of the protein. This could explain its altered binding mode found experimentally and suggest a propargyl substituent to be used in a putative ligand fused through a central pyrrolidine ring.
As a result, F903 adopts a very similar binding position as fragment 75 (Figure 4b). Thus, F903 confirms the spatial orientation observed for the scaffold in 75. In the case of fragment 283, the crystal structure with the related compound F717 suggests a different binding orientation. While 283 forms a direct H-bond to Asp35 and is oriented in the S1’ pocket, F717 is found in a significantly different position. It is no longer in direct contact with the catalytic dyad but occupies the S6 pocket with its terminal pyridine moiety. The central part formed by its thiazole moiety partially occupies the S4 pocket, and the terminal piperidine is located in the S3 pocket. The electron density suggests that the ligand adopts two similar conformations in the protein-bound state. Superimposing F717 onto the binding position of 283 provides an answer as to why F717 is found with a different orientation than 283 (Figure 4d). The slender propargyl substituent of 283 fits optimally into a small crevice between two adjacent loops formed by residues 79–81 and 300–302. In contrast, the bulky pyridyl side chain of F717 will hardly fit into this crevice without steric clashes. Therefore, decoration with the slender propargyl substituent found at 283 seems advisable for the design of a fused ligand.
Another design aspect concerns the central carbocycle to be selected for the fusion of 75 with 283. The mutual superposition suggests that either the terminal pyrrolidine or piperidine ring can be used as the joining element of the two fragments. Remarkably, the piperidin-4-ol ring moiety, with the hydroxy group in an axial position, adopts a chair conformation in 283, whereas a twist boat geometry is found in F717 (Figure 4c). Only in the chair conformation can optimal interaction with the aspartyl residues in the catalytic site be achieved. Without sophisticated quantum chemical calculations taking into account the protein environment, it is difficult to estimate whether the boat conformation in F717, in which the piperidine nitrogen is not involved in charge-assisted hydrogen bonding, or the chair conformation in 283, in which the nitrogen is in contact with the dyad, is energetically more favorable. Five- and six-membered rings have very different conformational properties [50,51]. While a six-membered ring is rather rigid and limited to a small set of energetically favorable conformations, a five-membered ring has a much higher intrinsic flexibility and allows for a much larger conformational multiplicity. Since the fusion of the two fragments is a crucial design step, we believe that a five-membered pyrrolidine moiety will be the better choice as a bridging element, as this ring system exhibits a higher conformational flexibility and thus a better adaptability to achieve the required binding pose.
2.5. Binding Affinities of AnalytiCon Ligands by Isothermal Titration Calorimetry (ITC)
To obtain a rough estimate of the affinity of the different ligands, we performed ITC measurements. The affinities of 11 of the 15 fragments and all 10 follow-up compounds could be determined by displacement ITC [52,53]. Due to inefficient water solubility or distant binding of the fragment from the active site, no competitive displacement of the reference ligand SAP114 could be achieved for the following ligands: 62, 175, 203, and 245. The observed Kd values for 75, 140, 164, 166, and 270 are in the high millimolar range above >10 mM (Table S4). Single-digit millimolar values were obtained for 56, 80, 81, 134, 274, and 283. These results confirm the expected weak binding affinities of the crystallographic fragment hits. In comparison, the Kd values of the follow-up compounds were generally somewhat lower than those of the fragments. While most of the follow-up compounds also fall in the low millimolar range, four had affinities in the three-digit micromolar range. This is consistent with the expectation that follow-up compounds will have higher binding affinities due to more functional groups binding to the protein. However, the ligand efficiencies of all compounds studied remained below 1 kJ mol−1 atom−1 (Table S4). Thus, they did not exceed the threshold of 1.25 kJ mol−1 atom−1, which is considered promising for further drug development [54,55]. This may be due to the large binding cleft of EP, which facilitates the flexibility of binding possibilities.
