Abstract
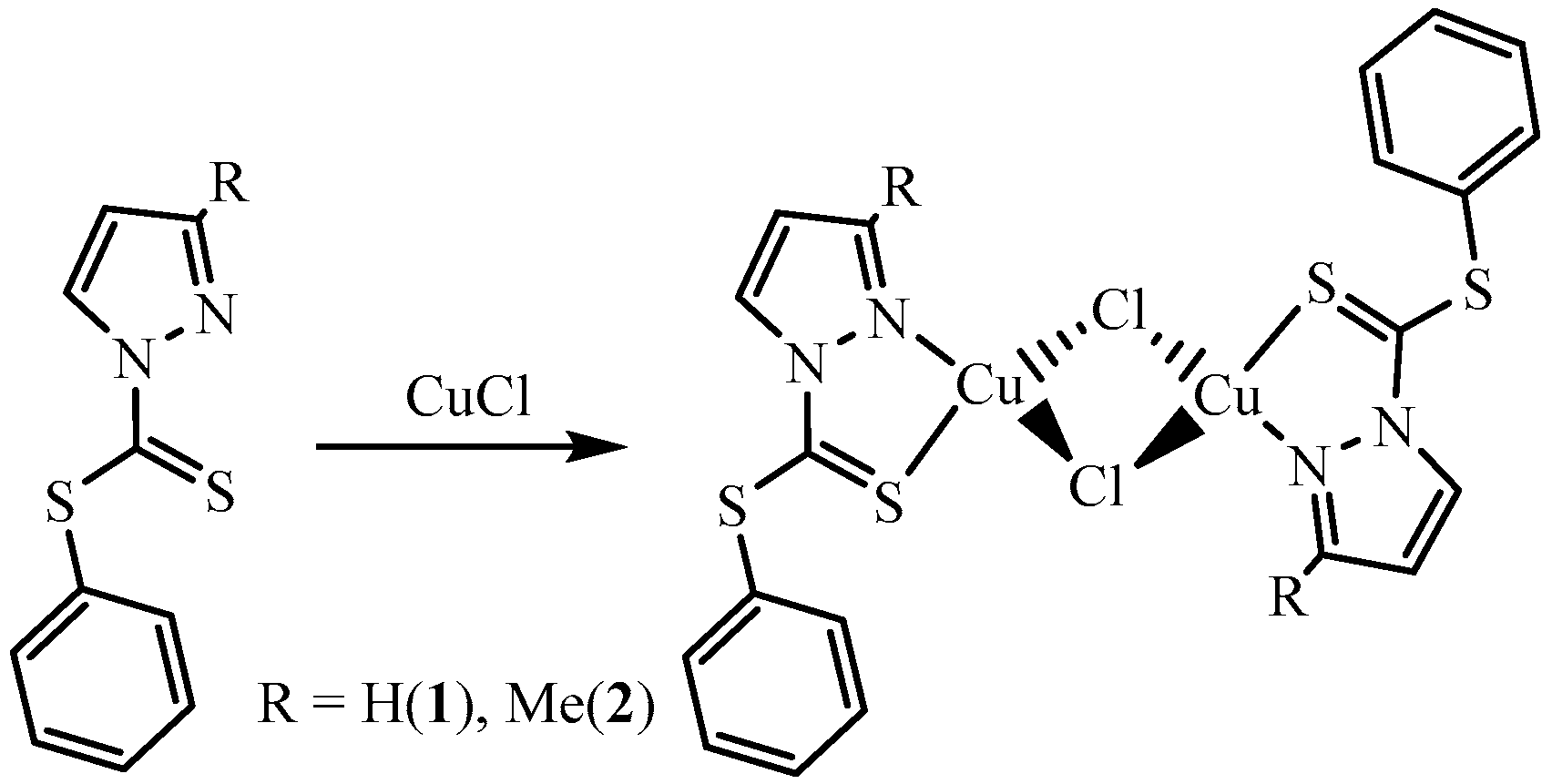
The CuCl binuclear complexes were synthesized with phenyl-1H-pyrazole-1-carbodithioate (L1) and phenyl-3-methyl-1H-pyrazole-1-carbodithioate (L2) ligands. The complexes were isolated as crystalline material in a reasonable quantity. The complexes were crystallized in acetonitrile (MeCN) and characterized for their single crystal, using X-ray diffraction. The two units with the general formula LCuCl are bridged together via chlorido ligands, affording (LCuCl)2-type complexes. The complexes, [Cu2(μ-Cl)2(L1)2] 1 and [Cu2(μ-Cl)2(L2)2] 2 are monoclinic and triclinic with space group P21/n and Pi, respectively. The crystal packing is stabilized by C1(p)⋯C(p) and S⋯C(p) interactions extended in 2D fashion in complex 1, while complex 2 is stabilized by C(p)⋯S interactions extended in a 1D fashion. Structural features and secondary interactions present in both complexes discussed in this article.
1. Introduction
The coordination chemistry of copper(I) with sulfur- and nitrogen-containing ligands is of great interest, due to structural variations and large-scale biological applications [1,2,3]. Copper itself is an essential element, and its importance cannot be neglected from health perspective [4,5], as its absence may cause severe health problems in human beings, i.e., reduced immunity, augmented vulnerability to various oral and systemic diseases, impaired physical and mental growth, and reduced efficiency [6]. The analysis proved that it is found in human milk as a component of metabolic enzymes, hormones and antioxidants [7]. The carbodithioate moiety containing ligands are valuable metal-coordination compounds, due to their extra stability. That is why chemists are interested in synthesizing these complexes for pharmaceutical and medicinal uses. Ziram (zincdimethyldithiocabamate) and zineb (zinc ethylene-1,2-bisdithiocarbamate) are well known fungicides [8]. The complexes of copper are important for their biological importance in certain activities. The biological activities of sulfur- and nitrogen-based copper complexes have been studied thoroughly by different researchers, and the complexes were fairly effective against gram-negative and gram-positive bacteria [9]. Hydrazine carbodithioate Schiff bases were prepared, and treated with copper salts. The complexes were investigated for antibacterial activities, with moderate results [10]. Transition-metal complexes of copper with ligands having heteroatom nitrogen and sulfur show carcinostatic, antitumor, antiviral, antifungal and antibacterial activities [11]. The complexes of benzoylpyridine Schiff-base ligands were found to be sufficiently active towards certain microorganisms [12]. The copper complexes of S-benzyl dithiocarbazate were synthesized and studied for analgesic activities; the complexes show good anti-inflammatory activity compared to standard Indomethacine [13]. The copper complexes of S-allyldithiocarbazate were synthesized and tested for anticancer activity. It was found that the respected complexes are more toxic to cancer cells while less toxic to normal cells [14]. The complexes with carbodithioate moiety also exhibit good antioxidant activity [15]. The thiosemicarbazone complexes of copper were also tested for ribonucleotide-reductase-R2 inhibition, as this enzyme plays a crucial role in nucleic-acid metabolism, so it is the best target for anticancer therapy. Investigation showed that copper complexes of the respective ligands are effective anti-proliferative agents, being tested for several cancer cells [16]. The halide-bridged complexes of Cu(I) and Cu(II) with piperidine-1-carbodithioato ligand were prepared and tested successfully for magnetic and conductive properties. They were tested as the sensitizing material in dye-sensitized solar cells (DSSCs) [17].
Copper complexes with ligands containing the carbodithioate moiety are also of considerable interest in several inter-disciplinary areas such as bioinorganic chemistry, catalysis, photochemistry and magnetochemistry. These compounds may also have many potential applications in pharmaceutical chemistry, including use as antibacterial, antifungal, anticancer and anti-inflammatory agents. Here we report the synthesis and crystal structure of bi-nuclear copper(I) complexes with phenyl-1H-pyrazole-1-carbodithioate and phenyl 3-methyl-1H-pyrazole-1-carbodithioate ligands.
2. Materials and Methods
2.1. Synthesis of Ligands and General Considerations
The ligands phenyl-1H-pyrazole-1-carbodithioate and phenyl 3-methyl-1H-pyrazole-1-carbodithioate were prepared, according to the reported method [18,19]. Equimolar amounts of phenylchlorodithioformate and pyrazole or its derivative were treated together in dry toluene and in the presence of an equimolar amount of triethyl amine. The formation of white precipitated ammonium salt Et3N·HCl indicates the reaction completion. The reaction was continued for 48 h, and after completion the product was washed with water, and the final product was dried over sodium sulfate. Solvents and all other volatile material were evaporated under reduced pressure, and a viscous orange oil of both ligand L1 and ligand L2 was obtained. Chemicals used in this study such as CuCl, MeOH and acetonitrile were purchased from Sigma Aldrich. All chemical reagents used in this study were used without further purification. Elemental analyses (CHN) were performed on a Vario EL III instrument. For the single crystal of both the complexes, X-ray-diffraction data were collected through STOE-IPDS II equipped with an Oxford Cryostream low-temperature unit, and graphite-monochromator Mo-Kα radiation (λ = 0.71073 Å). Structure solutions and refinements of complexes were accomplished using SIR97 [20], SHELXL2018/3 [21], WinGX [22], SAINT [23] and PLATON [24].
