Morphological Investigation of Protein Crystals by Atomic Force Microscopy
Abstract
1. Introduction on Atomic Force Microscope
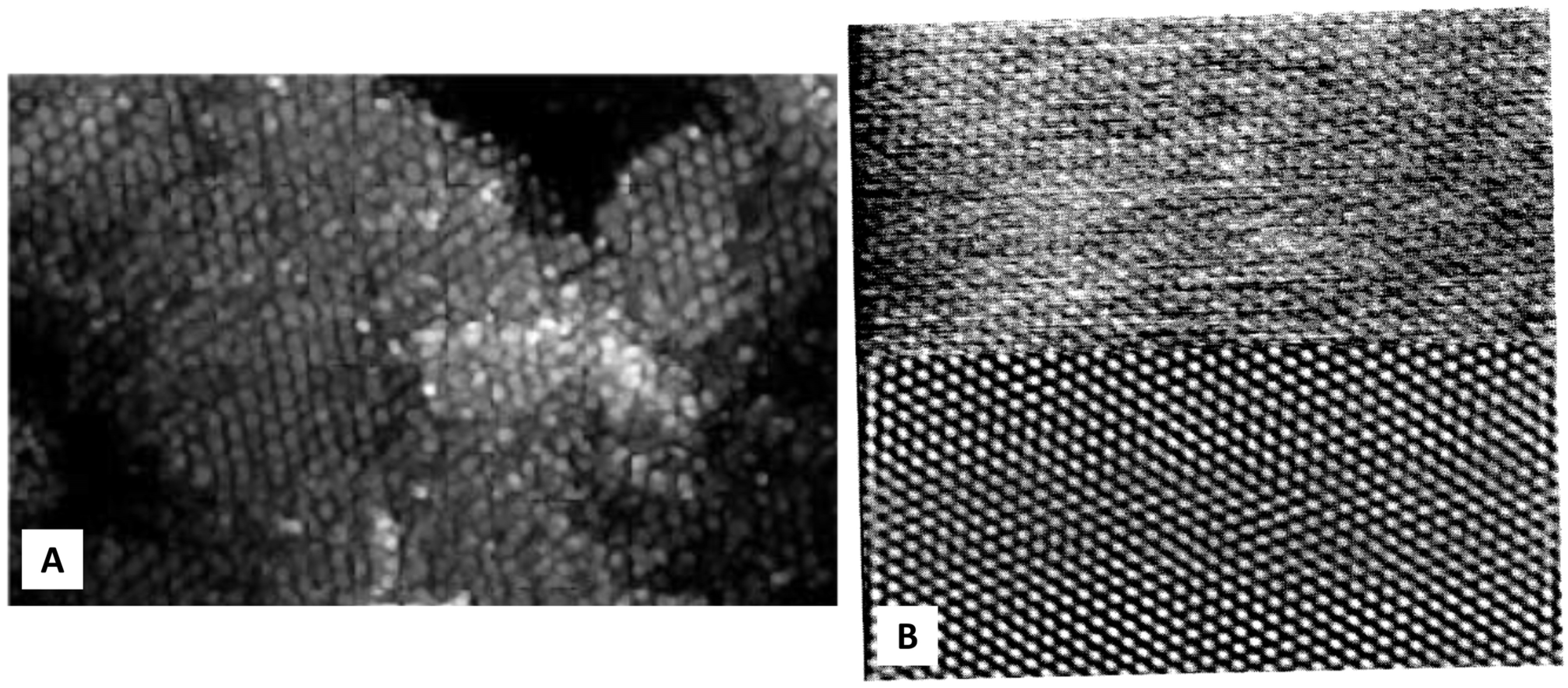
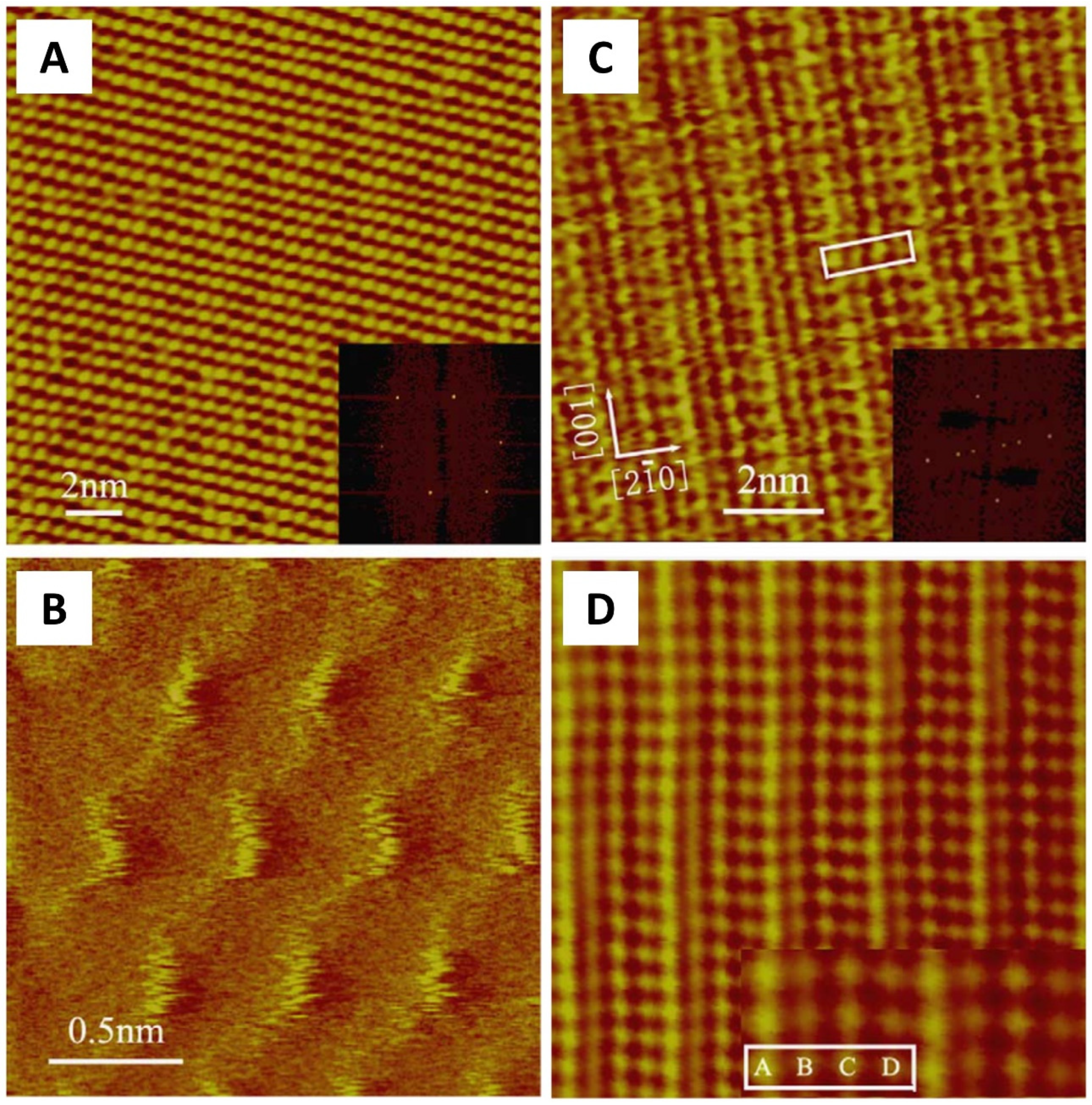
2. Protein Crystal: From Molecule Distribution to Surface Morphology with AFM
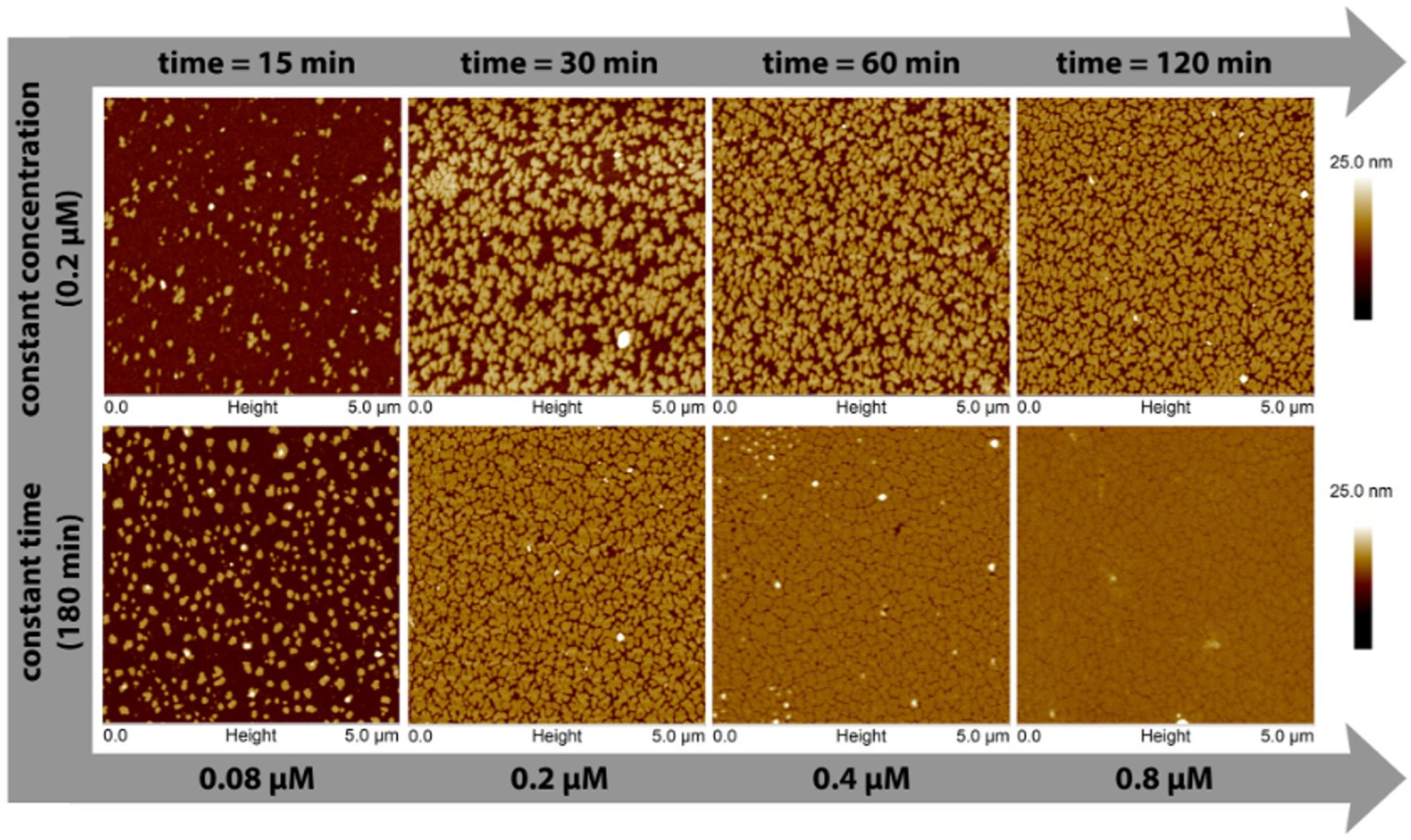
3. S-Layers
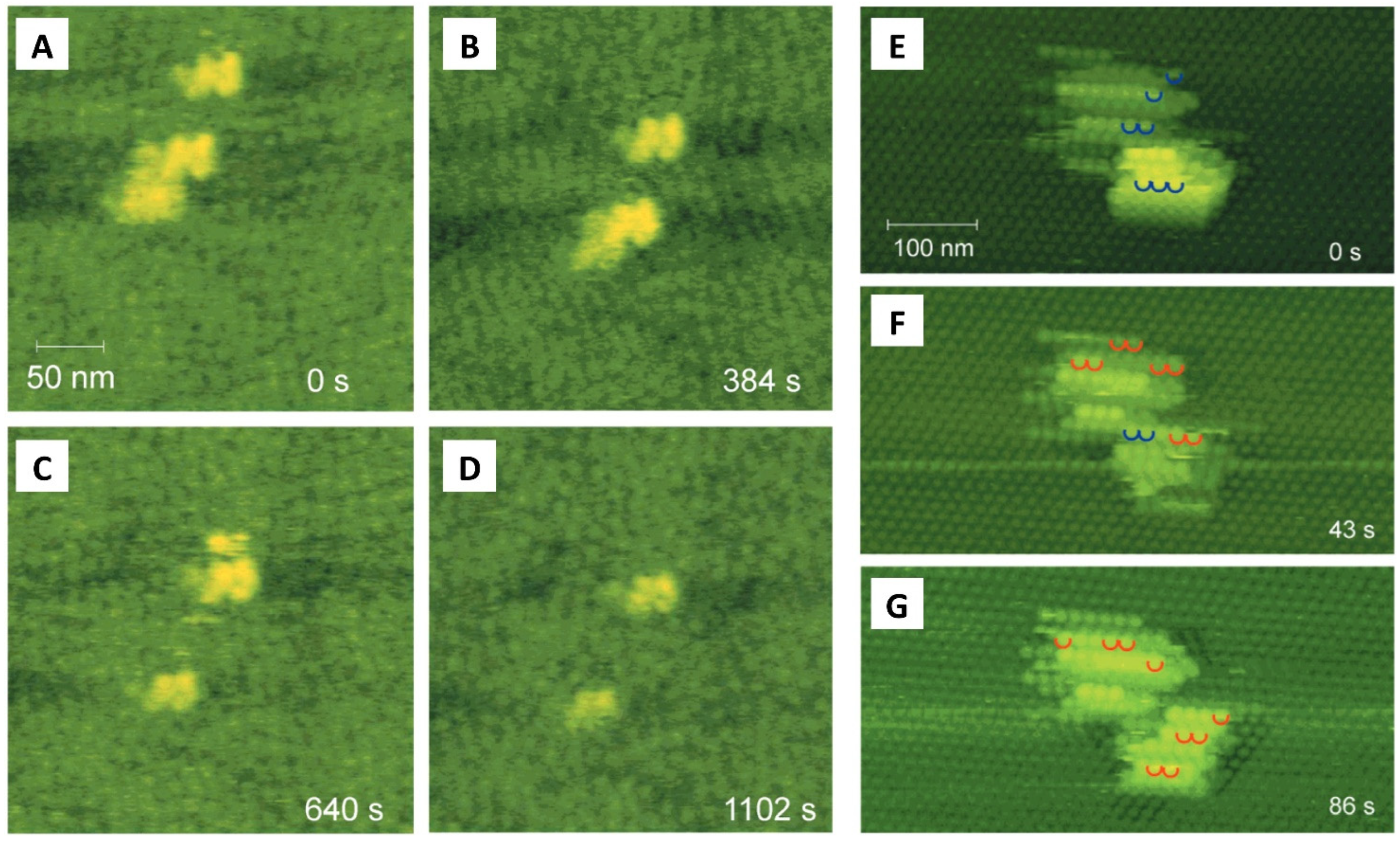
4. Real-Time Visualization of Biomolecular Dynamic: High-Speed AFM
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef]
- Durbin, S.D.; Carlson, W.E. Lysozyme Crystal Growth Studied by Atomic Force Microscopy. J. Cryst. Growth 1992, 122, 71–79. [Google Scholar] [CrossRef]
- Durbin, S.D.; Carlson, W.E.; Saros, M.T. In Situ Studies of Protein Crystal Growth by Atomic Force Microscopy. J. Phys. D Appl. Phys. 1993, 26, B128. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic Force Microscopy as a Multifunctional Molecular Toolbox in Nanobiotechnology. Nat. Nanotechnol. 2008, 3, 261–269. [Google Scholar] [CrossRef]
- Rotondi, S.M.C.; Canepa, P.; Angeli, E.; Canepa, M.; Cavalleri, O. DNA Sensing Platforms: Novel Insights into Molecular Grafting Using Low Perturbative AFM Imaging. Sensors 2023, 23, 4557. [Google Scholar] [CrossRef]
- Aragno, I.; Odetti, P.; Altamura, F.; Cavalleri, O.; Rolandi, R. Structure of Rat Tail Tendon Collagen Examined by Atomic Force Microscope. Experientia 1995, 51, 1063–1067. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.-S.; Rao, J.; Gimzewski, J.K. Nanomechanical Analysis of Cells from Cancer Patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Svaldo-Lanero, T.; Krol, S.; Magrassi, R.; Diaspro, A.; Rolandi, R.; Gliozzi, A.; Cavalleri, O. Morphology, Mechanical Properties and Viability of Encapsulated Cells. Ultramicroscopy 2007, 107, 913–921. [Google Scholar] [CrossRef]
- Dorobantu, L.S.; Gray, M.R. Application of Atomic Force Microscopy in Bacterial Research. Scanning 2010, 32, 74–96. [Google Scholar] [CrossRef]
- Formosa, C.; Grare, M.; Duval, R.E.; Dague, E. Nanoscale Effects of Antibiotics on P. Aeruginosa. Nanomedicine 2012, 8, 12–16. [Google Scholar] [CrossRef]
- Aguayo, S.; Donos, N.; Spratt, D.; Bozec, L. Single-Bacterium Nanomechanics in Biomedicine: Unravelling the Dynamics of Bacterial Cells. Nanotechnology 2015, 26, 062001. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, O.H.; Snel, M.M.E.; Cambi, A.; Greve, J.; De Grooth, B.G.; Figdor, C.G. Biomolecular Interactions Measured by Atomic Force Microscopy. Biophys. J. 2000, 79, 3267–3281. [Google Scholar] [CrossRef] [PubMed]
- Canepa, P.; Firpo, G.; Mattera, L.; Canepa, M.; Cavalleri, O. Calcium and Phosphorous Enrichment of Porous Niobium and Titanium Oxides for Biomaterial Applications. Surf. Coat. Technol. 2020, 389, 125634. [Google Scholar] [CrossRef]
- McPherson, A.; Kuznetsov, Y.G.; Malkin, A.J.; Plomp, M. Macromolecular Crystal Growth Investigations Using Atomic Force Microscopy. J. Synchrotron Radiat. 2004, 11, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video Imaging of Walking Myosin V by High-Speed Atomic Force Microscopy. Nature 2010, 468, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, I.Y.; Firtel, M.; Henderson, G.S. In Situ High-resolution Atomic Force Microscope Imaging of Biological Surfaces. J. Vac. Sci. Technol. A 1996, 14, 674–678. [Google Scholar] [CrossRef]
- Pedraz, P.; Casado, S.; Rodriguez, V.; Giordano, M.C.; de Mongeot, F.B.; Ayuso-Sacido, A.; Gnecco, E. Adhesion Modification of Neural Stem Cells Induced by Nanoscale Ripple Patterns. Nanotechnology 2016, 27, 125301. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, J.; Xue, W.; Wu, W.; Wang, Y.; Mei, K.; Chen, Y.; Rao, D.; Yan, T.; Wang, J.; et al. Nanomechanical Vibration Profiling of Oocytes. Nano Res. 2023, 16, 2672–2681. [Google Scholar] [CrossRef]
- Haase, K.; Pelling, A.E. Investigating Cell Mechanics with Atomic Force Microscopy. J. R. Soc. Interface 2015, 12, 20140970. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y. Application of AFM in Microbiology: A Review. Scanning 2010, 32, 61–73. [Google Scholar] [CrossRef]
- Pelling, A.E.; Sehati, S.; Gralla, E.B.; Valentine, J.S.; Gimzewski, J.K. Local Nanomechanical Motion of the Cell Wall of Saccharomyces cerevisiae. Science 2004, 305, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wu, H.; Cai, P.; Fein, J.B.; Chen, W. Atomic Force Microscopy Measurements of Bacterial Adhesion and Biofilm Formation onto Clay-Sized Particles. Sci. Rep. 2015, 5, 16857. [Google Scholar] [CrossRef] [PubMed]
- Doktycz, M.J.; Sullivan, C.J.; Hoyt, P.R.; Pelletier, D.A.; Wu, S.; Allison, D.P. AFM Imaging of Bacteria in Liquid Media Immobilized on Gelatin Coated Mica Surfaces. Ultramicroscopy 2003, 97, 209–216. [Google Scholar] [CrossRef]
- Parisse, P.; Rago, I.; Ulloa Severino, L.; Perissinotto, F.; Ambrosetti, E.; Paoletti, P.; Ricci, M.; Beltrami, A.P.; Cesselli, D.; Casalis, L. Atomic Force Microscopy Analysis of Extracellular Vesicles. Eur. Biophys. J. 2017, 46, 813–820. [Google Scholar] [CrossRef]
- Vorselen, D.; Piontek, M.C.; Roos, W.H.; Wuite, G.J.L. Mechanical Characterization of Liposomes and Extracellular Vesicles, a Protocol. Front. Mol. Biosci. 2020, 7, 139. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.; Dante, S.; Rotondi, S.M.C.; Canepa, P.; Cavalleri, O.; Canepa, M. Spectroscopic Ellipsometry Investigation of a Sensing Functional Interface: DNA SAMs Hybridization. Adv. Mater. Interfaces 2022, 9, 2200364. [Google Scholar] [CrossRef]
- Canepa, P.; Gonella, G.; Pinto, G.; Grachev, V.; Canepa, M.; Cavalleri, O. Anchoring of Aminophosphonates on Titanium Oxide for Biomolecular Coupling. J. Phys. Chem. C 2019, 123, 16843–16850. [Google Scholar] [CrossRef]
- Liu, J.; Xu, R.; Zhu, Y.; Yang, D.-Q.; Nie, H.-Y.; Lau, W.M. AFM/XPS Analysis of the Growth and Architecture of Oriented Molecular Monolayer by Spin Cast Process and Its Cross-Linking Induced by Hyperthermal Hydrogen. Appl. Sci. 2022, 12, 6233. [Google Scholar] [CrossRef]
- Pinto, G.; Parisse, P.; Solano, I.; Canepa, P.; Canepa, M.; Casalis, L.; Cavalleri, O. Functionalizing Gold with Single Strand DNA: Novel Insight into Optical Properties via Combined Spectroscopic Ellipsometry and Nanolithography Measurements. Soft Matter 2019, 15, 2463–2468. [Google Scholar] [CrossRef]
- Canepa, P.; Gregurec, D.; Liessi, N.; Rotondi, S.M.C.; Moya, S.E.; Millo, E.; Canepa, M.; Cavalleri, O. Biofunctionalization of Porous Titanium Oxide through Amino Acid Coupling for Biomaterial Design. Materials 2023, 16, 784. [Google Scholar] [CrossRef]
- Solano, I.; Parisse, P.; Gramazio, F.; Cavalleri, O.; Bracco, G.; Castronovo, M.; Casalis, L.; Canepa, M. Spectroscopic Ellipsometry Meets AFM Nanolithography: About Hydration of Bio-Inert Oligo(Ethylene Glycol)-Terminated Self Assembled Monolayers on Gold. Phys. Chem. Chem. Phys. 2015, 17, 28774–28781. [Google Scholar] [CrossRef] [PubMed]
- Barrena, E.; Ocal, C.; Salmeron, M. Structure and Stability of Tilted-Chain Phases of Alkanethiols on Au(111). J. Chem. Phys. 2001, 114, 4210–4214. [Google Scholar] [CrossRef]
- Toccafondi, C.; Prato, M.; Maidecchi, G.; Penco, A.; Bisio, F.; Cavalleri, O.; Canepa, M. Optical Properties of Yeast Cytochrome c Monolayer on Gold: An in Situ Spectroscopic Ellipsometry Investigation. J. Colloid Interface Sci. 2011, 364, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Kobayashi, K.; Yamada, H. Investigation of Local Hydration Structures of Alkanethiol Self-Assembled Monolayers with Different Molecular Structures by FM-AFM. Langmuir 2018, 34, 15189–15194. [Google Scholar] [CrossRef]
- Pillai, S.; Pai, R.K. Controlled Growth and Formation of SAMs Investigated by Atomic Force Microscopy. Ultramicroscopy 2009, 109, 161–166. [Google Scholar] [CrossRef]