3. Conclusions and Outlook
The extraordinarily high hit rate and the possibility of using follow-up library entries for fragment hit validation and expansion make a crystallographic fragment screening campaign based on compounds derived from the natural product libraries highly attractive. Since three fragments shared a common bicyclic parent scaffold (56, 80, 81), we decided to first validate how well this parent scaffold could be grown into larger follow-up ligands. Two of the fragments bind directly with their presumably protonated amino group to the pivotal position between the two aspartates and displace the catalytic water molecule. The third fragment contacts the catalytic residues through the catalytic water. Follow-up ligands that carry an additional substituent on the critical amino group were selected from the NATx library. Although this increases the steric bulk at this position, a number of examples have been reported in the literature [56,57], in which basic amino nitrogen embedded in a chain can still interact directly with the aspartates of the dyad. In order to study the effect of the basic character of this nitrogen on the binding, we have also included ligand F290 with urea-type nitrogen in the series. In conclusion, the binding mode of the initial fragments was not conserved in this series of extended derivatives. They contact the dyad exclusively via the catalytic water molecule and the presumably uncharged urea derivative even binds without direct interaction with this catalytic water molecule. Considering the affinity improvement of the enlarged follow-up compounds, only a slight increase in binding affinity is observed. In our opinion, this was far too little to highlight this scaffold as a promising candidate for further development.
We therefore considered fusing two fragments (75, 283), which in their individual crystal structures, showed a promising overlap of two terminal heterocycles. To validate the chances of success, we selected two related ligands from the NATx library and determined their binding modes (F903 and F717). While F903 confirms the unchanged binding of 75, F717 deviates from the binding position observed for the starting fragment 283. Remarkably, by fitting the remotely bound ligand to the one near the catalytic center, it became clear that the pyridyl substituent of F717 is sterically too large to allow a similar binding mode to that observed for 283. The latter places its slender propargyl substituent in a narrow crevice of the enzyme. In addition, we observed conformational ambiguities in the terminal six-membered ring of 283 and F717, which led us to propose using a conformationally more flexible five-membered pyrrolidine ring as a bridging element between the two fragments to be fused. Considering the binding data from ITC, fragment 283 is a single-digit millimolar binder, while 75 binds even weaker. Furthermore, 283 requires the slender propargyl group for binding, further expansion at this site may be critical. Thus, in terms of further optimization of a fused follow-up compound, the focus should probably be on the side originally occupied by 75 rather than the side occupied by 283.
The ligands in the AnalytiCon libraries are all synthetic molecules inspired by natural product motifs (NATx libraries). A subset of smaller intermediates of typical fragment size are collected in the FRGx library. In addition to the high hit rates obtained with these fragments, the AnalytiCon libraries contain large sets of closely related analogs with modulated substitution patterns. This makes the evaluation of the obtained fragment hits extremely efficient and helps to select the best candidates for further optimization by adding alternative substituents or merging and fusing fragments into larger lead candidates.
4. Materials and Methods
4.1. Ligand Selection for the Initial Crystallographic Fragment Screening
An initial set of 73 fragments was selected from AnalytiCon’s FRGx library (Fragments from Nature—Inherent Synthetic Tractability, documented and publicly available at https://ac-discovery.com/wp-content/uploads/AnalytiCon_Discovery_NATx_Product_Information.pdf (assessed on 24 August 2024)). The selection was based on compound availability and diversity of the fragments.
After a successful first screening campaign using crystallography, we wanted to confirm the observed binding poses by studying slightly expanded ligands with similar chemical composition. We therefore retrieved promising candidates from the NATx library (Semi-Synthetic Screening Compound Library based on Natural Product Scaffolds) of AnalytiCon discovery (https://ac-discovery.com/screening-libraries/ (assessed on 24 August 2024)) and tested their binding using a biochemical assay performed as described in Köster et al. [58,59]
4.2. Crystallization of EP
Purified EP [58,59] was crystallized as described in Huschmann et al. [11]. EP crystals were grown using streak seeding in 400 nL sitting drops that contained 50% reservoir solution (0.1 M NaAc pH 4.6, 0.1 M NH4Ac pH 4.6 and 28% (w/v) PEG 4000) and 50% protein solution (4.8–5.1 mg/mL protein in 0.1 M NaAc pH 4.6). Crystallization experiments were carried out in INTELLI-PLATE 96-3 low-profile plates (MiTeGen/Art Robbins Instruments, Sunnyvale, CA, USA) using a Gryphon robot (Art Robbins Instruments, Sunnyvale, CA, USA). Crystals used for the soaking of ligand 283 were grown similarly using 2 µL drops and a reservoir with 24% (w/v) PEG8000 instead of 28% (w/v) PEG8000 on Cryschem plates (Hampton Research, Aliso Viejo, CA, USA) set up manually.