2.2. Synthesis of Di-μ-chlorido-bis(phenyl-1H-pyrazole-1-carbodithioato)dicopper(I) (1) and Its Analogous Complex Di-μ-chlorido-bis(phenyl-3-methyl-1H-pyrazole-1-carbodithioato)dicopper(I) (2)
A solution of copper salt was prepared in 5 mL analytical-grade methanol (CuCl 100 mg; 1.0 mmol) and was mixed with a separately prepared equimolar solution of ligand L1 (ligand contents 220 mg; 1.0 mmol, solvent methanol). Immediately after mixing both the solutions, the color turned green; stirring was continued for 30 min which led to the formation of green precipitates of the desired complex 1. The reaction mixture was allowed to stir further for 60 min, so that the completion of the reaction was ensured. The green solid was separated from the mother liquor, and was dried in open air. The powder product was redissolved in methanol and kept undisturbed, for slow evaporation. After one day, green-colored crystals of the complex appeared, and a crystal of suitable dimensions was selected for single-crystal X-ray-diffraction analysis, and its structure was determined. Complex 2 was prepared following the same experimental procedure, by mixing salt solution and L2 in equimolar amounts (100 mg; 1.00 mmol of CuCl and 230 mg; 1.0 mmol of phenyl-3-methyl-1H-pyrazol-1-carbodithioate)). The complexes were also studied for their elemental ratios of CHN.
1. Yield: 70%; Color of crystalline material: green; Chemical formula of complex: C24H24Cl2Cu2N4S4, Found/Cal (%): C 38.8/41.49, H 3.5/3.48, N 8.2/8.06.
2. Yield: 73%; Color of crystalline material: green; Chemical formula of complex: C22H20Cl2Cu2N4S4, Found/Cal (%): C 37.8/39.63, H 3.2/3.02, N 7.9/8.40.
2.3. Hirshfeld Surface Calculations
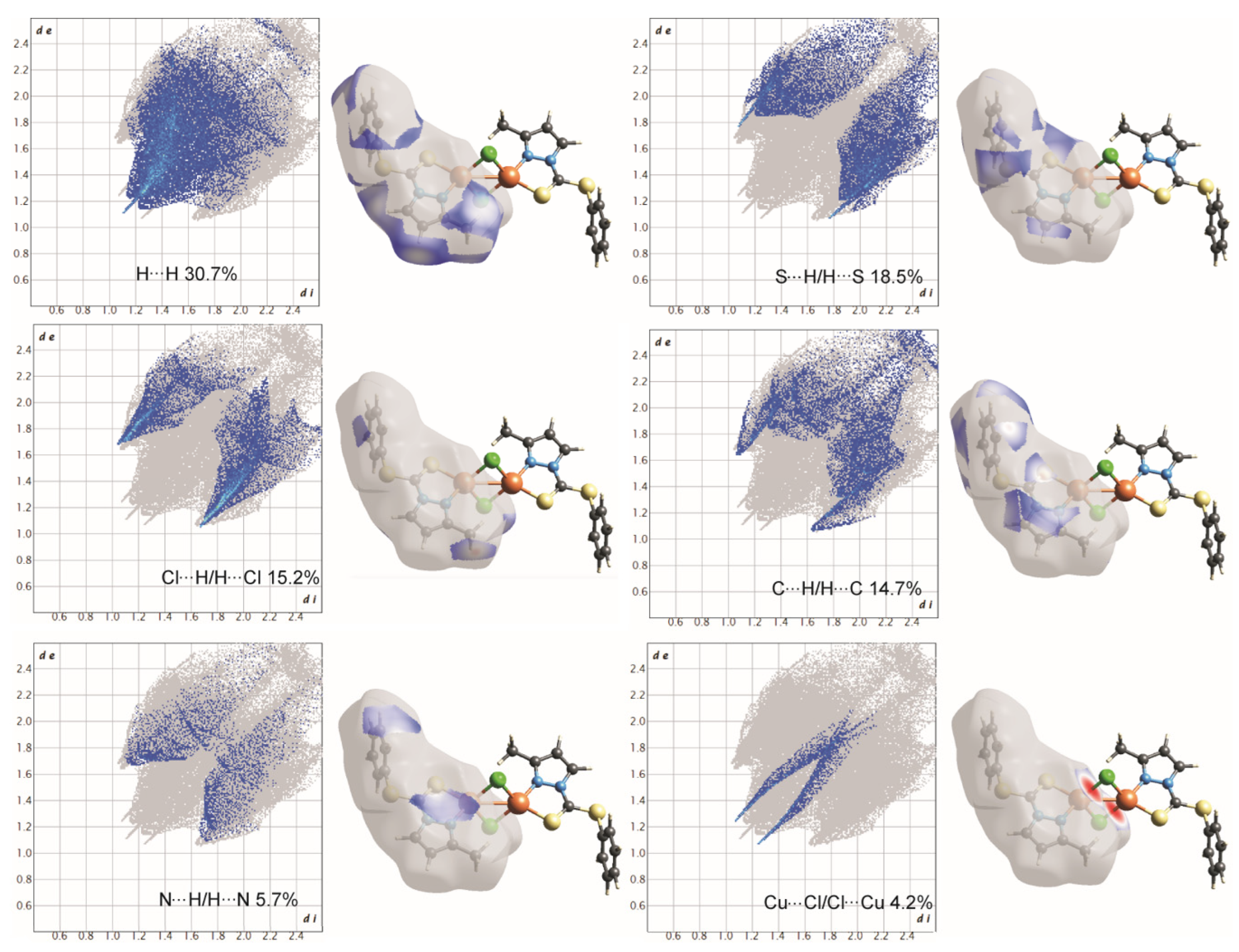
The Hirshfeld surfaces and subsequent two-dimensional (2D) fingerprint plots of the herein reported compounds were calculated using Crystal Explorer 17.50 [25]. Crystallographic information were obtained for each X-ray single crystal from their crystallographic information files (.cif) and were imported to Crystal Explorer to generate the Hirshfeld surfaces, with the following settings being applied: property: none; resolution: high (standard). For fingerprint generation (di vs. de plot) we applied range: standard; filter: by elements; fingerprint-filter options are both inside–outside elements, including reciprocal contacts. The interactions with normalized contact distance in crystal structures shorter than the sum of the corresponding van der Waals radii of the atoms are visualized by red spots, and those with longer contacts having the positive dnorm value are shown in blue [26].
3. Results and Discussion
The complexes 1 and 2 were synthesized by the reaction of CuCl with L1 and L2 in equimolar ratios. The reaction for the synthesis of complexes is summarized in Scheme 1. Based on the molar ratio of metal to ligand, it was expected to obtain final products with the general formula LCuCl. The metal (Cu(I)) being coordinatively unsaturated afforded binuclear Cu(I) complexes. The binuclear complexes bridged together via chloride, in each complex. The metal atom was bonded to two bridging chlorido ligands, a nitrogen atom of the pyrazolyl moiety and the thionyl sulfur of the ligand. The elemental ratio of the compounds was determined (elemental/CHN analysis), and the structure was confirmed for the single crystal of each complex, using X-ray diffraction, CCDC numbers are provided below in section “supplementary material”.
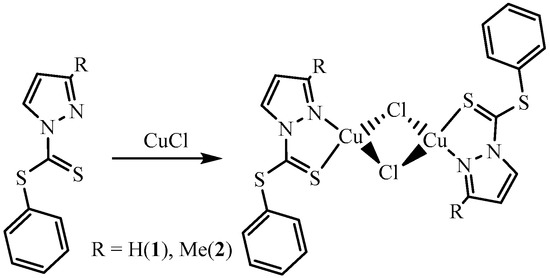
Scheme 1.
Synthesis of complex 1 and 2; reactions were carried out in MeOH and crystals were grown in MeCN.