- Bueno, O.V.M.; Benítez, J.J.; San-Miguel, M.A. Understanding Segregation Processes in SAMs Formed by Mixtures of Hydroxylated and Non-Hydroxylated Fatty Acids. RSC Adv. 2019, 9, 39252–39263. [Google Scholar] [CrossRef] [PubMed]
- Canepa, P.; Solano, I.; Uttiya, S.; Gemme, G.; Rolandi, R.; Canepa, M.; Cavalleri, O. Phosphonate Molecular Layers on TiO 2 Surfaces. MATEC Web Conf. 2017, 98, 03001. [Google Scholar] [CrossRef]
- Buzio, R.; Gerbi, A.; Barra, M.; Chiarella, F.; Gnecco, E.; Cassinese, A. Subnanometer Resolution and Enhanced Friction Contrast at the Surface of Perylene Diimide PDI8-CN2 Thin Films in Ambient Conditions. Langmuir 2018, 34, 3207–3214. [Google Scholar] [CrossRef]
- Chi, L.F.; Anders, M.; Fuchs, H.; Johnston, R.R.; Ringsdorf, H. Domain Structures in Langmuir-Blodgett Films Investigated by Atomic Force Microscopy. Science 1993, 259, 213–216. [Google Scholar] [CrossRef]
- Gupta, R.K.; Suresh, K.A. AFM Studies on Langmuir-Blodgett Films of Cholesterol. Eur. Phys. J. E 2004, 14, 35–42. [Google Scholar] [CrossRef]
- Hagedorn, S.; Drolle, E.; Lorentz, H.; Srinivasan, S.; Leonenko, Z.; Jones, L. Atomic Force Microscopy and Langmuir–Blodgett Monolayer Technique to Assess Contact Lens Deposits and Human Meibum Extracts. J. Optom. 2015, 8, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manyes, S.; Domènech, Ò.; Sanz, F.; Montero, M.T.; Hernandez-Borrell, J. Atomic Force Microscopy and Force Spectroscopy Study of Langmuir–Blodgett Films Formed by Heteroacid Phospholipids of Biological Interest. Biochim. Biophys. Acta (BBA)—Biomembr. 2007, 1768, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Rispoli, P.; Carzino, R.; Svaldo-Lanero, T.; Relini, A.; Cavalleri, O.; Fasano, A.; Liuzzi, G.M.; Carlone, G.; Riccio, P.; Gliozzi, A.; et al. A Thermodynamic and Structural Study of Myelin Basic Protein in Lipid Membrane Models. Biophys. J. 2007, 93, 1999–2010. [Google Scholar] [CrossRef]
- Lv, Z.; Banerjee, S.; Zagorski, K.; Lyubchenko, Y.L. Supported Lipid Bilayers for Atomic Force Microscopy Studies. Methods Mol. Biol. 2018, 1814, 129–143. [Google Scholar] [CrossRef]
- Morandat, S.; Azouzi, S.; Beauvais, E.; Mastouri, A.; El Kirat, K. Atomic Force Microscopy of Model Lipid Membranes. Anal. Bioanal. Chem. 2013, 405, 1445–1461. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Boland, T.; Schneider, J.W.; Barger, W.R.; Lee, G.U. Characterization of the Physical Properties of Model Biomembranes at the Nanometer Scale with the Atomic Force Microscope. Faraday Discuss. 1999, 111, 79–94. [Google Scholar] [CrossRef]
- Das, C.; Sheikh, K.H.; Olmsted, P.D.; Connell, S.D. Nanoscale Mechanical Probing of Supported Lipid Bilayers with Atomic Force Microscopy. Phys. Rev. E 2010, 82, 041920. [Google Scholar] [CrossRef]
- Mingeot-Leclercq, M.-P.; Deleu, M.; Brasseur, R.; Dufrêne, Y.F. Atomic Force Microscopy of Supported Lipid Bilayers. Nat. Protoc. 2008, 3, 1654–1659. [Google Scholar] [CrossRef]
- Emanuele, M.; Esposito, A.; Camerini, S.; Antonucci, F.; Ferrara, S.; Seghezza, S.; Catelani, T.; Crescenzi, M.; Marotta, R.; Canale, C.; et al. Exogenous Alpha-Synuclein Alters Pre- and Post-Synaptic Activity by Fragmenting Lipid Rafts. EBioMedicine 2016, 7, 191–204. [Google Scholar] [CrossRef]
- Canepa, P.; Canale, C.; Cavalleri, O.; Marletta, G.; Messina, G.M.L.; Messori, M.; Novelli, R.; Mattioli, S.L.; Apparente, L.; Detta, N.; et al. Adsorption of the RhNGF Protein on Polypropylene with Different Grades of Copolymerization. Materials 2023, 16, 2076. [Google Scholar] [CrossRef]
- Morris, V.J.; Mackie, A.R.; Wilde, P.J.; Kirby, A.R.; Mills, E.C.N.; Patrick Gunning, A. Atomic Force Microscopy as a Tool for Interpreting the Rheology of Food Biopolymers at the Molecular Level. LWT—Food Sci. Technol. 2001, 34, 3–10. [Google Scholar] [CrossRef]
- Vannozzi, L.; Gouveia, P.; Pingue, P.; Canale, C.; Ricotti, L. Novel Ultrathin Films Based on a Blend of PEG- b -PCL and PLLA and Doped with ZnO Nanoparticles. ACS Appl. Mater. Interfaces 2020, 12, 21398–21410. [Google Scholar] [CrossRef] [PubMed]
- Guida, P.; Piscitelli, E.; Marrese, M.; Martino, V.; Cirillo, V.; Guarino, V.; Angeli, E.; Cocola, C.; Pelucchi, P.; Repetto, L.; et al. Integrating Microstructured Electrospun Scaffolds in an Open Microfluidic System for in Vitro Studies of Human Patient-Derived Primary Cells. ACS Biomater. Sci. Eng. 2020, 6, 3649–3663. [Google Scholar] [CrossRef] [PubMed]
- Canepa, P.; Ghiara, G.; Spotorno, R.; Canepa, M.; Cavalleri, O. Structural vs. Electrochemical Investigation of Niobium Oxide Layers Anodically Grown in a Ca and P Containing Electrolyte. J. Alloys Compd. 2021, 851, 156937. [Google Scholar] [CrossRef]
- Irie, Y.; Maruo, Y.; Kataoka, N.; Tanaka, H.; Kinoshita, K.; Kishida, S. AFM and XPS Study from Surfaces of Native Oxide/Al-Metal. Procedia Eng. 2017, 216, 182–189. [Google Scholar] [CrossRef]
- Egbu, J.; Ohodnicki, J.; Paul, R.; Baltrus, J.P.; Talaat, A.; Wright, R.F.; McHenry, M.E. Analysis of Surface Roughness and Oxidation of FeNi-Based Metal Amorphous Nanocomposite Alloys. J. Alloys Compd. 2022, 912, 165155. [Google Scholar] [CrossRef]
- Canepa, P.; Firpo, G.; Gatta, E.; Spotorno, R.; Giannoni, P.; Quarto, R.; Canepa, M.; Cavalleri, O. A Two-Step Approach to Tune the Micro and Nanoscale Morphology of Porous Niobium Oxide to Promote Osteointegration. Materials 2022, 15, 473. [Google Scholar] [CrossRef]
- Bhatnagar, M.; Giordano, M.C.; Mennucci, C.; Chowdhury, D.; Mazzanti, A.; Valle, G.D.; Martella, C.; Tummala, P.; Lamperti, A.; Molle, A.; et al. Ultra-Broadband Photon Harvesting in Large-Area Few-Layer MoS2 Nanostripe Gratings. Nanoscale 2020, 12, 24385–24393. [Google Scholar] [CrossRef] [PubMed]
- Polfus, J.M.; Muñiz, M.B.; Ali, A.; Barragan-Yani, D.A.; Vullum, P.E.; Sunding, M.F.; Taniguchi, T.; Watanabe, K.; Belle, B.D. Temperature-Dependent Adhesion in van Der Waals Heterostructures. Adv. Mater. Interfaces 2021, 8, 2100838. [Google Scholar] [CrossRef]
- Magnozzi, M.; Pflug, T.; Ferrera, M.; Pace, S.; Ramó, L.; Olbrich, M.; Canepa, P.; Ağircan, H.; Horn, A.; Forti, S.; et al. Local Optical Properties in CVD-Grown Monolayer WS2 Flakes. J. Phys. Chem. C 2021, 125, 16059–16065. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, J.; Wang, Y.; Liu, R.; Huai, X.; Jiang, J.; Anfuso, C. Atomic Force Microscopy for Two-Dimensional Materials: A Tutorial Review. Opt. Commun. 2018, 406, 3–17. [Google Scholar] [CrossRef]
- Sikora, A.; Woszczyna, M.; Friedemann, M.; Ahlers, F.J.; Kalbac, M. AFM Diagnostics of Graphene-Based Quantum Hall Devices. Micron 2012, 43, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhou, Y.; Padture, N.P.; Huey, B.D. Anomalous 3D Nanoscale Photoconduction in Hybrid Perovskite Semiconductors Revealed by Tomographic Atomic Force Microscopy. Nat. Commun. 2020, 11, 3308. [Google Scholar] [CrossRef] [PubMed]
- Buzio, R.; Calvini, P.; Ferroni, A.; Valbusa, U. Surface Analysis of Paper Documents Damaged by Foxing. Appl. Phys. A 2004, 79, 383–387. [Google Scholar] [CrossRef]
- Huang, Z.; Stolichnov, I.; Bernand-Mantel, A.; Schott, M.; Auffret, S.; Gaudin, G.; Pizzini, S.; Ranno, L.; Setter, N. Non-Volatile Polarization Switch of Magnetic Domain Wall Velocity. Appl. Phys. Lett. 2015, 107, 252902. [Google Scholar] [CrossRef]
- Ekar, J.; Kovač, J. AFM Study of Roughness Development during ToF-SIMS Depth Profiling of Multilayers with a Cs+ Ion Beam in a H2 Atmosphere. Langmuir 2022, 38, 12871–12880. [Google Scholar] [CrossRef]
- Buzio, R.; Toma, A.; Chincarini, A.; de Mongeot, F.B.; Boragno, C.; Valbusa, U. Atomic Force Microscopy and X-Ray Photoelectron Spectroscopy Characterization of Low-Energy Ion Sputtered Mica. Surf. Sci. 2007, 601, 2735–2739. [Google Scholar] [CrossRef]