4.3. Soaking of Crystals
The soaking procedure was based on the original protocol of [59] in the way that the cryo-protection was included. Following the approach by Huschmann et al. [11], only one fragment was soaked per crystal. The stock solutions of fragments and follow-up compounds were dissolved to 1 M DMSO. If necessary, heat and sonication were applied for proper dissolving. For the soaking experiment, all soaking buffers containing 7.5 µL reservoir solution, 2.5 µL glycerol, and 1 µL of the fragment or follow-up compound stock solution in DMSO were prepared in a deep well block and shaken overnight. With a multichannel pipette 30 µL soaking solution without fragments and 1 µL drops of soaking solution containing one fragment per well were pipetted in INTELLI-PLATE 96-3 low profile plates. At least two crystals per fragment or follow-up compound were incubated in 1 µL of soaking buffer for a few minutes to 48 h, harvested with a MiTeGen loop, and flash-cooled in liquid nitrogen. For soaking of 283, the procedure was slightly altered. The soaking buffer contained 25% (v/v) MPD as cryo-protection instead of glycerol and was prepared manually. Soaking was performed on Cryschem M plates (Hampton Research, Aliso Viejo, CA, USA) with empty reservoirs. Two crystals were incubated in 2 µL of soaking buffer for 24 h, harvested with a nylon loop, and flash-cooled in liquid nitrogen.
4.4. Data Collection and Processing
EP diffraction data sets were collected at 13.5 keV at beamline BL14.1 at the BESSY II storage ring [60,61] using a PILATUS 6M detector (DECTRIS). To ensure high completeness and reduce radiation damage to a minimum, a data collection strategy was calculated for each data set using iMOSFLM [62]. From the calculated rotation starting point, 200° of data were measured from each crystal in 0.1° increments and with an exposure time of 0.1–0.3 s. The processing of all data sets was conducted with the expert system and graphical user interface XDSAPP [63,64,65]. Occasionally, minor manual adjustments to the automated processing protocol were needed. Data for 283 were collected on P11 at DESY using an EIGER2 detector (DECTRIS). No data collection strategy was calculated for 283, but instead, 180° of data in 0.1° increments with an exposure time of 10 ms was collected. Detailed data collection and processing parameters are given in Table S1.
4.5. Structure Refinement and Fragment Binding Analysis
The reflection files (mtz format) resulting from XDSAPP were subjected to an automated refinement pipeline that uses PHENIX.REFINE [66]. The starting model for this pipeline was PDB-code 4Y3J [11]. Electron density maps and coordinate files of each experiment obtained from the automated refinement were inspected manually, and the presence or absence of the expected fragment was assigned according to the difference electron density map. Subsequently, the identified hit structures were subjected to several cycles of alternating model building in COOT [67] and refinement in PHENIX [66]. Occupancy refinement was carried out for all fragments. Detailed refinement and validation statistics are shown in Table S2.
Overall, quality is assured by the following relevant data quality indicators: the Isa values [68], with an average of 33.5, the data resolution (1.12–1.76 Å), and the Rfree values (14.7–23.7%). The fragments and follow-up compounds have refined occupancies between 50 and 99%, with corresponding real space correlation coefficients (RSCC [69]) ranging from 0.72 to 0.95 (Table S3). All fragments were defined unambiguously in the electron density. Nine of the twenty-five ligands were in the top category (RSCC 1.0-0.9; in cases of several ligand molecules the one with the higher RSCC was considered), and fifteen ligands were in the second category (RSCC 0.9-0.8), according to the validation categorization by [70]. Thus, many of the ligands fit the density well, while the second category ligands fit the density at least partially [70]. Although the fragments are rather weak binders, only one of the observed ligands (F717) showed an RSCC below 0.72, which may be due to the high flexibility of the fragment, which only forms a stacking interaction with its pyridine ring and additionally had to be modeled in two conformations. Two additional fragment molecules (F144, F283) are bound multiple times, with the lower populated fragment showing RSCC values of 0.74 and 0.75, respectively. However, the more highly populated fragment has RSCC values of 0.90 and 0.87.