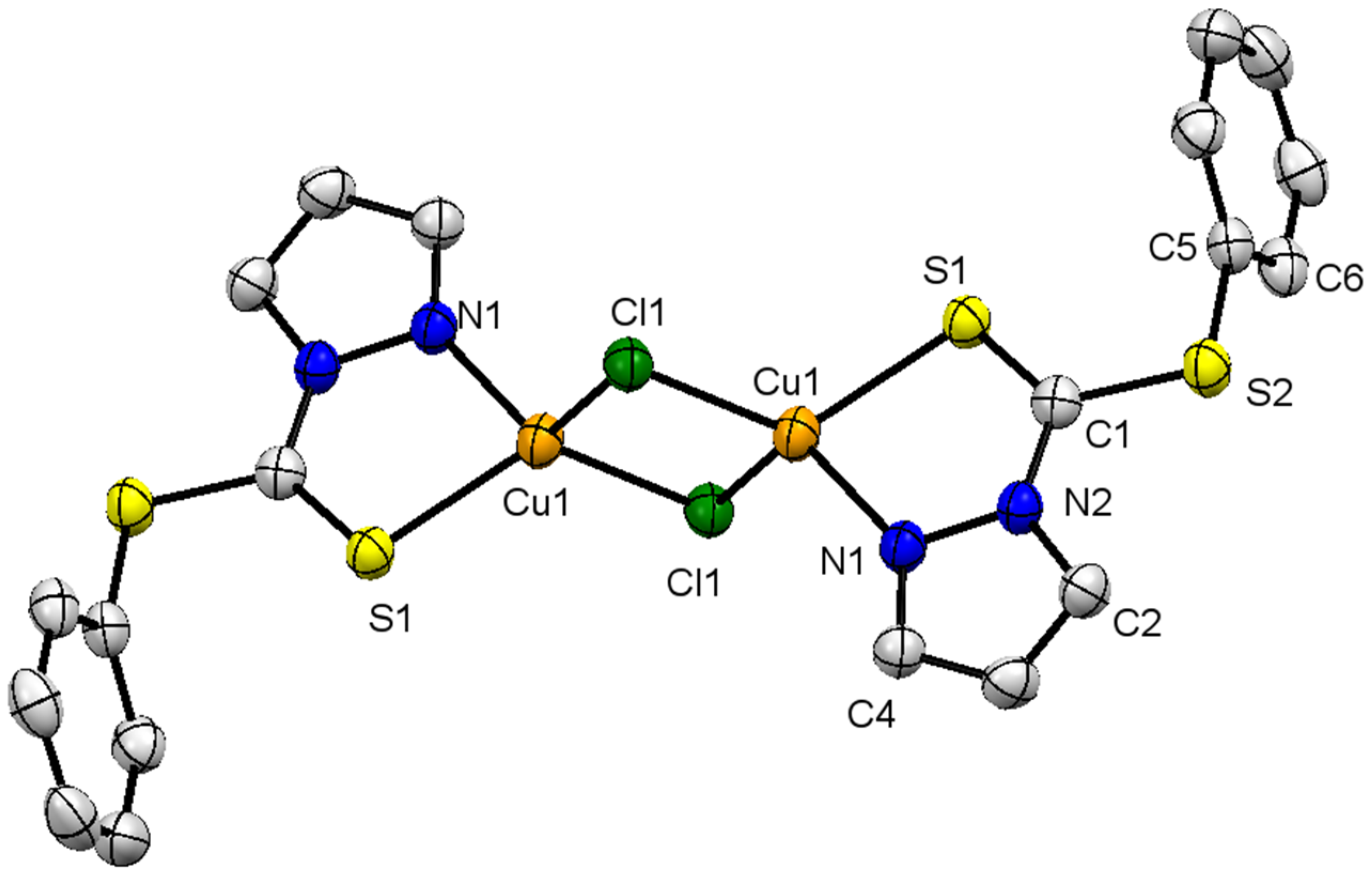
The crystal structure with selected bond lengths and angles of complex 1, is shown in Figure 1. It has a monoclinic crystal system with space group P21/n. Two copper ions in the complex are bridged together with the help of chlorido ligands. The central metal ion in the monomer unit is trigonal planar, wherein the metal is coordinatively unsaturated and interacts with a neighboring unit to make the dimer through the Cl− bridge. The metal in the bimetallic molecule adopts distorted tetrahedral geometry, bonded to two chlorido and a N and S of the chelating ligand. The pyrazolyl moiety contains two N atoms wherein C=N is more basic than the C-N, as has been proved in the literature for certain metal complexes [27]. Similarly, the two S in the nucleophilic center are also chemically different, and C=S is softer and more basis in comparison to -S-. The presence of N and S in the ligand L1 makes a stable chelating ligand, to afford a 5-membered metallacycle of the C2NSCu type.
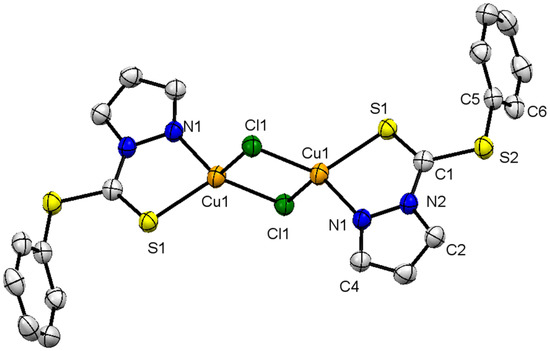
Figure 1.
X-ray single-crystal structure of compound 1; partial numbering scheme for carbon and non-carbon atoms, ellipsoids drawn at 50% probability level, all hydrogen atoms are omitted for clarity. Color scheme: Cu = orange; Cl: green; S = yellow; and N = blue. Selected structural features: bond lengths (Å) and angles (°); Cu(1)-N(1) 2.057(2), Cu(1)-S(1) 2.287(7), Cu(1)-Cl(1) 2.318(7), Cu(1)-Cl(1) 2.325(7), S(2)-C(1) 1.749(3), S(2)-C(5) 1.770(3), S(1)-C(1) 1.637(3); N(1)-Cu(1)-S(1) 85.44(6), N(1)-Cu(1)-Cl(1) 114.19(6), S(1)-Cu(1)-Cl(1) 123.14(3), N(1)-Cu(1)-Cl(1) 117.94(7), S(1)-Cu(1)-Cl(1) 116.51(3),Cl(1)-Cu(1)-Cl(1) 100.66(2), Cu(1)-Cl(1)-Cu(1) 79.34(2), C(1)-S(2)-C(5) 100.87(12), C(1)-S(1)-Cu(1) 98.12(9), C(2)-N(2)-N(1) 110.1(2), C(2)-N(2)-C(1) 130.1(2), N(1)-N(2)-C(1) 119.7(2), C(4)-N(1)-N(2) 104.9(2), C(4)-N(1)-Cu(1) 140.63(18), N(2)-N(1)-Cu(1) 114.44(16), N(2)-C(1)-S(1) 122.21(18), N(2)-C(1)-S(2) 112.25(17), S(1)-C(1)-S(2) 125.54(16).
The geometry around the Cu(I) as discussed above is distorted tetrahedral, with observed bond angles ∠N(1)-Cu(1)-Cl(1) 114.19°, ∠S(1)-Cu(1)-Cl(1) 123.14°, ∠N(1)-Cu(1)-Cl(1) 117.94° and an acute angle ∠N(1)-Cu(1)-S(1) 85.44°. The acute angle is obvious, due to the bidentate (chelating) behavior of the ligand to afford the metallacycle. The open angles are wider as compared to normal angles of tetrahedrally arranged monodentate ligands around a copper ion [28]. The sp2 hybridized C(1) adopts trigonal-planar geometry, with all atoms being coplanar with slight deviation, and all angles close to 120°, i.e., ∠N(2)-C(1)-S(1) 122.21°, ∠N(2)-C(1)-S(2) 112.25°, ∠S(1)-C(1)-S(2) 125.54°. The smallest angle ∠N(2)-C(1)-S(2) 112.25° is because of N(2) and S(2), as part of the 5-membered metallacylic ring. All other bond angles are similar to those observed for 5-membered metallacycles containing Cu as the central metal ion [29]. The Cu(1)-N(1) and Cu(1)-S(1) bond distances are of 2.057 Å and 2.29 Å, respectively, and are within the expected range, as has been reported for Cu complexes with N, S-donor ligands [29].