- Hu, C.; Li, Z. A Review on the Mechanical Properties of Cement-Based Materials Measured by Nanoindentation. Constr. Build. Mater. 2015, 90, 80–90. [Google Scholar] [CrossRef]
- Blangiardo, A.; Lagomarsino, G.; Basso, A.; Canepa, P.; Cavalleri, O.; Rossi, S.; Monticelli, O. Preparation, Application and Recycling of a Catalytic Microflow Reactor Based on Polylactic Acid. Appl. Surf. Sci. 2021, 569, 151019. [Google Scholar] [CrossRef]
- Bisio, F.; Palombo, M.; Prato, M.; Cavalleri, O.; Barborini, E.; Vinati, S.; Franchi, M.; Mattera, L.; Canepa, M. Optical Properties of Cluster-Assembled Nanoporous Gold Films. Phys. Rev. B 2009, 80, 205428. [Google Scholar] [CrossRef]
- Bellotti, R.; Picotto, G.B.; Ribotta, L. AFM Measurements and Tip Characterization of Nanoparticles with Different Shapes. Nanomanuf. Metrol. 2022, 5, 127–138. [Google Scholar] [CrossRef]
- Friedrich, S.; Cappella, B. Friction and Mechanical Properties of AFM-Scan-Induced Ripples in Polymer Films. Front. Mech. Eng. 2021, 7, 672898. [Google Scholar] [CrossRef]
- Lin, Y.-T.; He, H.; Kaya, H.; Liu, H.; Ngo, D.; Smith, N.J.; Banerjee, J.; Borhan, A.; Kim, S.H. Photothermal Atomic Force Microscopy Coupled with Infrared Spectroscopy (AFM-IR) Analysis of High Extinction Coefficient Materials: A Case Study with Silica and Silicate Glasses. Anal. Chem. 2022, 94, 5231–5239. [Google Scholar] [CrossRef] [PubMed]
- Chada, N.; Sigdel, K.P.; Gari, R.R.S.; Matin, T.R.; Randall, L.L.; King, G.M. Glass Is a Viable Substrate for Precision Force Microscopy of Membrane Proteins. Sci. Rep. 2015, 5, 12550. [Google Scholar] [CrossRef]
- Kempe, A.; Schopf, J.W.; Altermann, W.; Kudryavtsev, A.B.; Heckl, W.M. Atomic Force Microscopy of Precambrian Microscopic Fossils. Proc. Natl. Acad. Sci. USA 2002, 99, 9117–9120. [Google Scholar] [CrossRef]
- Benítez, J.J.; Guzman-Puyol, S.; Domínguez, E.; Heredia, A.; Heredia-Guerrero, J.A. Applications and Potentialities of Atomic Force Microscopy in Fossil and Extant Plant Cuticle Characterization. Rev. Palaeobot. Palynol. 2019, 268, 125–132. [Google Scholar] [CrossRef]
- Sapienza, L.; Liu, J.; Song, J.D.; Fält, S.; Wegscheider, W.; Badolato, A.; Srinivasan, K. Combined Atomic Force Microscopy and Photoluminescence Imaging to Select Single InAs/GaAs Quantum Dots for Quantum Photonic Devices. Sci. Rep. 2017, 7, 6205. [Google Scholar] [CrossRef]
- Hennessy, K.; Badolato, A.; Winger, M.; Gerace, D.; Atatüre, M.; Gulde, S.; Fält, S.; Hu, E.L.; Imamoğlu, A. Quantum Nature of a Strongly Coupled Single Quantum Dot–Cavity System. Nature 2007, 445, 896–899. [Google Scholar] [CrossRef]
- Müller, D.J.; Helenius, J.; Alsteens, D.; Dufrêne, Y.F. Force Probing Surfaces of Living Cells to Molecular Resolution. Nat. Chem. Biol. 2009, 5, 383–390. [Google Scholar] [CrossRef]
- Kerdegari, S.; Canepa, P.; Odino, D.; Oropesa-Nuñez, R.; Relini, A.; Cavalleri, O.; Canale, C. Insights in Cell Biomechanics through Atomic Force Microscopy. Materials 2023, 16, 2980. [Google Scholar] [CrossRef]
- Viljoen, A.; Mathelié-Guinlet, M.; Ray, A.; Strohmeyer, N.; Oh, Y.J.; Hinterdorfer, P.; Müller, D.J.; Alsteens, D.; Dufrêne, Y.F. Force Spectroscopy of Single Cells Using Atomic Force Microscopy. Nat. Rev. Methods Prim. 2021, 1, 63. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Martínez-Martín, D.; Medalsy, I.; Alsteens, D.; Müller, D.J. Multiparametric Imaging of Biological Systems by Force-Distance Curve-Based AFM. Nat. Methods 2013, 10, 847–854. [Google Scholar] [CrossRef]
- Jalili, N.; Laxminarayana, K. A Review of Atomic Force Microscopy Imaging Systems: Application to Molecular Metrology and Biological Sciences. Mechatronics 2004, 14, 907–945. [Google Scholar] [CrossRef]
- Lavanya, S.B.; Jayanth, G.R. Control of Interaction Force in Constant-Height Contact Mode Atomic Force Microscopy. Mechatronics 2022, 88, 102914. [Google Scholar] [CrossRef]
- Biczysko, P.; Dzierka, A.; Jóźwiak, G.; Rudek, M.; Gotszalk, T.; Janus, P.; Grabiec, P.; Rangelow, I.W. Contact Atomic Force Microscopy Using Piezoresistive Cantilevers in Load Force Modulation Mode. Ultramicroscopy 2018, 184, 199–208. [Google Scholar] [CrossRef]
- Zhong, Q.; Inniss, D.; Kjoller, K.; Elings, V.B. Fractured Polymer/Silica Fiber Surface Studied by Tapping Mode Atomic Force Microscopy. Surf. Sci. 1993, 290, L688–L692. [Google Scholar] [CrossRef]
- Garcia, R.; Herruzo, E.T. The Emergence of Multifrequency Force Microscopy. Nat. Nanotechnol. 2012, 7, 217–226. [Google Scholar] [CrossRef]
- Guo, S.; Solares, S.D.; Mochalin, V.; Neitzel, I.; Gogotsi, Y.; Kalinin, S.V.; Jesse, S. Multifrequency Imaging in the Intermittent Contact Mode of Atomic Force Microscopy: Beyond Phase Imaging. Small 2012, 8, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Dokukin, M.E.; Sokolov, I. Nanoscale Compositional Mapping of Cells, Tissues, and Polymers with Ringing Mode of Atomic Force Microscopy. Sci. Rep. 2017, 7, 11828. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Fu, J.; Miao, H.; Li, F. Contact Resonance Force Microscopy with Higher-Eigenmode for Nanoscale Viscoelasticity Measurements. J. Appl. Phys. 2014, 116, 034310. [Google Scholar] [CrossRef]
- Stan, G.; Solares, S.D.; Pittenger, B.; Erina, N.; Su, C. Nanoscale Mechanics by Tomographic Contact Resonance Atomic Force Microscopy. Nanoscale 2013, 6, 962–969. [Google Scholar] [CrossRef]
- Radmacher, M.; Cleveland, J.P.; Fritz, M.; Hansma, H.G.; Hansma, P.K. Mapping Interaction Forces with the Atomic Force Microscope. Biophys. J. 1994, 66, 2159–2165. [Google Scholar] [CrossRef]
- Martín-Rodríguez, A.J.; González-Orive, A.; Hernández-Creus, A.; Morales, A.; Dorta-Guerra, R.; Norte, M.; Martín, V.S.; Fernández, J.J. On the Influence of the Culture Conditions in Bacterial Antifouling Bioassays and Biofilm Properties: Shewanella Algae, a Case Study. BMC Microbiol. 2014, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Dupres, V.; Yunus, S.; Latgé, J.-P.; Heinisch, J.J.; Dufrêne, Y.F. High-Resolution Imaging of Chemical and Biological Sites on Living Cells Using Peak Force Tapping Atomic Force Microscopy. Langmuir 2012, 28, 16738–16744. [Google Scholar] [CrossRef]
- Pinto, G.; Canepa, P.; Canale, C.; Canepa, M.; Cavalleri, O. Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods. Materials 2020, 13, 2888. [Google Scholar] [CrossRef]
- Chopinet, L.; Formosa, C.; Rols, M.P.; Duval, R.E.; Dague, E. Imaging Living Cells Surface and Quantifying Its Properties at High Resolution Using AFM in QITM Mode. Micron 2013, 48, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Hansma, P.K.; Schitter, G.; Fantner, G.E.; Prater, C. High-Speed Atomic Force Microscopy. Science 2006, 314, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Viani, M.B.; Schäffer, T.E.; Paloczi, G.T.; Pietrasanta, L.I.; Smith, B.L.; Thompson, J.B.; Richter, M.; Rief, M.; Gaub, H.E.; Plaxco, K.W.; et al. Fast Imaging and Fast Force Spectroscopy of Single Biopolymers with a New Atomic Force Microscope Designed for Small Cantilevers. Rev. Sci. Instrum. 1999, 70, 4300–4303. [Google Scholar] [CrossRef]
- Humphris, A.D.L.; Miles, M.J.; Hobbs, J.K. A Mechanical Microscope: High-Speed Atomic Force Microscopy. Appl. Phys. Lett. 2005, 86, 034106. [Google Scholar] [CrossRef]
- Sulchek, T.; Yaralioglu, G.G.; Quate, C.F.; Minne, S.C. Characterization and Optimization of Scan Speed for Tapping-Mode Atomic Force Microscopy. Rev. Sci. Instrum. 2002, 73, 2928–2936. [Google Scholar] [CrossRef]