4.6. Isothermal Titration Calorimetry (ITC)
All crystallographic hits were subjected to displacement ITC titration experiments, as described in [11]. Experiments were carried out using a MicroCalTM ITC200 system (GE Healthcare, Atlanta, GA, USA) at 25 °C. The sample cell was filled with ca. 300 µL of purified EP [59] at a concentration of 50 µM in 0.1 M NaAc buffer at pH 4.6, 3% (v/v) DMSO, and 2 mM fragment or follow-up compound. The same solution without any fragment or follow-up compound was used to determine the thermodynamic profile of the reference ligand SAP114 (molecule 9 in [71]). SAP114 was present at a concentration of 500 µM in the same buffer and titrated into the sample cell with 1.2–1.5 µL injections (22–31 individual injections) and 180 s spacings. The temperature difference/heat release was measured and integrated using the software Nitpic, version 1.0.3 [72]. Isotherm fitting was then performed using Sedphat, version 10.58d [73]. The association constant Ka is directly accessible from the slope of the binding curve. Based on this value, the dissociation constant Kd, as well as the Gibbs free energy ΔG°, can be determined using the equations ΔG° = RT × ln(Kd), where R is the gas constant (8.3144 J mol−1 K−1) and T is the temperature of the experiment in Kelvin (298.15 K). Furthermore, we determined the ligand efficiency of each fragment (LE) via the equation LE = −ΔG°/non-H-atom-count.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cryst14090755/s1: Description of crystal structures of the complexes with the follow-up compounds; Table S1: Data collection and processing statistics; Table S2: Structure refinement and validation statistics; Table S3: Occupancies and RSCC of the 25 ligand hits; Table S4: ITC analysis of fragment and follow-up compound binding to EP; Table S5: Chemical formulas of AnalytiCon discovery hit structures.
Author Contributions
Conceptualization: A.H. and G.K.; ITC measurements: A.M; crystallographic data collection and evaluation: F.U.H., J.M. and A.H.; crystal growth and soaking: F.U.H., M.R. and J.S.; data handling, computational analysis: J.S.; interpretation of data: F.U.H., A.M., A.H., S.G. and G.K.; writing—original draft: F.U.H., A.H. and G.K.; writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Federal Ministry of Education and Research (BMBF), BioChancePlus, FragScreen no. 0315161C and Frag2Xtal no. 05K13M1. European Research Council (ERC), Grant 268145-DrugProfilBind.
Data Availability Statement
The crystal structures have been deposited with the Protein Data Bank and can be accessed using the PDB-codes listed above in Section 2.2 and in Table S1.
Acknowledgments
The authors are grateful to AnalytiCon GmbH for making a subset of their NATx library available for this study. The present study was supported by the German Federal Ministry of Education and Research (BMBF) under the BioChancePlus program (Project FragScreen no. 0315161C) and the BMBF Project Frag2Xtal (no. 05K13M1). We acknowledge the support during data collection and helpful discussions on the fragment screening project with Manfred S. Weiss, Uwe Mueller, Karine Röwer, and Monika Ühlein at BESSY II (Berlin, Germany). We also thank the Helmholz-Zentrum Berlin, Germany, for the travel support. We acknowledge the provision of beam time at DESY and the support of the beamline staff of P11. We are grateful for the financial support from the European Research Council (ERC) of the European Union (grant 268145-DrugProfilBind).