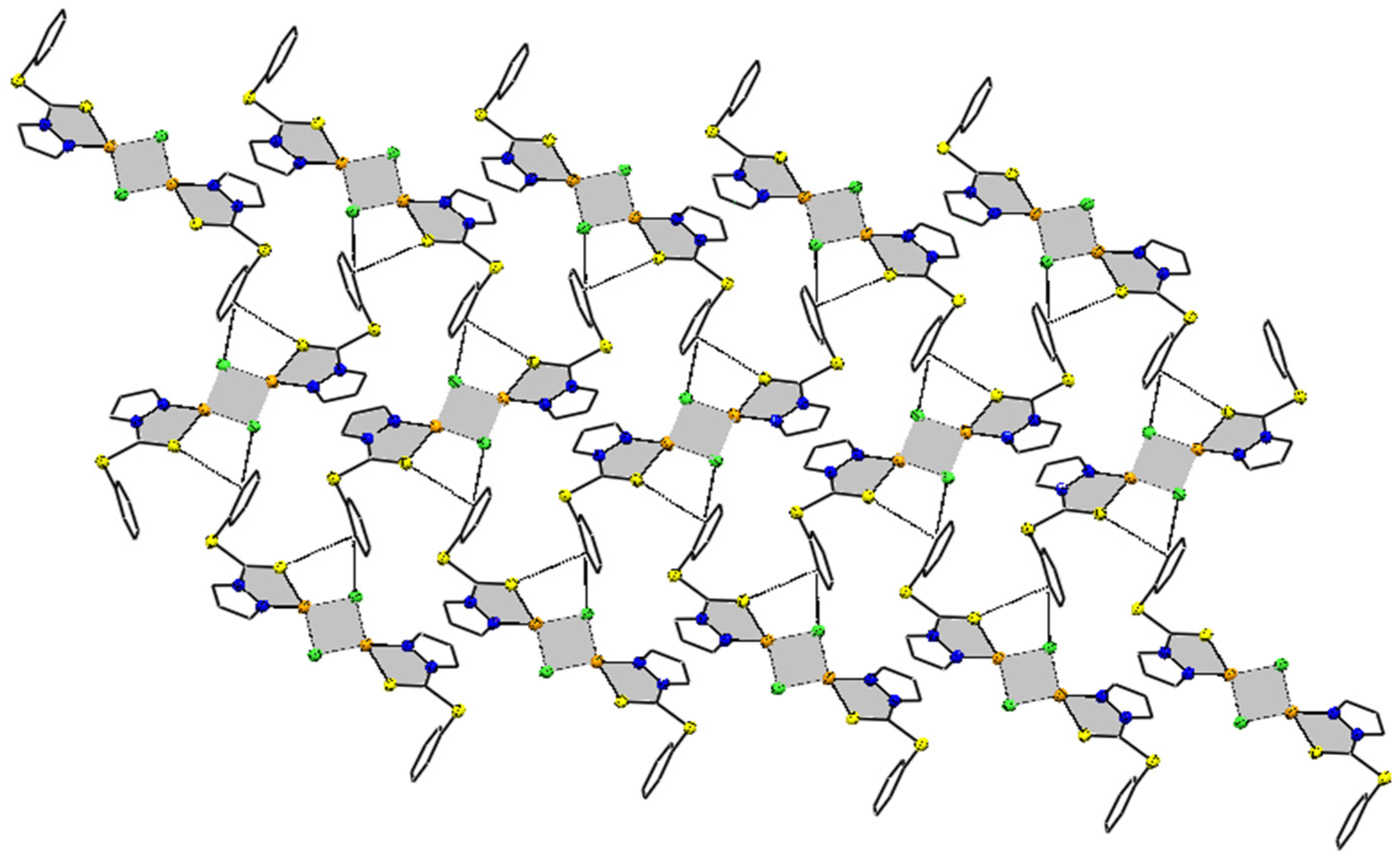
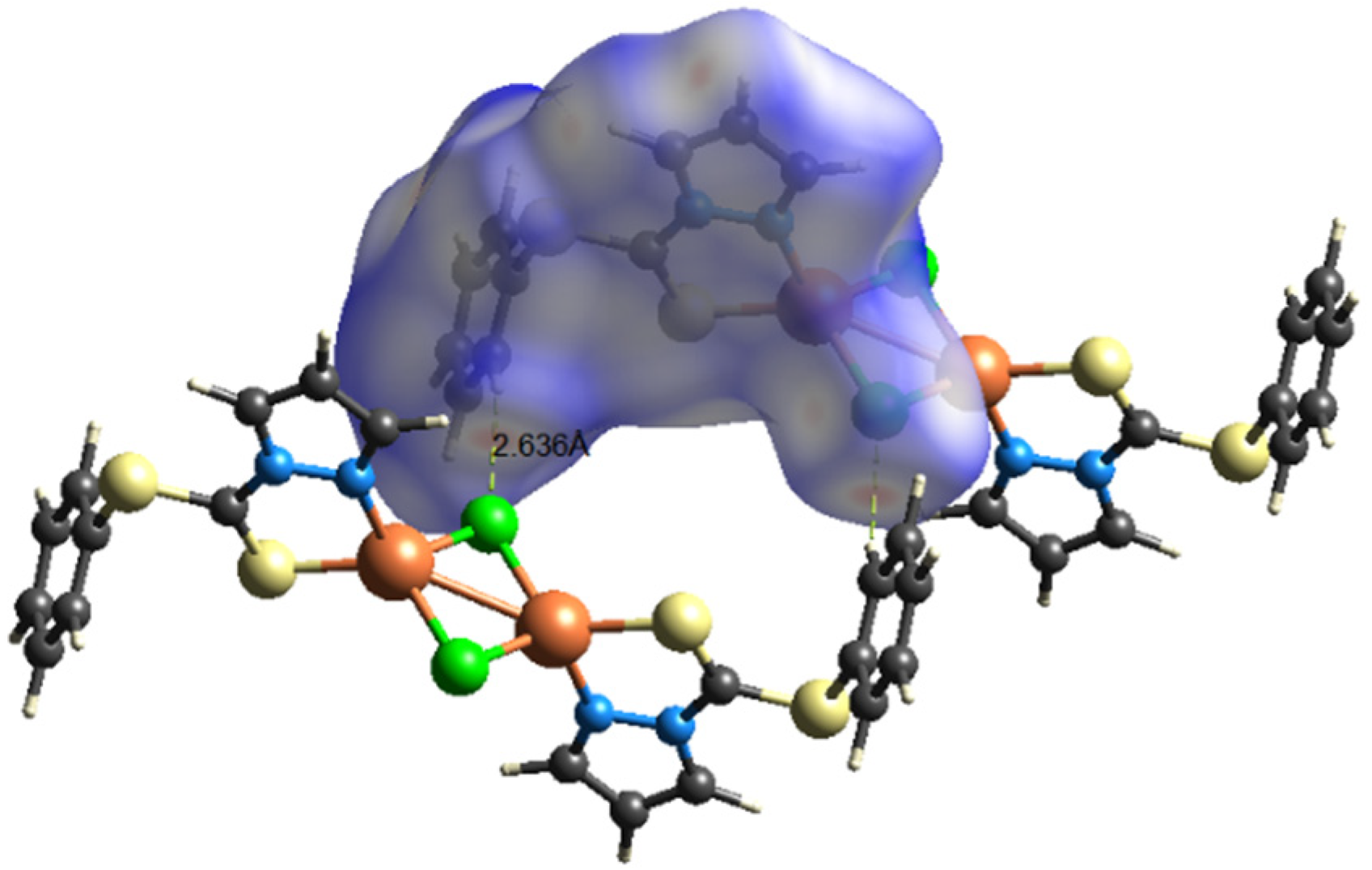
The supramolecular chemistry reveals that molecules of the complex in solid state are bonded to each other via intermolecular interactions of the phenyl ring C(6)(p)⋯Cl(1)(p) (3.688 Å) and C(6)(p)⋯S(1) (3.354 Å). This interaction arranges molecules in three parallel layers in such a way that alternate layers are exactly superimposable over each other. As shown in the 2D-supramolecular fragment of the molecule in Figure 2, Cl and S of the same molecule are active, to stabilize the supramolecular structure of the complex. There is no evidence of π-electrons of 5-membered pyrazole moiety including the N atoms of the ring. Other types of short-ranged interactions were also negligible.
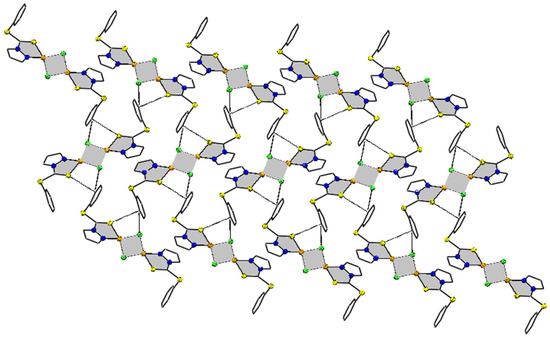
Figure 2.
Supramolecular fragment of compound 1 stabilized by C(6)(p)⋯Cl(1)(p) and S(1)⋯C(6)(p) interaction, with separation distance of 3.688 and 3.354 Å, respectively. The five- and four-membered rings containing Cu are shown shaded, to make the interactions reader friendly. Hanging contacts and all hydrogen atoms are deleted for simplicity.