- Manalis, S.R.; Minne, S.C.; Quate, C.F. Atomic Force Microscopy for High Speed Imaging Using Cantilevers with an Integrated Actuator and Sensor. Appl. Phys. Lett. 1996, 68, 871–873. [Google Scholar] [CrossRef]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A High-Speed Atomic Force Microscope for Studying Biological Macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Scheuring, S. Filming Biomolecular Processes by High-Speed Atomic Force Microscopy. Chem. Rev. 2014, 114, 3120–3188. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Uchihashi, T.; Fukuma, T. High-Speed Atomic Force Microscopy for Nano-Visualization of Dynamic Biomolecular Processes. Prog. Surf. Sci. 2008, 83, 337–437. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; Land, T.A.; DeYoreo, J.J.; McPherson, A. Mechanisms of Growth for Protein and Virus Crystals. Nat. Struct. Mol. Biol. 1995, 2, 956–959. [Google Scholar] [CrossRef]
- Malkin, A.J.; Land, T.A.; Kuznetsov, Y.G.; McPherson, A.; DeYoreo, J.J. Investigation of Virus Crystal Growth Mechanisms by In Situ Atomic Force Microscopy. Phys. Rev. Lett. 1995, 75, 2778–2781. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; McPherson, A. Incorporation of Microcrystals by Growing Protein and Virus Crystals. Proteins 1996, 24, 247–252. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; McPherson, A. An in Situ AFM Investigation of Catalase Crystallization. Surf. Sci. 1997, 393, 95–107. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; Glantz, W.; McPherson, A. Atomic Force Microscopy Studies of Surface Morphology and Growth Kinetics in Thaumatin Crystallization. J. Phys. Chem. 1996, 100, 11736–11743. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; McPherson, A. Defect Structure of Macromolecular Crystals. J. Struct. Biol. 1996, 117, 124–137. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Malkin, A.J.; Glantz, W.; McPherson, A. In Situ Atomic Force Microscopy Studies of Protein and Virus Crystal Growth Mechanisms. J. Cryst. Growth 1996, 168, 63–73. [Google Scholar] [CrossRef]
- Konnert, J.H.; D’Antonio, P.; Ward, K.B. Observation of Growth Steps, Spiral Dislocations and Molecular Packing on the Surface of Lysozyme Crystals with the Atomic Force Microscope. Acta Crystallogr. D 1994, 50, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.M.; Ward, M.D. Atomic Force Microscopy of Insulin Single Crystals: Direct Visualization of Molecules and Crystal Growth. Biophys. J. 1996, 71, 1071–1078. [Google Scholar] [CrossRef]
- Land, T.A.; De Yoreo, J.J.; Lee, J.D. An In-Situ AFM Investigation of Canavalin Crystallization Kinetics. Surf. Sci. 1997, 384, 136–155. [Google Scholar] [CrossRef]
- YuG, K.; Malkin, A.J.; Land, T.A.; DeYoreo, J.J.; Barba, A.P.; Konnert, J.; McPherson, A. Molecular Resolution Imaging of Macromolecular Crystals by Atomic Force Microscopy. Biophys. J. 1997, 72, 2357–2364. [Google Scholar] [CrossRef]
- Asherie, N.; Lomakin, A.; Benedek, G.B. Phase Diagram of Colloidal Solutions. Phys. Rev. Lett. 1996, 77, 4832–4835. [Google Scholar] [CrossRef]
- Liu, C.; Lomakin, A.; Thurston, G.M.; Hayden, D.; Pande, A.; Pande, J.; Ogun, O.; Asherie, N.; Benedek, G.B. Phase Separation in Multicomponent Aqueous-Protein Solutions. J. Phys. Chem. 1995, 99, 454–461. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Konnert, J.; Malkin, A.J.; McPherson, A. The Advancement and Structure of Growth Steps on Thaumatin Crystals Visualized by Atomic Force Microscopy at Molecular Resolution. Surf. Sci. 1999, 440, 69–80. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; McPherson, A. In Situ Atomic Force Microscopy Studies of Surface Morphology, Growth Kinetics, Defect Structure and Dissolution in Macromolecular Crystallization. J. Cryst. Growth 1999, 196, 471–488. [Google Scholar] [CrossRef]
- Mollica, V.; Borassi, A.; Relini, A.; Cavalleri, O.; Bolognesi, M.; Rolandi, R.; Gliozzi, A. An Atomic Force Microscopy Investigation of Protein Crystal Surface Topography. Eur. Biophys. J. 2001, 30, 313–318. [Google Scholar] [CrossRef]
- Li, H.; Nadarajah, A.; Pusey, M.L. Determining the Molecular-Growth Mechanisms of Protein Crystal Faces by Atomic Force Microscopy. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 1036–1045. [Google Scholar] [CrossRef]
- Durbin, S.D.; Feher, G. Studies of Crystal Growth Mechanisms of Proteins by Electron Microscopy. J. Mol. Biol. 1990, 212, 763–774. [Google Scholar] [CrossRef]
- Malkin, A.J.; McPherson, A. Light-Scattering Investigations of Nucleation Processes and Kinetics of Crystallization in Macromolecular Systems. Acta Crystallogr. Sect. D 1994, 50, 385–395. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Malkin, A.J.; Greenwood, A.; McPherson, A. Interferometric Studies of Growth Kinetics and Surface Morphology in Macromolecular Crystal Growth: Canavalin, Thaumatin, and Turnip Yellow Mosaic Virus. J. Struct. Biol. 1995, 114, 184–196. [Google Scholar] [CrossRef]
- Larson, S.B.; Koszelak, S.; Day, J.; Greenwood, A.; Dodds, J.A.; McPherson, A. Double-Helical RNA in Satellite Tobacco Mosaic Virus. Nature 1993, 361, 179–182. [Google Scholar] [CrossRef]
- Koszelak, S.; Day, J.; Leja, C.; Cudney, R.; McPherson, A. Protein and Virus Crystal Growth on International Microgravity Laboratory-2. Biophys. J. 1995, 69, 13–19. [Google Scholar] [CrossRef]
- Guo, H.M.; Liu, H.W.; Wang, Y.L.; Gao, H.J.; Gong, Y.; Jiang, H.Y.; Wang, W.Q. Surface Structures of Dl-Valine and l-Alanine Crystals Observed by Atomic Force Microscopy at a Molecular Resolution. Surf. Sci. 2004, 552, 70–76. [Google Scholar] [CrossRef]
- Wang, W.Q.; Gong, Y.; Liang, Z.; Sun, F.L.; Shi, D.X.; Gao, H.J.; Lin, X.; Jiang, P.; Wang, Z.M. Direct Observation of Surface Structure of D-Alanine and d-/l-Valine Crystals by Atomic Force Microscopy and Comparison with X-ray Diffraction Analysis. Surf. Sci. 2002, 512, L379–L384. [Google Scholar] [CrossRef]
- Ohnishi, S.; Hara, M.; Furuno, T.; Sasabe, H. Imaging the Ordered Arrays of Water-Soluble Protein Ferritin with the Atomic Force Microscope. Biophys. J. 1992, 63, 1425–1431. [Google Scholar] [CrossRef]
- Furuno, T.; Sasabe, H.; Ikegami, A. Imaging Two-Dimensional Arrays of Soluble Proteins by Atomic Force Microscopy in Contact Mode Using a Sharp Supertip. Ultramicroscopy 1998, 70, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, S.; Hara, M.; Furuno, T.; Okada, T.; Sasabe, H. Direct Visualization of Polypeptide Shell of Ferritin Molecule by Atomic Force Microscopy. Biophys. J. 1993, 65, 573–577. [Google Scholar] [CrossRef]
- Pum, D.; Toca-Herrera, J.L.; Sleytr, U.B. S-Layer Protein Self-Assembly. Int. J. Mol. Sci. 2013, 14, 2484–2501. [Google Scholar] [CrossRef] [PubMed]
- Houwink, A.L. A Macromolecular Mono-Layer in the Cell Wall of Spirillum Spec. Biochim. Biophys. Acta 1953, 10, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Messner, P.; Abdul Mazid, M.; Unger, F.M.; Sleytr, U.B. Artificial Antigens. Synthetic Carbohydrate Haptens Immobilized on Crystalline Bacterial Surface Layer Glycoproteins. Carbohydr. Res. 1992, 233, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Messner, P.; Pum, D.; Sára, M. Crystalline Bacterial Cell Surface Layers (S Layers): From Supramolecular Cell Structure to Biomimetics and Nanotechnology. Angew. Chem. Int. Ed. 1999, 38, 1034–1054. [Google Scholar] [CrossRef]
- Claus, H.; Akça, E.; Debaerdemaeker, T.; Evrard, C.; Declercq, J.-P.; Harris, J.R.; Schlott, B.; König, H. Molecular Organization of Selected Prokaryotic S-Layer Proteins. Can. J. Microbiol. 2005, 51, 731–743. [Google Scholar] [CrossRef]
- Albers, S.-V.; Meyer, B.H. The Archaeal Cell Envelope. Nat. Rev. Microbiol. 2011, 9, 414–426. [Google Scholar] [CrossRef]
- Hynönen, U.; Palva, A. Lactobacillus Surface Layer Proteins: Structure, Function and Applications. Appl. Microbiol. Biotechnol. 2013, 97, 5225–5243. [Google Scholar] [CrossRef]