Conflicts of Interest
Authors Moritz Ruf, Johanna Senst, and Serghei Glinca were employed by the company CrystalsFirst GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Hajduk, P.J.; Sheppard, G.; Nettesheim, D.G.; Olejniczak, E.T.; Shuker, S.B.; Meadows, R.P.; Steinman, D.H.; Carrera, G.M., Jr.; Marcotte, P.A.; Severin, J.; et al. Discovery of Potent Nonpeptide Inhibitors of Stromelysin Using SAR by NMR. J. Am. Chem. Soc. 1997, 119, 5818–5827. [Google Scholar] [CrossRef]
- Blundell, T.L.; Jhoti, H.; Abell, C. High-Throughput Crystallography for Lead Discovery in Drug Design. Nat. Rev. Drug Discov. 2002, 1, 45–54. [Google Scholar] [CrossRef]
- Erlanson, D.A.; McDowell, R.S.; O’Brien, T. Fragment-based drug discovery. J. Med. Chem. 2004, 47, 3463–3482. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Greer, J. A Decade of Fragment-based Drug Design: Strategic Advances and Lessons Learned. Nat. Rev. Drug Discov. 2007, 6, 211–219. [Google Scholar] [CrossRef]
- Baker, M. Fragment-based lead discovery grows up. Nat. Rev. Drug Discov. 2013, 12, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Hou, J.; Zimmerman, M.D.; Wlodawer, A.; Minor, W. The future of crystallography in drug discovery. Expert Opin. Drug Discov. 2013, 9, 125–137. [Google Scholar] [CrossRef]
- Bijak, V.; Szczygiel, M.; Lenkiewicz, J.; Gucwa, M.; Cooper, D.R.; Murzyn, K.; Minor, W. The current role and evolution of X-ray crystallography in drug discovery and development. Expert Opin. Drug Discov. 2023, 18, 1221–1230. [Google Scholar] [CrossRef]
- Woodhead, A.J.; Erlanson, D.A.; de Esch, I.J.P.; Holvey, R.S.; Jahnke, W.; Pathuri, P. Fragment-to-Lead Medicinal Chemistry Publications in 2022. J. Med. Chem. 2024, 67, 2287–2304. [Google Scholar] [CrossRef]
- Schiebel, J.; Radeva, N.; Krimmer, S.G.; Wang, X.; Stieler, M.; Ehrmann, F.R.; Fu, K.; Huschmann, F.U.; Metz, A.; Weiss, M.S.; et al. Six Biophysical Screening Methods Miss a Large Proportion of Crystallographically Discovered Fragment Hits: A Case Study. ACS Chem. Biol. 2016, 11, 1693–1701. [Google Scholar] [CrossRef]
- Huschmann, F.U.; Linnik, J.; Sparta, K.; Ühlein, M.; Wang, X.; Metz, A.; Schiebel, J.; Heine, A.; Klebe, G.; Weiss, M.S.; et al. Structures of endothiapepsin-fragment complexes by crystallographic fragment-screening using a novel, diverse and affordable 96-compound-fragment library. Acta Crystallogr. F 2016, F72, 346–355. [Google Scholar] [CrossRef]
- Füsser, F.T.; Wollenhaupt, J.; Weiss, M.S.; Kümmel, D.; Koch, O. Novel starting points for fragment-based drug design against mycobacterial thioredoxin reductase identified using crystallographic fragment screening. Acta Crystallogr. Sect. D 2023, D79, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Walsh, M.A.; et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhang, R.; Groves, M.R. Fragment Screening in the Development of a Novel Anti-Malarial. Crystals 2023, 13, 1610. [Google Scholar] [CrossRef]
- de Souza Neto, L.R.; Montoya, B.O.; Brandão-Neto, J.; Verma, A.; Bowyer, S.; Moreira-Filho, J.T.; Dantas, R.F.; Neves, B.J.; Andrade, C.H.; von Delft, F.; et al. Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni. Sci. Rep. 2024, 14, 1582. [Google Scholar] [CrossRef]
- Neumann, P.; Heidemann, J.L.; Wollenhaupt, J.; Dickmanns, A.; Agthe, M.; Weiss, M.S.; Ficner, R. A small step towards an important goal: Fragment screen of the c-di-AMP-synthesizing enzyme Cda. Acta Crystallogr. Sect. D 2024, D80, 350–361. [Google Scholar] [CrossRef]
- Douangamath, A.; Powell, A.; Fearon, D.; Collins, P.M.; Talon, R.; Krojer, T.; Skyner, R.; Brandao-Neto, J.; Dunnett, L.; Dias, A.; et al. Achieving Efficient Fragment Screening at XChem Facility at Diamond Light Source. J. Vis. Exp. 2021, 29, e62414. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, D.P.; Steuber, J.; Fritz, G.; Wojdyla, J.A.; Sharpe, M.E. Fast fragment and compound screening pipeline at the Swiss Light Source. Methods Enzymol. 2023, 690, 235–284. [Google Scholar]
- Metz, A.; Stegmann, D.P.; Panepucci, E.H.; Buehlmann, S.; Huang, C.-Y.; McAuley, K.E.; Wang, M.; Wojdyla, J.A.; Sharpea, M.E.; Smitha, K.M.L. HEIDI: An experiment-management platform enabling high-throughput fragment and compound screening. Acta Crystallogr. Sect. D 2024, D80, 328–335. [Google Scholar] [CrossRef]
- Keserű, G.M.; Erlanson, D.A.; Ferenczy, G.G.; Hann, M.M.; Murray, C.W.; Pickett, S.D. Design Principles for Fragment Libraries: Maximizing the Value of Learnings from Pharma Fragment-Based Drug Discovery (FBDD) Programs for Use in Academia. J. Med. Chem. 2016, 59, 8189–8206. [Google Scholar] [CrossRef]
- Troelsen, N.S.; Clausen, M.H. Library Design Strategies To Accelerate Fragment-Based Drug Discovery. Chemistry 2020, 26, 11391–11403. [Google Scholar] [CrossRef]
- Available online: https://practicalfragments.blogspot.com/2023/12/review-of-2023-reviews.html (accessed on 24 August 2024).