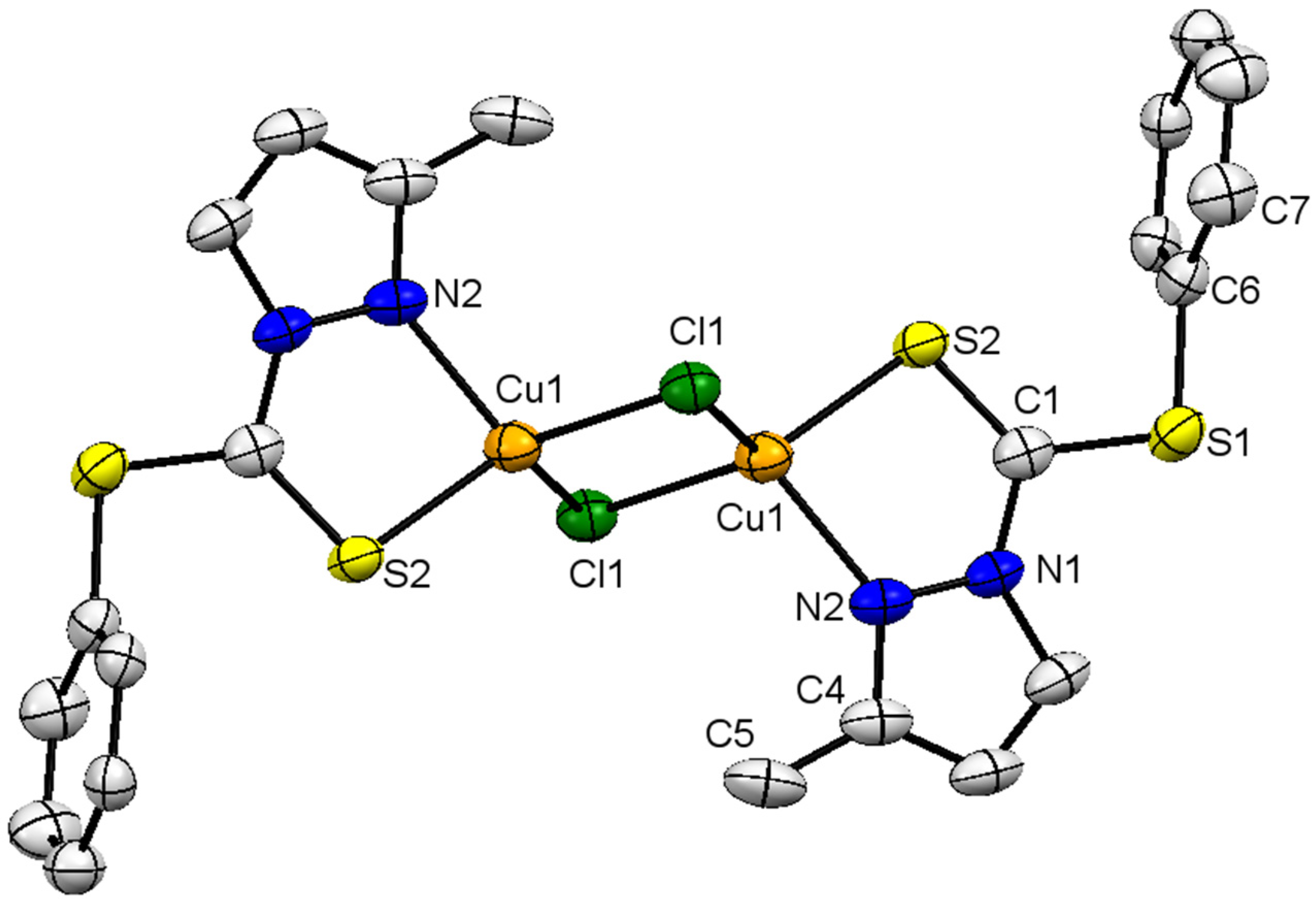
The molecular structure of complex 2 is depicted in Figure 3, along with structural features as the footnote. Crystal-structure solution and refinement parameters are summarized in Table 1. The molecule is triclinic-bearing-P1 space group. The complex is homo structural to that of complex 1, i.e., bimetallic. The geometry around the Cu(I) ion is distorted tetrahedral, and bond angles around the metal ion are within the expected range, between 85.44° and 121.67°. The C(1) is similar in geometry to that present in complex 1, which is trigonal planar (with sp2 hybridization) with negligible deviation from the planarity. All angles around C(1) are approximately equal to 360° (∠S(1)-C(1)-N(1), ∠S(1)-C(1)-S(2) and ∠S(2)-C(1)-N(1)), measuring 112.40, 126.01 and 121.50°, respectively. As expected, the endocyclic bond angle ∠N(1)-C(1)-S(1) is relatively smaller than the exocyclic angles (126.01° and 121.5°). The Cu(1)-N(2) and Cu(1)-S(1) bond lengths are 2.054 Å and 2.281 Å, respectively, and are comparable to other Cu complexes stabilized by N, S-donor ligands [30]. The Cu(1)-Cl(1) bond lengths (2.317 Å and 2.338 Å) are different with respect to each other, which reveals the difference in nature of interaction (bond) with the same metal ion, i.e., the Cu-Cl and Cl→Cu bond, respectively. The coordinate covalent bond is slightly longer than the covalent bond, but the electron density is still considerably distributed over the Cl(1)-Cu(1)-Cl(1) fragment. The difference in Cu-Cl-bond lengths also indicates the electron-density delocalization and the nature of the bond being formed between Cl and Cu ions. The difference in bond lengths of the Cu-Cl bond in complex 2 is less (0.021 Å) than the difference in the Cu-Clbridged bond reported in the literature (Δ = 0.452 Å). The Cu-Clbridged bond is longer than the Cu-Clterminal bond, the latter being 2.2307 Å [31]. The S(2)-C(1) bond length of 1.643 Å and S(1)-C(1) bond length of 1.737 Å clearly show the C=S double- and C-S single-bond characters. The elongation of the C=S bond in the complex is because of the flow of electron density towards metal from the ligand (C=S bond length for the uncoordinated thiourea compound is 1.662–1.698 Å [32,33,34,35]. The C=S bond in complex 2 is slightly stronger than the bond between the same ligand and the Pd(II) metal ion, being 1.643 and 1.662 Å, respectively [18].
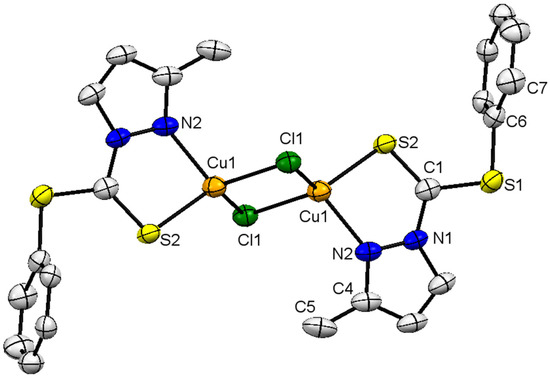
Figure 3.
Solid-state molecular structure of 2; partial-numbering scheme, thermal ellipsoids are drawn at 50% probability level, for clarity reasons all hydrogen atoms are omitted. Color scheme: Cu = orange; Cl: green; S = yellow; and N = blue. Selected structural features: bond lengths (Å) and angles (°); Cu(1)-N(2) 2.054(3), Cu(1)-S(1) 2.281(9), Cu(1)-Cl(1) 2.317(9), Cu(1)-Cl(1) 2.338(9), Cu(1)-Cu(1) 2.901(9),Cl(1)-Cu(1) 2.317(9), S(2)-C(1) 1.643(3), S(1)-C(1) 1.737(4), S(2)-C(6) 1.768(3); N(2)-Cu(1)-S(2) 85.44(8), N(2)-Cu(1)-Cl(1) 121.67(7), S(2)-Cu(1)-Cl(1) 114.11(3), N(2)-Cu(1)-Cl(1) 106.36(8), S(2)-Cu(1)-Cl(1) 127.03(3), Cl(1)-Cu(1)-Cl(1) 102.92(3), Cu(1)-Cl(1)-Cu(1) 77.08(3), C(1)-S(2)-Cu(1) 98.25(12), C(1)-S(2)-C(6) 101.93(15), N(1)-C(1)-S(2) 121.50(3), N(1)-C(1)-S(1) 112.40(2), S(2)-C(1)-S(1) 126.01(19), C(4)-N(2)-Cu(1) 138.2(3), N(1)-N(2)-Cu(1) 114.50(18).

Table 1.
Data pertinent to crystal-structure determination and refinements of compound 1 and 2.
The extended, supramolecular structure of compound 2 is shown in Figure 4, and is considerably different from the supramolecular structure of 1. In complex 2, molecules are extended in a one-dimensional fashion, making the 1D straight chain. The C(1)-S(1) and C(6)-S(2) separation distances indicate that the lone pair of electrons is somehow shifted to C(1), making S(2) partially electron deficient. The electron deficiency thus generated on the sulfur S(2) causes the production of an interaction with π-electrons of the aromatic ring, extending the structure as an 1D chain. The molecules within the same chain are separated by a distance 3.493 Å, with respect to each other.
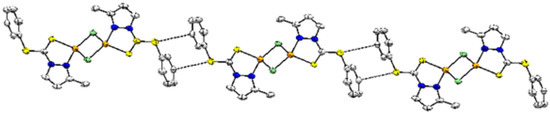
Figure 4.
1D Supramolecular structure of 2 stabilized by C(p)⋯S interaction, with a separation distance of 3.491 Å. Color scheme: Cu = orange; Cl: green; S = yellow; and N = blue.