- Sleytr, U.B. Self-Assembly of the Hexagonally and Tetragonally Arranged Subunits of Bacterial Surface Layers and Their Reattachment to Cell Walls. J. Ultrastruct. Res. 1976, 55, 360–377. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.-M.; Pum, D. S-Layers: Principles and Applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Beveridge, T.J. Bacterial S-Layers. Trends Microbiol. 1999, 7, 253–260. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The Unseen Majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- Egelseer, E.-M.; Sára, M.; Pum, D.; Schuster, B.; Sleytr, U.B. Genetically Engineered S-Layer Proteins and S-Layer-Specific Heteropolysaccharides as Components of a Versatile Molecular Construction Kit for Applications in Nanobiotechnology. In NanoBioTechnology: BioInspired Devices and Materials of the Future; Shoseyov, O., Levy, I., Eds.; Humana Press: Totowa, NJ, USA, 2008; pp. 55–86. ISBN 978-1-59745-218-2. [Google Scholar]
- Schuster, B.; Sleytr, U.B. Composite S-Layer Lipid Structures. J. Struct. Biol. 2009, 168, 207–216. [Google Scholar] [CrossRef]
- Breitwieser, A.; Siedlaczek, P.; Lichtenegger, H.; Sleytr, U.B.; Pum, D. S-Layer Protein Coated Carbon Nanotubes. Coatings 2019, 9, 492. [Google Scholar] [CrossRef]
- Egelseer, E.M.; Ilk, N.; Pum, D.; Messner, P.; Schäffer, C.; Schuster, B.; Sleytr, U.B. S-Layers, Microbial, Biotechnological Applications. In Encyclopedia of Industrial Biotechnology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009; pp. 1–25. ISBN 978-0-470-05458-1. [Google Scholar]
- Ilk, N.; Egelseer, E.M.; Sleytr, U.B. S-Layer Fusion Proteins—Construction Principles and Applications. Curr. Opin. Biotechnol. 2011, 22, 824–831. [Google Scholar] [CrossRef]
- Schuster, B.; Pum, D.; Sára, M.; Sleytr, U.B. S-Layer Proteins as Key Components of a Versatile Molecular Construction Kit for Biomedical Nanotechnology. Mini Rev. Med. Chem. 2006, 6, 909–920. [Google Scholar] [CrossRef]
- Schuster, B.; Sleytr, U.B. Nanotechnology with S-Layer Proteins. In Protein Nanotechnology: Protocols, Instrumentation, and Applications; Gerrard, J.A., Domigan, L.J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 195–218. ISBN 978-1-4939-9869-2. [Google Scholar]
- Damiati, S.; Peacock, M.; Mhanna, R.; Søpstad, S.; Sleytr, U.B.; Schuster, B. Bioinspired Detection Sensor Based on Functional Nanostructures of S-Proteins to Target the Folate Receptors in Breast Cancer Cells. Sens. Actuators B Chem. 2018, 267, 224–230. [Google Scholar] [CrossRef]
- Schuster, B.; Sleytr, U.B. S-Layer Ultrafiltration Membranes. Membranes 2021, 11, 275. [Google Scholar] [CrossRef]
- Schuster, B. S-Layer Protein-Based Biosensors. Biosensors 2018, 8, 40. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Glauert, A.M. Analysis of Regular Arrays of Subunits on Bacterial Surfaces; Evidence for a Dynamic Process of Assembly. J. Ultrastruct. Res. 1975, 50, 103–116. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Messner, P. Crystalline Surface Layers on Bacteria. Annu. Rev. Microbiol. 1983, 37, 311–339. [Google Scholar] [CrossRef]
- Pavkov-Keller, T.; Howorka, S.; Keller, W. Chapter 3—The Structure of Bacterial S-Layer Proteins. In Progress in Molecular Biology and Translational Science; Howorka, S., Ed.; Molecular Assembly in Natural and Engineered Systems; Academic Press: Cambridge, MA, USA, 2011; Volume 103, pp. 73–130. [Google Scholar]
- Sára, M.; Sleytr, U.B. Crystalline Bacterial Cell Surface Layers (S-Layers): From Cell Structure to Biomimetics. Prog. Biophys. Mol. Biol. 1996, 65, 83–111. [Google Scholar] [CrossRef]
- Thornley, M.J.; Glauert, A.M.; Sleytr, U.B.; Markham, R.; Horne, R.W.; Hicks, R.M. Structure and Assembly of Bacterial Surface Layers Composed of Regular Arrays of Subunits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997, 268, 147–153. [Google Scholar] [CrossRef]
- Müller, D.J.; Baumeister, W.; Engel, A. Conformational Change of the Hexagonally Packed Intermediate Layer of Deinococcus Radiodurans Monitored by Atomic Force Microscopy. J. Bacteriol. 1996, 178, 3025–3030. [Google Scholar] [CrossRef]
- Müller, D.J.; Baumeister, W.; Engel, A. Controlled Unzipping of a Bacterial Surface Layer with Atomic Force Microscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 13170–13174. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Flores, S.; Kasry, A.; Butt, H.-J.; Vavilala, C.; Schmittel, M.; Pum, D.; Sleytr, U.B.; Toca-Herrera, J.L. From Native to Non-Native Two-Dimensional Protein Lattices through Underlying Hydrophilic/Hydrophobic Nanoprotrusions. Angew. Chem. Int. Ed. 2008, 47, 4707–4710. [Google Scholar] [CrossRef]
- Ebner, A.; Kienberger, F.; Huber, C.; Kamruzzahan, A.S.M.; Pastushenko, V.P.; Tang, J.; Kada, G.; Gruber, H.J.; Sleytr, U.B.; Sára, M.; et al. Atomic-Force-Microscopy Imaging and Molecular-Recognition-Force Microscopy of Recrystallized Heterotetramers Comprising an S-Layer-Streptavidin Fusion Protein. ChemBioChem 2006, 7, 588–591. [Google Scholar] [CrossRef]
- López, A.E.; Pum, D.; Sleytr, U.B.; Toca-Herrera, J.L. Influence of Surface Chemistry and Protein Concentration on the Adsorption Rate and S-Layer Crystal Formation. Phys. Chem. Chem. Phys. 2011, 13, 11905–11913. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Cencerrado, A.; Iturri, J.; Toca-Herrera, J.L. In-Situ 2D Bacterial Crystal Growth as a Function of Protein Concentration: An Atomic Force Microscopy Study. Microsc. Res. Tech. 2018, 81, 1095–1104. [Google Scholar] [CrossRef]
- Yau, S.-T.; Vekilov, P.G. Direct Observation of Nucleus Structure and Nucleation Pathways in Apoferritin Crystallization. J. Am. Chem. Soc. 2001, 123, 1080–1089. [Google Scholar] [CrossRef]
- Tang, J.; Krajcikova, D.; Zhu, R.; Ebner, A.; Cutting, S.; Gruber, H.J.; Barak, I.; Hinterdorfer, P. Atomic Force Microscopy Imaging and Single Molecule Recognition Force Spectroscopy of Coat Proteins on the Surface of Bacillus Subtilis Spore. J. Mol. Recognit. 2007, 20, 483–489. [Google Scholar] [CrossRef]
- Chung, S.; Shin, S.-H.; Bertozzi, C.R.; De Yoreo, J.J. Self-Catalyzed Growth of S Layers via an Amorphous-to-Crystalline Transition Limited by Folding Kinetics. Proc. Natl. Acad. Sci. USA 2010, 107, 16536–16541. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.P.; Him, J.L.K.; Tessier, B.; Tessier, C.; Brisson, A.R. On the Kinetics of Adsorption and Two-Dimensional Self-Assembly of Annexin A5 on Supported Lipid Bilayers. Biophys. J. 2005, 89, 3372–3385. [Google Scholar] [CrossRef]
- Scheuring, S.; Stahlberg, H.; Chami, M.; Houssin, C.; Rigaud, J.-L.; Engel, A. Charting and Unzipping the Surface Layer of Corynebacterium Glutamicum with the Atomic Force Microscope. Mol. Microbiol. 2002, 44, 675–684. [Google Scholar] [CrossRef]
- Peyret, J.L.; Bayan, N.; Joliff, G.; Gulik-Krzywicki, T.; Mathieu, L.; Shechter, E.; Leblon, G. Characterization of the CspB Gene Encoding PS2, an Ordered Surface-Layer Protein in Corynebacterium Glutamicum. Mol. Microbiol. 1993, 9, 97–109. [Google Scholar] [CrossRef]
- Bahl, H.; Scholz, H.; Bayan, N.; Chami, M.; Leblon, G.; Gulik-Krzywicki, T.; Shechter, E.; Fouet, A.; Mesnage, S.; Tosi-Couture, E.; et al. IV. Molecular Biology of S-Layers. FEMS Microbiol. Rev. 1997, 20, 47–98. [Google Scholar] [CrossRef] [PubMed]
- Chami, M.; Bayan, N.; Peyret, J.L.; Gulik-Krzywicki, T.; Leblon, G.; Shechter, E. The S-Layer Protein of Corynebacterium Glutamicum Is Anchored to the Cell Wall by Its C-Terminal Hydrophobic Domain. Mol. Microbiol. 1997, 23, 483–492. [Google Scholar] [CrossRef]
- Müller, D.J.; Fotiadis, D.; Scheuring, S.; Müller, S.A.; Engel, A. Electrostatically Balanced Subnanometer Imaging of Biological Specimens by Atomic Force Microscope. Biophys. J. 1999, 76, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Hoh, J.H.; Lal, R.; John, S.A.; Revel, J.-P.; Arnsdorf, M.F. Atomic Force Microscopy and Dissection of Gap Junctions. Science 1991, 253, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Schabert, F.A.; Henn, C.; Engel, A. Native Escherichia Coli OmpF Porin Surfaces Probed by Atomic Force Microscopy. Science 1995, 268, 92–94. [Google Scholar] [CrossRef]