- Available online: https://www.cambridgemedchemconsulting.com/ (accessed on 24 August 2024).
- Congreve, M.; Carr, R.; Murray, C.; Jhoti, H.A. ‘Rule of Three’ for fragment-based lead discovery. Drug Discov. Today 2003, 8, 876–877. [Google Scholar] [CrossRef]
- Jhoti, H.; Williams, G.; Rees, D.C.; Murray, C.W. The ‘rule of three’ for fragment-based drug discovery: Where are we now? Nat. Rev. Drug Discov. 2013, 12, 644–645. [Google Scholar] [CrossRef]
- Over, B.; Wetzel, S.; Grütter, C.; Nakai, Y.; Renner, S.; Rauh, D.; Waldmann, H. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 2013, 5, 21–28. [Google Scholar] [CrossRef]
- Prescher, H.; Koch, G.; Schuhmann, T.; Ertl, P.; Bussenault, A.; Glick, M.; Dix, I.; Petersen, F.; Lizos, D.E. Construction of a 3D-shaped, natural product like fragment library by fragmentation and diversification of natural products. Bioorg. Med. Chem. 2017, 25, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Pascolutti, M.; Campitelli, M.; Nguyen, B.; Pham, N.; Gorse, A.-D.; Quinn, R.J. Capturing Nature’s Diversity. PLoS ONE 2015, 10, e0120942. [Google Scholar] [CrossRef]
- Chopra, B.; Dhingra, A.K. Natural products: A lead for drug discovery and development. Phytother. Res. 2021, 35, 4660–4702. [Google Scholar] [CrossRef] [PubMed]
- Grigalunas, M.; Burhop, A.; Zinken, S.; Pahl, A.; Gally, J.M.; Wild, N.; Mantel, Y.; Sievers, S.; Foley, D.J.; Scheel, R.; et al. Natural product fragment combination to performance-diverse pseudo-natural products. Nat. Commun. 2021, 12, 1883. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.F.; Hamilton, D.J.; de Esch, I.J.P.; Wijtmans, M.; O’Brien, P. Escape from planarity in fragment-based drug discovery: A synthetic strategy analysis of synthetic 3D fragment libraries. Drug Discov. Today 2022, 27, 2484–2496. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Available online: https://ac-discovery.com/wp-content/uploads/AnalytiCon_Discovery_NATx_Product_Information.pdf (accessed on 24 August 2024).
- Haustedt, L.O.; Siems, K. The Role of Natural Products in Drug Discovery. In Small Molecule Medicinal Chemistry: Strategies and Technologies; Czechtizky, W., Hamley, P., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; Chapter 14; ISBN 97811187716002016. [Google Scholar]
- Available online: https://ac-discovery.com/fragments-nature/ (accessed on 24 August 2024).