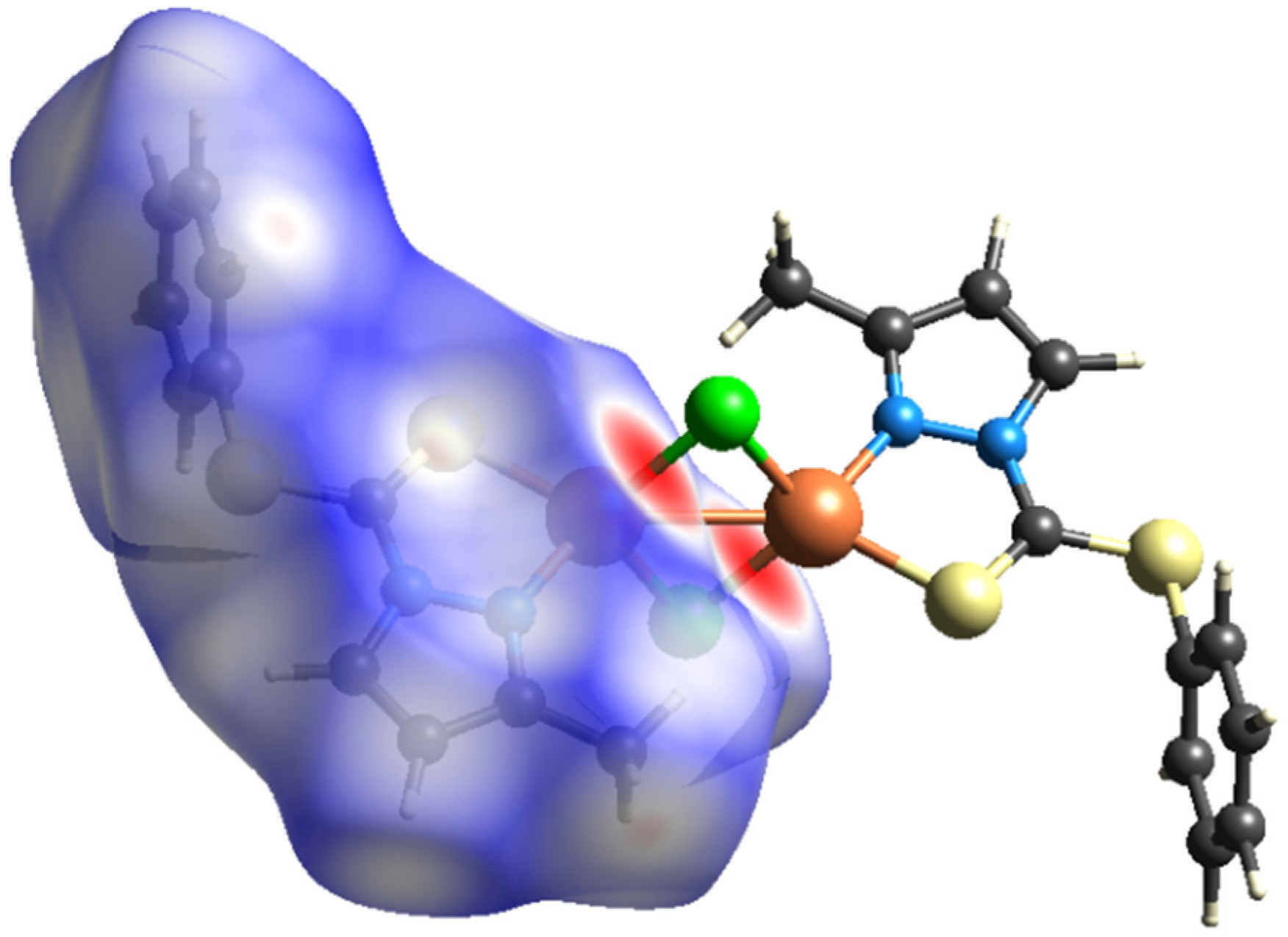
To further quantify and obtain insights about the strengths as well as the differences of intermolecular interactions in 1 and 2, Hirshfeld-surface analyses were performed. The interactions with a normalized contact distance shorter than the sum of the corresponding van der Waals radii of the atoms, have been visualized by red spots, and the longer contacts having the positive dnorm value are shown in blue (Figure 5 and Figure 6). The corresponding intermolecular interactions with significant contributions are shown in Figure 7 and Figure 8.
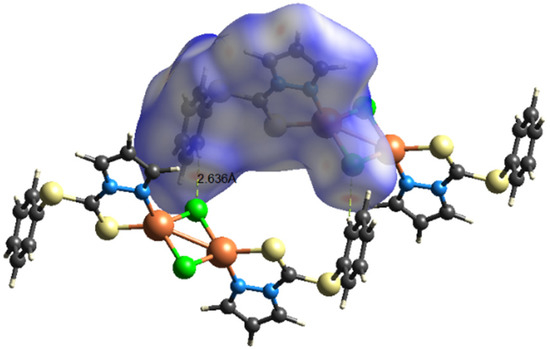
Figure 5.
Visualization of the three-dimensional Hirshfeld surface of 1 mapped over dnorm in the range −0.5175 to 1.1515. Color scheme: Cu = orange; Cl: green; S = yellow; and N = blue.
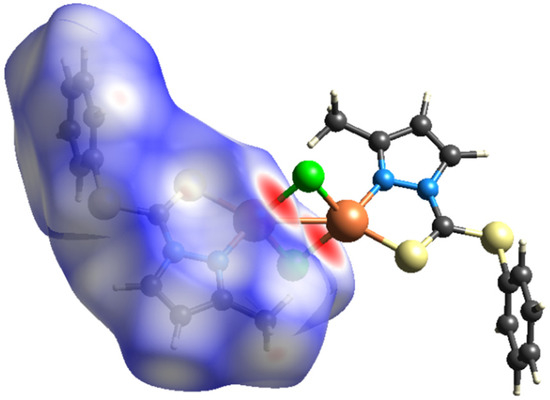
Figure 6.
Visualization of the three-dimensional Hirshfeld surface of 2 mapped over norm in the range −0.5221 to 1.1503. Color scheme: Cu = orange; Cl: green; S = yellow; and N = blue.
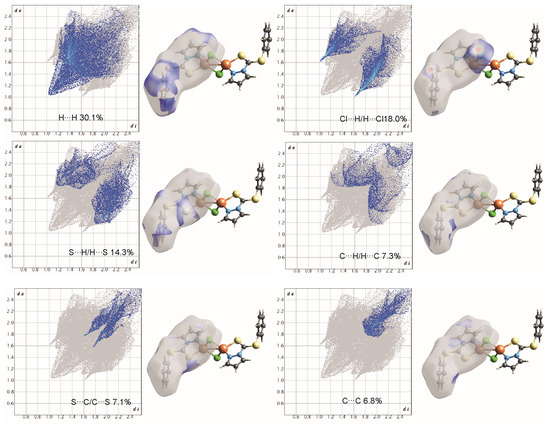
Figure 7.
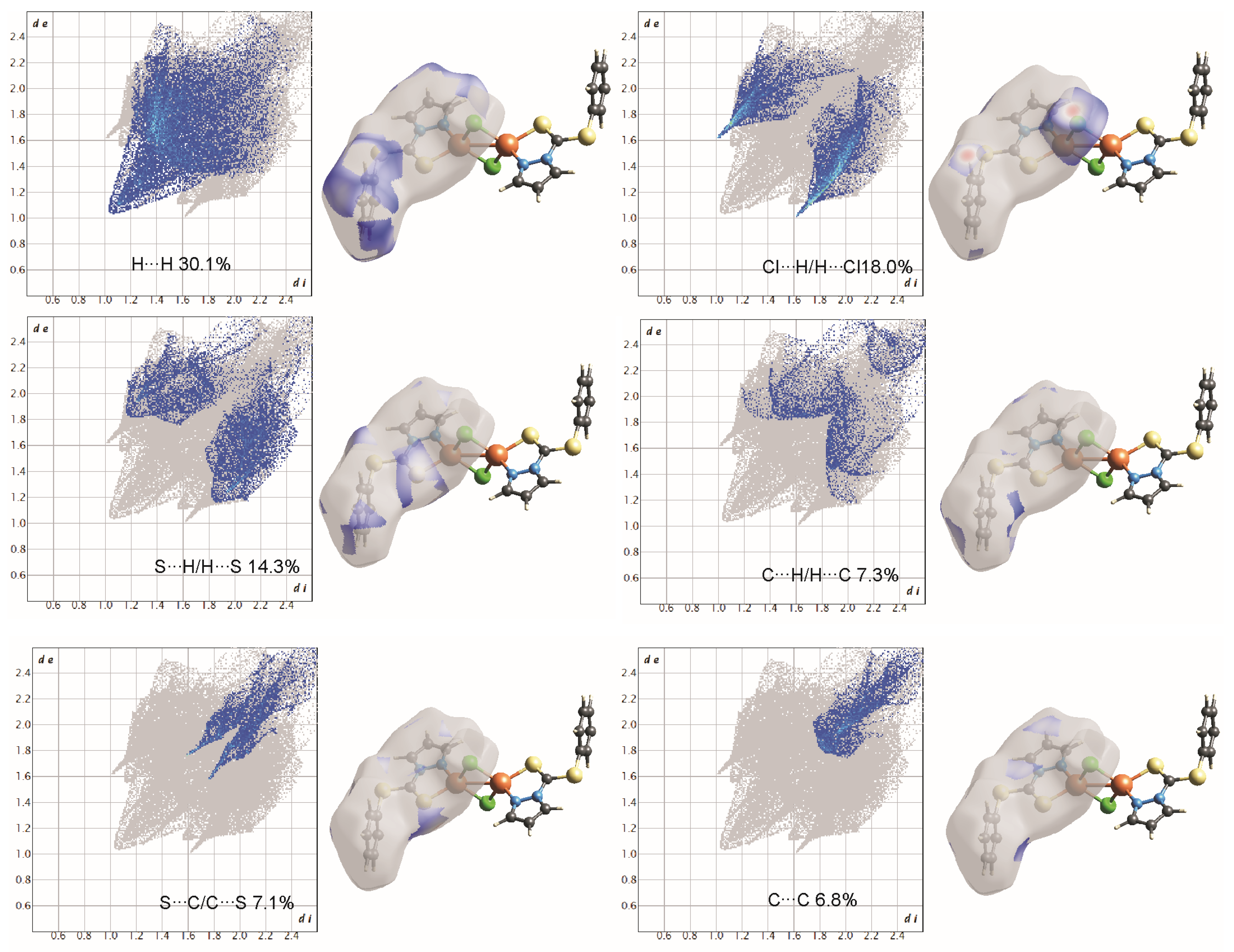
Two-dimensional fingerprint plots of complex 1, all intermolecular contacts are included. The percentage contribution for each contact is specified at the lower part of each subfigure.