- Fotiadis, D.; Hasler, L.; Müller, D.J.; Stahlberg, H.; Kistler, J.; Engel, A. Surface Tongue-and-Groove Contours on Lens MIP Facilitate Cell-to-Cell Adherence. J. Mol. Biol. 2000, 300, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-H.; Chung, S.; Sanii, B.; Comolli, L.R.; Bertozzi, C.R.; De Yoreo, J.J. Direct Observation of Kinetic Traps Associated with Structural Transformations Leading to Multiple Pathways of S-Layer Assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 12968–12973. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Sára, M.; Pum, D.; Schuster, B. Characterization and Use of Crystalline Bacterial Cell Surface Layers. Prog. Surf. Sci. 2001, 68, 231–278. [Google Scholar] [CrossRef]
- Lopez, A.E.; Moreno-Flores, S.; Pum, D.; Sleytr, U.B.; Toca-Herrera, J.L. Surface Dependence of Protein Nanocrystal Formation. Small 2010, 6, 396–403. [Google Scholar] [CrossRef]
- Györvary, E.S.; Stein, O.; Pum, D.; Sleytr, U.B. Self-Assembly and Recrystallization of Bacterial S-Layer Proteins at Silicon Supports Imaged in Real Time by Atomic Force Microscopy. J. Microsc. 2003, 212, 300–306. [Google Scholar] [CrossRef]
- Radmacher, M.; Fritz, M.; Hansma, P.K. Imaging Soft Samples with the Atomic Force Microscope: Gelatin in Water and Propanol. Biophys. J. 1995, 69, 264–270. [Google Scholar] [CrossRef]
- Harrison, P.M.; Arosio, P. The Ferritins: Molecular Properties, Iron Storage Function and Cellular Regulation. Biochim. Biophys. Acta (BBA) Bioenerg. 1996, 1275, 161–203. [Google Scholar] [CrossRef]
- Hempstead, P.D.; Yewdall, S.J.; Fernie, A.R.; Lawson, D.M.; Artymiuk, P.J.; Rice, D.W.; Ford, G.C.; Harrison, P.M. Comparison of the Three-Dimensional Structures of Recombinant Human H and Horse L Ferritins at High Resolution11Edited by R. Huber. J. Mol. Biol. 1997, 268, 424–448. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.M.; Artymiuk, P.J.; Yewdall, S.J.; Smith, J.M.A.; Livingstone, J.C.; Treffry, A.; Luzzago, A.; Levi, S.; Arosio, P.; Cesareni, G.; et al. Solving the Structure of Human H Ferritin by Genetically Engineering Intermolecular Crystal Contacts. Nature 1991, 349, 541–544. [Google Scholar] [CrossRef]
- Thomas, B.R.; Carter, D.; Rosenberger, F. Effect of Microheterogeneity on Horse Spleen Apoferritin Crystallization. J. Cryst. Growth 1998, 187, 499–510. [Google Scholar] [CrossRef]
- Yau, S.-T.; Thomas, B.R.; Vekilov, P.G. Molecular Mechanisms of Crystallization and Defect Formation. Phys. Rev. Lett. 2000, 85, 353–356. [Google Scholar] [CrossRef]
- Hurle, D.T.J. (Ed.) Handbook of Crystal Growth; North-Holland: Amsterdam, The Netherlands; New York, NY, USA, 1993; ISBN 978-0-444-88908-9. [Google Scholar]
- Morgenstern, K.; Lægsgaard, E.; Stensgaard, I.; Besenbacher, F. Transition from One-Dimensional to Two-Dimensional Island Decay on an Anisotropic Surface. Phys. Rev. Lett. 1999, 83, 1613–1616. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Malkin, A.J.; McPherson, A. Atomic-Force-Microscopy Studies of Phase Separations in Macromolecular Systems. Phys. Rev. B 1998, 58, 6097–6103. [Google Scholar] [CrossRef]
- Georgalis, Y.; Umbach, P.; Zielenkiewicz, A.; Utzig, E.; Zielenkiewicz, W.; Zielenkiewicz, P.; Saenger, W. Microcalorimetric and Small-Angle Light Scattering Studies on Nucleating Lysozyme Solutions. J. Am. Chem. Soc. 1997, 119, 11959–11965. [Google Scholar] [CrossRef]
- Ando, T. High-Speed Atomic Force Microscopy Coming of Age. Nanotechnology 2012, 23, 062001. [Google Scholar] [CrossRef]
- Hansma, P.K.; Cleveland, J.P.; Radmacher, M.; Walters, D.A.; Hillner, P.E.; Bezanilla, M.; Fritz, M.; Vie, D.; Hansma, H.G.; Prater, C.B.; et al. Tapping Mode Atomic Force Microscopy in Liquids. Appl. Phys. Lett. 1994, 64, 1738–1740. [Google Scholar] [CrossRef]
- Kodera, N.; Yamashita, H.; Ando, T. Active Damping of the Scanner for High-Speed Atomic Force Microscopy. Rev. Sci. Instrum. 2005, 76, 053708. [Google Scholar] [CrossRef]
- Kokavecz, J.; Tóth, Z.; Horváth, Z.L.; Heszler, P.; Mechler, Á. Novel Amplitude and Frequency Demodulation Algorithm for a Virtual Dynamic Atomic Force Microscope. Nanotechnology 2006, 17, S173. [Google Scholar] [CrossRef]
- Uchihashi, T.; Kodera, N.; Ando, T. Guide to Video Recording of Structure Dynamics and Dynamic Processes of Proteins by High-Speed Atomic Force Microscopy. Nat. Protoc. 2012, 7, 1193–1206. [Google Scholar] [CrossRef]
- Uchihashi, T.; Scheuring, S. Applications of High-Speed Atomic Force Microscopy to Real-Time Visualization of Dynamic Biomolecular Processes. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 229–240. [Google Scholar] [CrossRef]
- Yokokawa, M.; Wada, C.; Ando, T.; Sakai, N.; Yagi, A.; Yoshimura, S.H.; Takeyasu, K. Fast-Scanning Atomic Force Microscopy Reveals the ATP/ADP-Dependent Conformational Changes of GroEL. EMBO J. 2006, 25, 4567–4576. [Google Scholar] [CrossRef]
- Ruan, Y.; Miyagi, A.; Wang, X.; Chami, M.; Boudker, O.; Scheuring, S. Direct Visualization of Glutamate Transporter Elevator Mechanism by High-Speed AFM. Proc. Natl. Acad. Sci. USA 2017, 114, 1584–1588. [Google Scholar] [CrossRef]
- Rangl, M.; Miyagi, A.; Kowal, J.; Stahlberg, H.; Nimigean, C.M.; Scheuring, S. Real-Time Visualization of Conformational Changes within Single MloK1 Cyclic Nucleotide-Modulated Channels. Nat. Commun. 2016, 7, 12789. [Google Scholar] [CrossRef]
- Noi, K.; Yamamoto, D.; Nishikori, S.; Arita-Morioka, K.; Kato, T.; Ando, T.; Ogura, T. High-Speed Atomic Force Microscopic Observation of ATP-Dependent Rotation of the AAA+ Chaperone P97. Structure 2013, 21, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Inoue, K.; Shibata, M.; Uchihashi, T.; Sasaki, J.; Kandori, H.; Ando, T. Role of Trimer–Trimer Interaction of Bacteriorhodopsin Studied by Optical Spectroscopy and High-Speed Atomic Force Microscopy. J. Struct. Biol. 2013, 184, 2–11. [Google Scholar] [CrossRef]
- Shibata, M.; Yamashita, H.; Uchihashi, T.; Kandori, H.; Ando, T. High-Speed Atomic Force Microscopy Shows Dynamic Molecular Processes in Photoactivated Bacteriorhodopsin. Nat. Nanotechnol. 2010, 5, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Yokokawa, M.; Takeyasu, K. Motion of the Ca2+-Pump Captured. FEBS J. 2011, 278, 3025–3031. [Google Scholar] [CrossRef]
- Heath, G.R.; Scheuring, S. Advances in High-Speed Atomic Force Microscopy (HS-AFM) Reveal Dynamics of Transmembrane Channels and Transporters. Curr. Opin. Struct. Biol. 2019, 57, 93–102. [Google Scholar] [CrossRef]
- Sakiyama, Y.; Mazur, A.; Kapinos, L.E.; Lim, R.Y.H. Spatiotemporal Dynamics of the Nuclear Pore Complex Transport Barrier Resolved by High-Speed Atomic Force Microscopy. Nat. Nanotechnol. 2016, 11, 719–723. [Google Scholar] [CrossRef]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-Speed Atomic Force Microscopy Reveals Rotary Catalysis of Rotorless F1-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef]
- Shibata, M.; Uchihashi, T.; Yamashita, H.; Kandori, H.; Ando, T. Structural Changes in Bacteriorhodopsin in Response to Alternate Illumination Observed by High-Speed Atomic Force Microscopy. Angew. Chem. Int. Ed. 2011, 50, 4410–4413. [Google Scholar] [CrossRef] [PubMed]
- Eeftens, J.M.; Katan, A.J.; Kschonsak, M.; Hassler, M.; de Wilde, L.; Dief, E.M.; Haering, C.H.; Dekker, C. Condensin Smc2-Smc4 Dimers Are Flexible and Dynamic. Cell Rep. 2016, 14, 1813–1818. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Sumitomo, K.; Tsuda, M.; Koizumi, S.; Inoue, K.; Torimitsu, K. Direct Observation of ATP-Induced Conformational Changes in Single P2X4 Receptors. PLoS Biol. 2009, 7, e1000103. [Google Scholar] [CrossRef]