- Pearl, L.; Blundell, T. The active site of aspartic proteinases. FEBS Lett. 1984, 174, 96–101. [Google Scholar] [CrossRef]
- Dash, C.; Kulkarni, A.; Dunn, B.; Rao, M. Aspartic Peptidase Inhibitors: Implications in Drug Development. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 89–119. [Google Scholar] [CrossRef]
- Eder, J.; Hommel, U.; Cumin, F.; Martoglio, B.; Gerhartz, B. Aspartic Proteases in Drug Discovery. Curr. Pharm. Des. 2007, 13, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K. (Ed.) Aspartic acid proteases as therapeutic targets. In Methods and Principles in Medicinal Chemistry; Wiley-VCH: Weinheim, Germany, 2010; Volume 45. [Google Scholar]
- Schiebel, J.; Krimmer, S.G.; Sparta, K.; Knörlein, A.; Wang, X.; Park, A.Y.; Stieler, M.; Ehrmann, F.R.; Fu, K.; Radeva, N.; et al. High-throughput Crystallography: Reliable and Efficient Identification of Fragment Hits. Structure 2016, 24, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Radeva, N.; Krimmer, S.G.; Stieler, M.; Schiebel, J.; Fu, K.; Wang, X.; Ehrmann, F.R.; Metz, A.; Huschmann, F.U.; Weiss, M.; et al. Remote Interplay of Small Molecules with Endothiapepsin—Hot spot analysis. J. Med. Chem. 2016, 59, 7561–7575. [Google Scholar] [CrossRef]
- Radeva, N.; Schiebel, J.; Wang, X.; Krimmer, S.G.; Fu, K.; Stieler, M.; Ehrmann, F.R.; Metz, A.; Rickmeyer, T.; Betz, M.; et al. Active Site Mapping of an Aspartic Protease by Multiple Fragment Crystal Structures: Versatile Warheads to Address a Catalytic Dyad. J. Med. Chem. 2016, 59, 9743–9759. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Bancet, A.; Raingeval, C.; Lomberget, T.; Le Borgne, M.; Guichou, J.F.; Krimm, I. Fragment Linking Strategies for Structure-Based Drug Design. J. Med. Chem. 2020, 63, 11420–11435. [Google Scholar] [CrossRef]
- Grenier, D.; Audebert, S.; Preto, J.; Guichou, J.F.; Krimm, I. Linkers in fragment-based drug design: An overview of the literature. Expert Opin. Drug Discov. 2023, 18, 987–1009. [Google Scholar] [CrossRef] [PubMed]
- Bedwell, E.V.; McCarthy, W.J.; Coyne, A.G.; Abell, C. Development of potent inhibitors by fragment-linking strategies. Chem. Biol. Drug Des. 2022, 100, 469–486. [Google Scholar] [CrossRef]
- Murray, C.W.; Verdonk, M.L. The consequences of translational and rotational entropy lost by small molecules on binding to proteins. J. Comput. Aided Mol. Des. 2002, 16, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Nazaré, M.; Matter, H.; Will, D.W.; Wagner, M.; Urmann, M.; Czech, J.; Schreuder, H.; Bauer, A.; Ritter, K.; Wehner, V. Fragment deconstruction of small, potent factor Xa inhibitors: Exploring the superadditivity energetics of fragment linking in protein-ligand complexes. Angew. Chem. Int. Ed. Engl. 2012, 51, 905–911. [Google Scholar] [CrossRef]
- Borsi, V.; Calderone, V.; Fragai, M.; Luchinat, C.; Sarti, N. Entropic contribution to the linking coefficient in fragment-based drug design: A case study. J. Med. Chem. 2010, 53, 4285–4289. [Google Scholar] [CrossRef]
- Chan, L.; Hutchison, G.R.; Morris, G.M. Understanding Ring Puckering in Small Molecules and Cyclic Peptides. J. Chem. Inf. Model. 2021, 61, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Dragojlovic, V. Conformational analysis of cycloalkanes. ChemTexts 2015, 1, 14. [Google Scholar] [CrossRef]
- Rühmann, E.; Betz, M.; Fricke, M.; Heine, A.; Schäfer, M.; Klebe, G. Thermodynamic Signatures of Fragment Binding: Validation of Direct versus Displacement ITC Titrations. Biochim. Biophys. Acta 2015, 1850, 647–656. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Zhang, Z.-Y. Low-Affinity Binding Determined by Titration Calorimetry Using a High-Affinity Coupling Ligand: A Thermodynamic Study of Ligand Binding to Protein Tyrosine Phosphatase 1B. Anal. Biochem. 1998, 261, 139–148. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Chen, K.; Sharp, K.A.; Kollman, P.A. The maximal affinity of ligands. Proc. Natl. Acad. Sci. USA 1999, 96, 9997–10002. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Groom, C.R.; Alex, A. Ligand efficiency: A useful metric for lead selection. Drug Discov. Today 2004, 9, 430–431. [Google Scholar] [CrossRef]