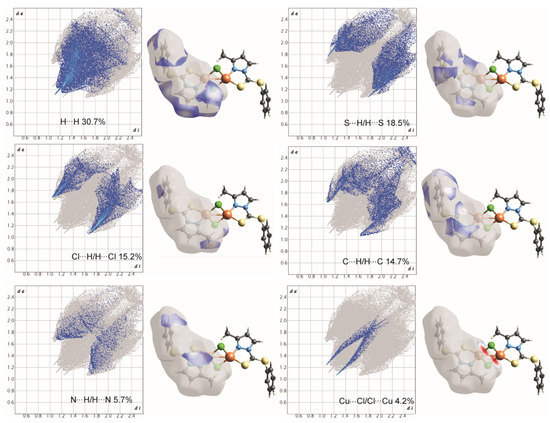
Figure 8.
2D-fingerprint plots for all intermolecular contacts in complex 2. The percentage of contribution for each contact is specified as shown at the bottom of each subfigure.
On the Hirshfeld surfaces, the H⋯H, Cl⋯H/H⋯Cl and S⋯H/H⋯S interactions appear as the largest regions (30.1, 18.01 and 14.3% (1) and 30.7, 15.2 and 18.5% (2), respectively) of the fingerprint plot. Interestingly, the contributions due to Cl⋯H/H⋯Cl and S⋯H/H⋯S interactions have been swapped, as 1 shows stronger Cl⋯H/H⋯Cl hydrogen bonding (2.636 Å), which could be observed as dark red spots and sharp spikes in the fingerprint plots, and could be attributed to the lower bulk of the stabilizing ligand. This might also be the reason for larger C⋯H/H⋯C π interactions in 2 (14.7%) than in 1 (7.3%). On the other hand the S···C/C···S and C…C π interactions are more pronounced in 1 (7.1 and 6.5%, respectively), than in 2 (2.0 and 3.5%, respectively). The other interatomic contacts that make comparatively less contribution to crystal packing are nearly identical, and are due to Cu⋯Cl/Cl⋯Cu, N⋯H/H⋯N and N⋯C/C⋯N and π interactions (4.3, 4.1 and 2.1% (1) and 4.2, 5.7 and 1.9% (2), respectively).
There are few examples of carbodithioate-type ligands containing the N(S)C=S fragment. The ligands have different chemistry, and the CuL2-type complexes have been isolated and structurally characterized in the solid state for a single crystal. The complex in this case was mononuclear Cu(I), while with other metal ions such as Cd and Zn, the chemistry was different [36]. In another report, the Cu(II) ion also affords a mononuclear complex with two chelating ligands, in the same way as in the cases of ligands discussed above [37]. The coordination chemistry of such ligands with CuCl/CuBr works in the same way, but with other metals such as Ni(II), and heavier halogen (I) the behavior is different [38]. An overview of the general reactivity of the carbodithioate ligands indicates that the solvent, reaction conditions and structure of the ligand play a role in determining the geometry of the final complex. Further studies in the field are required to obtain insights into the factors responsible for exact chemistry with the same and different metal ions. The chemistry of these ligands is interesting, and interesting results with other metal ions are very much expected.
4. Conclusions
Two ligands, namely phenyl-1H-pyrazole-1-carbodithioate and phenyl-3-methyl-1H-carbodithioate were synthesized and isolated as viscous oil. They were used for the syntheses of two copper(I) complexes. Complexes containing the Cu(I) ion were isolated in good purity. The binuclear copper(I) complexes bear two bridged-chlorido functions. The ligands act as a bidentate chelate, with N and S as potential donor sites. Each Cu(I) ion in the CuCl2NS fragment of the complex bears distorted-tetrahedral geometry. Structural features such as bond length and angles are in close agreement with structurally analogous complexes. The supramolecular chemistry of the complexes is quite different one from the other, owing to the presence of pyrazole and the methylpyrazol moiety in the ligand. The complex containing the pyrazole moiety affords 2D-supramolecular extension, while the methylpyrazole-containing complex affords the 1D chain. The ligands designed in this study are efficient chelators and useful for designing bimetallic complexes. The Hirshfeld-surface analysis of both the complexes reveals that H⋯H, Cl⋯H/H⋯Cl and S⋯H/H⋯S interactions appear. Complex 1 shows stronger Cl⋯H/H⋯Cl hydrogen bonding. Larger C⋯H/H⋯C π interactions in 2 (14.7%) than in 1 (7.3%) were observed. Other interactions, such as S⋯C/C⋯S and C⋯C π are more pronounced in 1 than in 2. Other interatomic contacts, Cu⋯Cl/Cl⋯Cu, N⋯H/H⋯N and N⋯C/C⋯N and π, contribute comparatively less towards crystal packing. The chemistry herein can be applied to the development of bimetallic complexes, for useful applications in the future.
Supplementary Materials
Crystallographic data for the structural analysis have been deposited with the Cambridge Crystallographic data center, CCDC Nos. 2238715 and 2238716 for 1 and 2, respectively. Copies of this information may be obtained free of charge from the +44(1223)336-033 or Email: deposit@ccdc.cam.ac.uk or http://www.ccdc.cam.ac.uk (accessed on 30 January 2023).
Author Contributions
Conceptualization, methodology, S.A.K. and E.K.; validation, visualization, E.K. and A.N.; formal analysis, investigation, S.A.K.; resources, E.K.; writing—original draft preparation, S.A.K.; writing—review and editing, E.K., A.N. and S.Q.; supervision, project administration, E.K.; funding acquisition, E.K. and A.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. 2716).