- Igarashi, K.; Uchihashi, T.; Koivula, A.; Wada, M.; Kimura, S.; Okamoto, T.; Penttilä, M.; Ando, T.; Samejima, M. Traffic Jams Reduce Hydrolytic Efficiency of Cellulase on Cellulose Surface. Science 2011, 333, 1279–1282. [Google Scholar] [CrossRef]
- Igarashi, K.; Koivula, A.; Wada, M.; Kimura, S.; Penttilä, M.; Samejima, M. High Speed Atomic Force Microscopy Visualizes Processive Movement of Trichoderma Reesei Cellobiohydrolase I on Crystalline Cellulose*. J. Biol. Chem. 2009, 284, 36186–36190. [Google Scholar] [CrossRef]
- Yokokawa, M.; Yoshimura, S.H.; Naito, Y.; Ando, T.; Yagi, A.; Sakai, N.; Takeyasu, K. Fast-Scanning Atomic Force Microscopy Reveals the Molecular Mechanism of DNA Cleavage by ApaI Endonuclease. IEE Proc. Nanobiotechnol. 2006, 153, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Nakayama, T.; Itami, M.; Kodera, N.; Ando, T.; Konno, H. High-Speed Atomic Force Microscopy Reveals Strongly Polarized Movement of Clostridial Collagenase along Collagen Fibrils. Sci. Rep. 2016, 6, 28975. [Google Scholar] [CrossRef]
- Crampton, N.; Yokokawa, M.; Dryden, D.T.F.; Edwardson, J.M.; Rao, D.N.; Takeyasu, K.; Yoshimura, S.H.; Henderson, R.M. Fast-Scan Atomic Force Microscopy Reveals That the Type III Restriction Enzyme EcoP15I Is Capable of DNA Translocation and Looping. Proc. Natl. Acad. Sci. USA 2007, 104, 12755–12760. [Google Scholar] [CrossRef]
- Watanabe-Nakayama, T.; Ono, K.; Itami, M.; Takahashi, R.; Teplow, D.B.; Yamada, M. High-Speed Atomic Force Microscopy Reveals Structural Dynamics of Amyloid Β1–42 Aggregates. Proc. Natl. Acad. Sci. USA 2016, 113, 5835–5840. [Google Scholar] [CrossRef]
- Milhiet, P.-E.; Yamamoto, D.; Berthoumieu, O.; Dosset, P.; Grimellec, C.L.; Verdier, J.-M.; Marchal, S.; Ando, T. Deciphering the Structure, Growth and Assembly of Amyloid-Like Fibrils Using High-Speed Atomic Force Microscopy. PLoS ONE 2010, 5, e13240. [Google Scholar] [CrossRef]
- Watanabe-Nakayama, T.; Sahoo, B.R.; Ramamoorthy, A.; Ono, K. High-Speed Atomic Force Microscopy Reveals the Structural Dynamics of the Amyloid-β and Amylin Aggregation Pathways. Int. J. Mol. Sci. 2020, 21, 4287. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-Molecule Imaging of Dynamic Motions of Biomolecules in DNA Origami Nanostructures Using High-Speed Atomic Force Microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef]
- Wickham, S.F.J.; Endo, M.; Katsuda, Y.; Hidaka, K.; Bath, J.; Sugiyama, H.; Turberfield, A.J. Direct Observation of Stepwise Movement of a Synthetic Molecular Transporter. Nat. Nanotechnol. 2011, 6, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Hashemi, M.; Warren, G.; Bianco, P.R.; Lyubchenko, Y.L. Dynamics of the Interaction of RecG Protein with Stalled Replication Forks. Biochemistry 2018, 57, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Casuso, I.; Sens, P.; Rico, F.; Scheuring, S. Experimental Evidence for Membrane-Mediated Protein-Protein Interaction. Biophys. J. 2010, 99, L47–L49. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Voïtchovsky, K.; Uchihashi, T.; Contera, S.A.; Ryan, J.F.; Ando, T. Dynamics of Bacteriorhodopsin 2D Crystal Observed by High-Speed Atomic Force Microscopy. J. Struct. Biol. 2009, 167, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, D.; Nagura, N.; Omote, S.; Taniguchi, M.; Ando, T. Streptavidin 2D Crystal Substrates for Visualizing Biomolecular Processes by Atomic Force Microscopy. Biophys. J. 2009, 97, 2358–2367. [Google Scholar] [CrossRef]
- Yamamoto, D.; Ando, T. Chaperonin GroEL–GroES Functions as Both Alternating and Non-Alternating Engines. J. Mol. Biol. 2016, 428, 3090–3101. [Google Scholar] [CrossRef]
- Ruan, Y.; Kao, K.; Lefebvre, S.; Marchesi, A.; Corringer, P.-J.; Hite, R.K.; Scheuring, S. Structural Titration of Receptor Ion Channel GLIC Gating by HS-AFM. Proc. Natl. Acad. Sci. USA 2018, 115, 10333–10338. [Google Scholar] [CrossRef]
- Munguira, I.; Casuso, I.; Takahashi, H.; Rico, F.; Miyagi, A.; Chami, M.; Scheuring, S. Glasslike Membrane Protein Diffusion in a Crowded Membrane. ACS Nano 2016, 10, 2584–2590. [Google Scholar] [CrossRef]
- Yilmaz, N.; Kobayashi, T. Visualization of Lipid Membrane Reorganization Induced by a Pore-Forming Toxin Using High-Speed Atomic Force Microscopy. ACS Nano 2015, 9, 7960–7967. [Google Scholar] [CrossRef]
- Casuso, I.; Khao, J.; Chami, M.; Paul-Gilloteaux, P.; Husain, M.; Duneau, J.-P.; Stahlberg, H.; Sturgis, J.N.; Scheuring, S. Characterization of the Motion of Membrane Proteins Using High-Speed Atomic Force Microscopy. Nat. Nanotechnol. 2012, 7, 525–529. [Google Scholar] [CrossRef]
- Colom, A.; Casuso, I.; Boudier, T.; Scheuring, S. High-Speed Atomic Force Microscopy: Cooperative Adhesion and Dynamic Equilibrium of Junctional Microdomain Membrane Proteins. J. Mol. Biol. 2012, 423, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Yamada, T.; Greimel, P.; Uchihashi, T.; Ando, T.; Kobayashi, T. Real-Time Visualization of Assembling of a Sphingomyelin-Specific Toxin on Planar Lipid Membranes. Biophys. J. 2013, 105, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Taoka, A.; Uchihashi, T.; Asano, T.; Ando, T.; Fukumori, Y. Single-Molecule Imaging on Living Bacterial Cell Surface by High-Speed AFM. J. Mol. Biol. 2012, 422, 300–309. [Google Scholar] [CrossRef]
- Yamamoto, D.; Uchihashi, T.; Kodera, N.; Ando, T. Anisotropic Diffusion of Point Defects in a Two-Dimensional Crystal of Streptavidin Observed by High-Speed Atomic Force Microscopy. Nanotechnology 2008, 19, 384009. [Google Scholar] [CrossRef]
- Chiaruttini, N.; Redondo-Morata, L.; Colom, A.; Humbert, F.; Lenz, M.; Scheuring, S.; Roux, A. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 2015, 163, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miyagi, A.; Redondo-Morata, L.; Scheuring, S. Temperature-Controlled High-Speed AFM: Real-Time Observation of Ripple Phase Transitions. Small 2016, 12, 6106–6113. [Google Scholar] [CrossRef]
- Rico, F.; Gonzalez, L.; Casuso, I.; Puig-Vidal, M.; Scheuring, S. High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations. Science 2013, 342, 741–743. [Google Scholar] [CrossRef]
- Takahashi, H.; Rico, F.; Chipot, C.; Scheuring, S. α-Helix Unwinding as Force Buffer in Spectrins. ACS Nano 2018, 12, 2719–2727. [Google Scholar] [CrossRef]
- Rigato, A.; Miyagi, A.; Scheuring, S.; Rico, F. High-Frequency Microrheology Reveals Cytoskeleton Dynamics in Living Cells. Nat. Phys. 2017, 13, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Smolyakov, G.; Formosa-Dague, C.; Severac, C.; Duval, R.E.; Dague, E. High Speed Indentation Measures by FV, QI and QNM Introduce a New Understanding of Bionanomechanical Experiments. Micron 2016, 85, 8–14. [Google Scholar] [CrossRef]
- Heath, G.R.; Scheuring, S. High-Speed AFM Height Spectroscopy Reveals Μs-Dynamics of Unlabeled Biomolecules. Nat. Commun. 2018, 9, 4983. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, A.; Gao, X.; Adaixo, R.; Rheinberger, J.; Stahlberg, H.; Nimigean, C.; Scheuring, S. An Iris Diaphragm Mechanism to Gate a Cyclic Nucleotide-Gated Ion Channel. Nat. Commun. 2018, 9, 3978. [Google Scholar] [CrossRef]
- Morgan, J.L.W.; Evans, E.G.B.; Zagotta, W.N. Functional Characterization and Optimization of a Bacterial Cyclic Nucleotide–Gated Channel. J. Biol. Chem. 2019, 294, 7503–7515. [Google Scholar] [CrossRef] [PubMed]
- Kaupp, U.B.; Seifert, R. Cyclic Nucleotide-Gated Ion Channels. Physiol. Rev. 2002, 82, 769–824. [Google Scholar] [CrossRef]
- Craven, K.B.; Zagotta, W.N. CNG AND HCN CHANNELS: Two Peas, One Pod. Annu. Rev. Physiol. 2006, 68, 375–401. [Google Scholar] [CrossRef]