- Blum, A.; Böttcher, J.; Heine, A.; Klebe, G.; Diederich, W.E. Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold. J. Med. Chem. 2008, 51, 2078–2087. [Google Scholar] [CrossRef]
- Blum, A.; Böttcher, J.; Sammet, B.; Luksch, T.; Heine, A.; Klebe, G.; Diederich, W.E. Achiral Oligoamines as Versatile Tool for the Development of Aspartic Protease Inhibitors. Bioorg. Med. Chem. 2008, 16, 8574–8586. [Google Scholar]
- Köster, H.; Craan, T.; Brass, S.; Herhaus, C.; Zentgraf, M.; Neumann, L.; Heine, A.; Klebe, G. A Small Nonrule of 3 Compatible Fragment Library Provides High Hit Rate of Endothiapepsin Crystal Structures with Various Fragment Chemotypes. J. Med. Chem. 2011, 54, 7784–7796. [Google Scholar] [CrossRef] [PubMed]
- Köster, H. Endothiapepsin und Proteinkinase A: Komplexstrukturen mit Neuartigen Inhibitoren, Durchmustern einer Fragmentbibliothek Sowie Inhibitordesign Ausgehend von Einer Sonde. Ph.D. Thesis, Philipps-Universität Marburg, Marburg, Germany, 2012. Available online: https://d-nb.info/102718376X/34 (accessed on 24 August 2024).
- Mueller, U.; Darowski, N.; Fuchs, M.R.; Förster, R.; Hellmig, M.; Paithankar, K.S.; Pühringer, S.; Steffien, M.; Zocher, G.; Weiss, M.S. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Rad. 2012, 19, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.; Förster, R.; Hellmig, M.; Huschmann, F.U.; Kastner, A.; Malecki, P.; Pühringer, S.; Röwer, M.; Sparta, K.; Steffien, M.; et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 2015, 130, 141–152. [Google Scholar] [CrossRef]
- Geoff, T.; Battye, G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Andrew, G.; Leslie, W. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst. 2011, D67, 271–281. [Google Scholar]
- Kabsch, W. XDS. Acta Crystallogr. Sect. 2010, D66, 125–132. [Google Scholar] [CrossRef]
- Sparta, K.M.; Krug, M.; Heinemann, U.; Mueller, U.; Weiss, M.S. XDSAPP2.0. J. Appl. Crystallogr. 2016, 49, 1085–1092. [Google Scholar] [CrossRef]
- Krug, M.; Weiss, M.S.; Heinemann, U.; Mueller, U. XDSAPP: A graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 2012, 45, 568–572. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D 2012, D68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 2010, D66, 486–501. [Google Scholar] [CrossRef]
- Diederichs, K. Quantifying instrument errors in macromolecular X-ray data sets. Acta Crystallogr. Sect. D 2010, D66, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Tickle, I.J. Statistical quality indicators for electron-density maps. Acta Crystallogr. Sect D 2012, D68, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Deller, M.C.; Rupp, B. Models of protein–ligand crystal structures: Trust, but verify. J. Comput. Aided Mol. Des. 2015, 29, 817–836. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, M.; Köster, H.; Bartholomäus, R.; Park, A.Y.; Shahim, A.; Heine, A.; Steuber, H.; Klebe, G.; Diederich, W.E. Tracing Binding Modes in Hit-to-Lead Optimization: Chameleon-Like Poses of Aspartic Protease Inhibitors. Angew. Chem. Int. Ed. Engl. 2015, 54, 2849–2853. [Google Scholar] [CrossRef]
- Keller, S.; Vargas, C.; Zhao, H.; Piszczek, G.; Brautigam, C.A.; Schuck, P. High-Precision Isothermal Titration Calorimetry with Automated Peak-Shape Analysis. Anal. Chem. 2012, 84, 5066–5073. [Google Scholar] [CrossRef]
- Houtman, J.C.; Brown, P.H.; Bowden, B.; Yamaguchi, H.; Appella, E.; Samelson, L.E.; Schuck, P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci. 2007, 16, 30–42. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).