- Robinson, R.B.; Siegelbaum, S.A. Hyperpolarization-Activated Cation Currents: From Molecules to Physiological Function. Annu. Rev. Physiol. 2003, 65, 453–480. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kornev, A.P. Protein Kinases: Evolution of Dynamic Regulatory Proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef]
- Eron, L.; Arditti, R.; Zubay, G.; Connaway, S.; Beckwith, J.R. An Adenosine 3′:5′-Cyclic Monophosphate-Binding Protein That Acts on the Transcription Process. Proc. Natl. Acad. Sci. USA 1971, 68, 215–218. [Google Scholar] [CrossRef]
- Pifferi, S.; Boccaccio, A.; Menini, A. Cyclic Nucleotide-Gated Ion Channels in Sensory Transduction. FEBS Lett. 2006, 580, 2853–2859. [Google Scholar] [CrossRef]
- DiFrancesco, J.C.; DiFrancesco, D. Dysfunctional HCN Ion Channels in Neurological Diseases. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Yu, F.H.; Yarov-Yarovoy, V.; Gutman, G.A.; Catterall, W.A. Overview of Molecular Relationships in the Voltage-Gated Ion Channel Superfamily. Pharm. Rev. 2005, 57, 387–395. [Google Scholar] [CrossRef]
- Schmidpeter, P.A.M.; Gao, X.; Uphadyay, V.; Rheinberger, J.; Nimigean, C.M. Ligand Binding and Activation Properties of the Purified Bacterial Cyclic Nucleotide–Gated Channel SthK. J. Gen. Physiol. 2018, 150, 821–834. [Google Scholar] [CrossRef]
- Brams, M.; Kusch, J.; Spurny, R.; Benndorf, K.; Ulens, C. Family of Prokaryote Cyclic Nucleotide-Modulated Ion Channels. Proc. Natl. Acad. Sci. USA 2014, 111, 7855–7860. [Google Scholar] [CrossRef]
- Guan, L. Structure and Mechanism of Membrane Transporters. Sci. Rep. 2022, 12, 13248. [Google Scholar] [CrossRef]
- Zerangue, N.; Kavanaugh, M.P. Flux Coupling in a Neuronal Glutamate Transporter. Nature 1996, 383, 634–637. [Google Scholar] [CrossRef]
- Takahashi, K.; Foster, J.B.; Lin, C.-L.G. Glutamate Transporter EAAT2: Regulation, Function, and Potential as a Therapeutic Target for Neurological and Psychiatric Disease. Cell. Mol. Life Sci. 2015, 72, 3489–3506. [Google Scholar] [CrossRef] [PubMed]
- Tzingounis, A.V.; Wadiche, J.I. Glutamate Transporters: Confining Runaway Excitation by Shaping Synaptic Transmission. Nat. Rev. Neurosci. 2007, 8, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C. Glutamate Uptake. Prog. Neurobiol. 2001, 65, 1–105. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, R.J.; Ryan, R.M. Mechanisms of Glutamate Transport. Physiol. Rev. 2013, 93, 1621–1657. [Google Scholar] [CrossRef]
- Reyes, N.; Ginter, C.; Boudker, O. Transport Mechanism of a Bacterial Homologue of Glutamate Transporters. Nature 2009, 462, 880–885. [Google Scholar] [CrossRef]
- Yernool, D.; Boudker, O.; Jin, Y.; Gouaux, E. Structure of a Glutamate Transporter Homologue from Pyrococcus Horikoshii. Nature 2004, 431, 811–818. [Google Scholar] [CrossRef]
- Akyuz, N.; Altman, R.B.; Blanchard, S.C.; Boudker, O. Transport Dynamics in a Glutamate Transporter Homologue. Nature 2013, 502, 114–118. [Google Scholar] [CrossRef]
- Akyuz, N.; Georgieva, E.R.; Zhou, Z.; Stolzenberg, S.; Cuendet, M.A.; Khelashvili, G.; Altman, R.B.; Terry, D.S.; Freed, J.H.; Weinstein, H.; et al. Transport Domain Unlocking Sets the Uptake Rate of an Aspartate Transporter. Nature 2015, 518, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Erkens, G.B.; Hänelt, I.; Goudsmits, J.M.H.; Slotboom, D.J.; van Oijen, A.M. Unsynchronised Subunit Motion in Single Trimeric Sodium-Coupled Aspartate Transporters. Nature 2013, 502, 119–123. [Google Scholar] [CrossRef]
- Georgieva, E.R.; Borbat, P.P.; Ginter, C.; Freed, J.H.; Boudker, O. Conformational Ensemble of the Sodium-Coupled Aspartate Transporter. Nat. Struct. Mol. Biol. 2013, 20, 215–221. [Google Scholar] [CrossRef]
- Hänelt, I.; Wunnicke, D.; Bordignon, E.; Steinhoff, H.-J.; Slotboom, D.J. Conformational Heterogeneity of the Aspartate Transporter GltPh. Nat. Struct. Mol. Biol. 2013, 20, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, K.; Lebrun, B.; Yasuda-Kamatani, Y.; Sakaitani, M.; Shigeri, Y.; Yumoto, N.; Nakajima, T. Dl-Threo-β-Benzyloxyaspartate, A Potent Blocker of Excitatory Amino Acid Transporters. Mol. Pharm. 1998, 53, 195–201. [Google Scholar] [CrossRef]
- Haupts, U.; Tittor, J.; Oesterhelt, D. CLOSING IN ON BACTERIORHODOPSIN: Progress in Understanding the Molecule. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 367–399. [Google Scholar] [CrossRef]
- Lanyi, J.K. Bacteriorhodopsin. Annu. Rev. Physiol. 2004, 66, 665–688. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Vassylyev, D.G.; Miyazawa, A.; Kidera, A.; Matsushima, M.; Mitsuoka, K.; Murata, K.; Hirai, T.; Fujiyoshi, Y. Surface of Bacteriorhodopsin Revealed by High-Resolution Electron Crystallography. Nature 1997, 389, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Luecke, H.; Schobert, B.; Richter, H.-T.; Cartailler, J.-P.; Lanyi, J.K. Structure of Bacteriorhodopsin at 1.55 Å Resolution 11Edited by D. C. Rees. J. Mol. Biol. 1999, 291, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Canena, D.; Sikora, M.; Klausberger, M.; Seferovic, H.; Mehdipour, A.R.; Hain, L.; Laurent, E.; Monteil, V.; Wirnsberger, G.; et al. Force-Tuned Avidity of Spike Variant-ACE2 Interactions Viewed on the Single-Molecule Level. Nat. Commun. 2022, 13, 7926. [Google Scholar] [CrossRef] [PubMed]


| Authors | Studied System |
|---|---|
| Kodera et al. (2010) [15] | Direct visualization of myosin V molecules walking along actin tracks. |
| Shibata et al. (2010) [201] | Visualization of the dynamic changes of the light-driven proton pump bacteriorhodopsin upon illumination. |
| Takahashi et al. (2016) [233] | Observation in real time of lipid bilayers dynamics phase transition from ripple phase to fluid phase reversibly using a newly developed and integrated temperature-control device. |
| Ruan et al. (2017) [197] | Observations of the transport dynamics of the glutamate transporter homolog in a membrane-reconstituted GltPh. |
| Heath et al. (2018) [238] | The introduction of HS-AFM height spectroscopy and its application to measure surface concentrations, diffusion rates, and oligomer sizes of annexin-V molecules during membrane binding and self-assembly. |
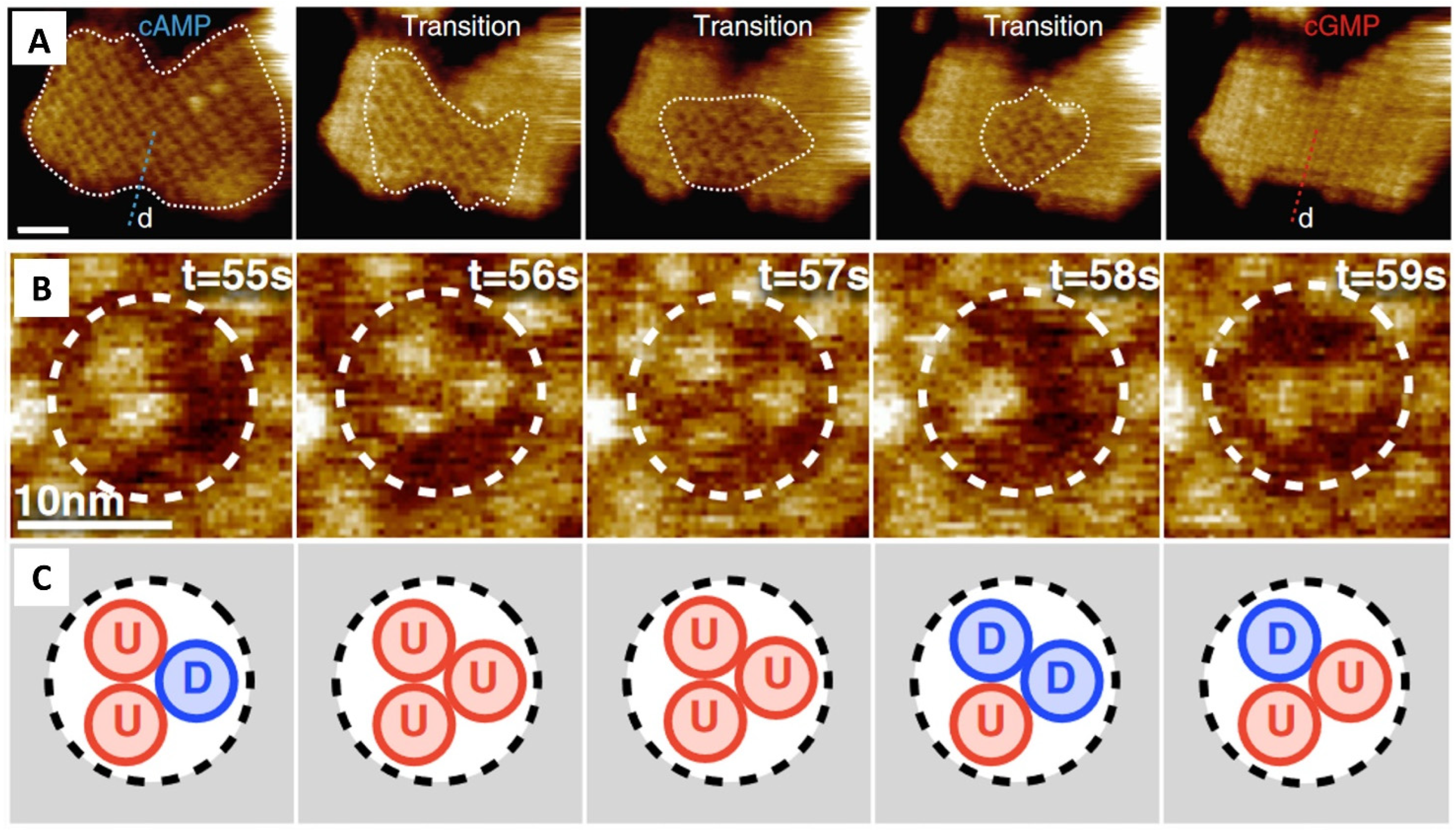
| Marchesi et al. (2018) [239] | Observation of the conformational reversible dynamics of a prokaryotic homolog of CNG channels, SthK, upon activation in response to cAMP-binding. |
| Ruan et al. (2018) [224] | The effect of buffer exchange on structural titration experiments to visualize GLIC gating at the single-molecule level under native conditions. |
| Zhu et al. (2022) [269] | Visualization of conformational dynamics of isolated spike trimers complexed with their essential entry receptor ACE2. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondi, S.M.C.; Ailuno, G.; Mattioli, S.L.; Pesce, A.; Cavalleri, O.; Canepa, P. Morphological Investigation of Protein Crystals by Atomic Force Microscopy. Crystals 2023, 13, 1149. https://doi.org/10.3390/cryst13071149
Rotondi SMC, Ailuno G, Mattioli SL, Pesce A, Cavalleri O, Canepa P. Morphological Investigation of Protein Crystals by Atomic Force Microscopy. Crystals. 2023; 13(7):1149. https://doi.org/10.3390/cryst13071149
Chicago/Turabian StyleRotondi, Silvia Maria Cristina, Giorgia Ailuno, Simone Luca Mattioli, Alessandra Pesce, Ornella Cavalleri, and Paolo Canepa. 2023. "Morphological Investigation of Protein Crystals by Atomic Force Microscopy" Crystals 13, no. 7: 1149. https://doi.org/10.3390/cryst13071149
APA StyleRotondi, S. M. C., Ailuno, G., Mattioli, S. L., Pesce, A., Cavalleri, O., & Canepa, P. (2023). Morphological Investigation of Protein Crystals by Atomic Force Microscopy. Crystals, 13(7), 1149. https://doi.org/10.3390/cryst13071149