Acknowledgments
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. 2716).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abd El-Wahab, H.; Abd El-Fattah, M.; El-Alfy, H.; Owda, M.; Lin, L.; Hamdy, I. Synthesis and characterisation of sulphonamide (Schiff base) ligand and its copper metal complex and their efficiency in polyurethane varnish as flame retardant and antimicrobial surface coating additives. Prog. Org. Coat. 2020, 142, 105577. [Google Scholar] [CrossRef]
- Dominelli, B.; Correia, J.D.G.; Kühn, F.E. Medicinal Applications of Gold(I/III)-Based Complexes Bearing N-Heterocyclic Carbene and Phosphine Ligands. J. Organomet. Chem. 2018, 866, 153–164. [Google Scholar] [CrossRef]
- Wang, S.; Shao, W.; Li, H.; Liu, C.; Wang, K.; Zhang, J. Synthesis, characterization and cytotoxicity of the gold(III) complexes of 4,5-dihydropyrazole-1-carbothioamide derivatives. Eur. J. Med. Chem. 2011, 46, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E. What are heavy metals? Long-standing controversy over the scientific use of the term ‘heavy metals’–proposal of a comprehensive definition. Toxicol. Environ. Chem. 2018, 100, 6–19. [Google Scholar] [CrossRef]
- Gul, Z.; Khan, S.; Khan, E. Organic Molecules Containing N, S and O Heteroatoms as Sensors for the Detection of Hg(II) Ion; Coordination and Efficiency toward Detection. Crit. Rev. Anal. Chem. 2022, 1–22. [Google Scholar] [CrossRef]
- Bhattacharya, P.T.; Misra, S.R.; Hussain, M. Nutritional aspects of essential trace elements in oral health and disease: An extensive review. Scientifica 2016, 2016, 5464373. [Google Scholar] [CrossRef]
- Mohd-Taufek, N.; Cartwright, D.; Davies, M.; Hewavitharana, A.K.; Koorts, P.; Shaw, P.N.; Sumner, R.; Lee, E.; Whitfield, K. The simultaneous analysis of eight essential trace elements in human milk by ICP-MS. Food Anal. Methods 2016, 9, 2068–2075. [Google Scholar] [CrossRef]
- Rathore, H.; Varshney, G.; Mojumdar, S.; Saleh, M. Synthesis, characterization and fungicidal activity of zinc diethyldithiocarbamate and phosphate. J. Therm. Anal. Calorim. 2007, 90, 681–686. [Google Scholar] [CrossRef]
- Balakrishnan, S.; Duraisamy, S.; Kasi, M.; Kandasamy, S.; Sarkar, R.; Kumarasamy, A. Syntheses, physicochemical characterization, antibacterial studies on potassium morpholine dithiocarbamate nickel (II), copper (II) metal complexes and their ligands. Heliyon 2019, 5, e01687. [Google Scholar] [CrossRef]
- Sultana, U.S.; Habib, M.; Amin, M.; Mahiuddin, M.; Zahan, M.; Islam, A. Synthesis, Structural Properties of Hydrazine Carbodithioate Schiff Base and Their Metal Complexes and Evaluation of Their Biological Activity. Egypt. J. Chem. 2020, 63, 3811–3816. [Google Scholar] [CrossRef]
- Egza, T.F.; Mitiku, A.A.; Lemma, T.S.; Eeranna, B.C. A review on pharmacological activity of some selected transition metal complexes with heterocyclic ligands. World J. Pharm. Res. 2020, 9, 100–127. [Google Scholar]
- Maity, D. Biological Applications of Schiff base Metal Complexes-A Review. Int. J. Res. Anal. Rev. 2019, 6, 471–478. [Google Scholar]
- Zangrando, E.; Islam, M.; Islam, M.A.-A.A.; Sheikh, M.; Tarafder, M.; Miyatake, R.; Zahan, R.; Hossain, M. Synthesis, characterization and bio-activity of nickel (II) and copper (II) complexes of a bidentate NS Schiff base of S-benzyl dithiocarbazate. Inorg. Chim. Acta 2015, 427, 278–284. [Google Scholar] [CrossRef]
- Nanjundan, N.; Narayanasamy, R.; Butcher, R.J.; Jasinski, J.P.; Velmurugan, K.; Nandhakumar, R.; Balakumaran, M.D.; Kalaichelvan, P.T.; Gnanasoundari, V.G. Synthesis, crystal structure, biomolecular interactions and anticancer properties of Ni (II), Cu (II) and Zn (II) complexes bearing S-allyldithiocarbazate. Inorg. Chim. Acta 2017, 455, 283–297. [Google Scholar] [CrossRef]
- Nawaz, H.; Abbasi, R.; Nafees, M.; ur Rehman, Z.; Ahmad, I.; Arshad, M.N.; Asiri, A.M.; Waseem, A. Structural chemistry and anticancer activity of new heteroleptic palladium (II) carbodithioates. J. Mol. Struct. 2021, 1225, 129058. [Google Scholar] [CrossRef]
- Zaltariov, M.F.; Hammerstad, M.; Arabshahi, H.J.; Jovanovic, K.; Richter, K.W.; Cazacu, M.; Shova, S.; Balan, M.; Andersen, N.H.; Radulović, S.A. New iminodiacetate–thiosemicarbazone hybrids and their copper (II) complexes are potential ribonucleotide reductase R2 inhibitors with high antiproliferative activity. Inorg. Chem. 2017, 56, 3532–3549. [Google Scholar] [CrossRef]
- Tanaka, N.; Okubo, T.; Anma, H.; Kim, K.H.; Inuzuka, Y.; Maekawa, M.; Kuroda-Sowa, T. Halido-Bridged 1D Mixed-Valence CuI–CuII Coordination Polymers Bearing a Piperidine-1-carbodithioato Ligand: Crystal Structure, Magnetic and Conductive Properties, and Application in Dye-Sensitized Solar Cells. Eur. J. Inorg. Chem. 2013, 2013, 3384–3391. [Google Scholar] [CrossRef]
- Xu, H.; Yu, K.; Su, Z.; Zhou, B.; Wang, C.; Wang, C.; Zhou, B. A 3D K–Cu heterometal–organic coordination polymer with luminescent properties constructed from two kinds of Cu-cyanide complex units and binuclear K oxo-cluster. Inorg. Chem. Commun. 2016, 65, 54–58. [Google Scholar] [CrossRef]
- Khan, S.A. Transition Metal Complexes, Precursors for Catalytic and Biological Applications. Ph.D. Thesis, University of Malakand, Department of Chemistry, Malakand, Pakistan, 2015; pp. 1–120. [Google Scholar]
- Altomare, A.; Burla, M.C.; Camalli, M.; Cascarano, G.L.; Giacovazzo, C.; Guagliardi, A.; Moliterni, A.G.; Polidori, G.; Spagna, R. SIR97: A new tool for crystal structure determination and refinement. J. Appl. Crystallogr. 1999, 32, 115–119. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2007, 64, 112–122. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Bruker; Apex2 and Saint Bruker Axs Inc.: Madison, WI, USA, 2007.
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E. Bioaccumulation of non-essential hazardous heavy metals and metalloids in freshwater fish. Risk to human health. Environ. Chem. Lett. 2018, 16, 903–917. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Ilahi, I. Environmental chemistry and ecotoxicology of hazardous heavy metals: Environmental persistence, toxicity, and bioaccumulation. J. Chem. 2019, 2019, 6730305. [Google Scholar] [CrossRef]
- Khan, E.; Ali Khan, S.; Zahoor, M.; Nawaz Tahir, M.; Noor, A.; Altaf, A.A. Cu(II) coordination polymers stabilized by pyridine-2,6-dicarboxylate anion and pyrazole derivatives through ligand hydrolysis. J. Coord. Chem. 2018, 71, 2658–2673. [Google Scholar] [CrossRef]
- Shahzad, A.; Khan, E.; Said, M.; Khan, G.S.; Syed, M.G.; Noor, A.; Zahoor, M.; Ullah, R.; Bari, A. Complexes of 1, 3-diisobutyl thiourea with copper (I), zinc (II) and mercury (II): Their antioxidant and antibacterial evaluation. Crystals 2021, 11, 989. [Google Scholar] [CrossRef]
- Helton, M.E.; Chen, P.; Paul, P.P.; Tyeklár, Z.; Sommer, R.D.; Zakharov, L.N.; Rheingold, A.L.; Solomon, E.I.; Karlin, K.D. Reaction of elemental sulfur with a copper (I) complex forming a trans-μ-1, 2 end-on disulfide complex: New directions in copper-sulfur chemistry. J. Am. Chem. Soc. 2003, 125, 1160–1161. [Google Scholar] [CrossRef]
- Macías, B.; Villa, M.V.; Chicote, E.; Martin-Velasco, S.; Castiñeiras, A.; Borras, J. Copper complexes with dithiocarbamates derived from natural occurring amino acids. Crystal and molecular structure of [Cu(en)(EtOH)(H2O)3][Cu(dtc-pro)2]]. Polyhedron 2002, 21, 1899–1904. [Google Scholar] [CrossRef]
- Roy, S.; Mitra, P.; Patra, A.K. Cu(II) complexes with square pyramidal (N2S)CuCl2 chromophore: Jahn–Teller distortion and subsequent effect on spectral and structural properties. Inorg. Chim. Acta 2011, 370, 247–253. [Google Scholar] [CrossRef]
- Arslan, H.; Flörke, U.; Külcü, N. The crystal and molecular structure of 1-(2-chloro-benzoyl)-3-p-tolyl-thiourea. Turk. J. Chem. 2004, 28, 673–678. [Google Scholar]
- Craciunescu, D.G.; Furlani, A.; Scarcia, V.; Doadrio, A. Synthesis, Cytostatic, and Antitumor Properties of New Rh(I) Thiazole Complexes. Biol. Trace Elem. Res. 1985, 8, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Karatepe, M.; Karatas, F. Antioxidant, pro-oxidant effect of the thiosemicarbazone derivative Schiff base (4-(1-phenylmethylcyclobutane-3-yl)-2-(2-hydroxybenzylidenehydrazino) thiazole) and its metal complexes on rats. Cell Biochem. Funct. 2006, 24, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Coluccia, M.; Nassi, A.; Loseto, F.; Boccarelli, A.; Mariggio, M.A.; Giordano, D.; Intini, F.P.; Caputo, P.; Natile, G. A trans-platinum complex showing higher antitumor activity than the cis congeners. J. Med. Chem. 1993, 36, 510–512. [Google Scholar] [CrossRef] [PubMed]
- Santra, A.; Mondal, G.; Acharjya, M.; Bera, P.; Panja, A.; Mandal, T.K.; Mitra, P.; Bera, P. Catechol oxidase mimetic activity of copper(I) complexes of 3,5-dimethyl pyrazole derivatives: Coordination behavior, X-ray crystallography and electrochemical study. Polyhedron 2016, 113, 5–15. [Google Scholar] [CrossRef]
- Mondal, G.; Bera, P.; Santra, A.; Jana, S.; Mandal, T.N.; Mondal, A.; Seok, S.I.; Bera, P. Precursor-driven selective synthesis of hexagonal chalcocite (Cu2S) nanocrystals: Structural, optical, electrical and photocatalytic properties. New J. Chem. 2014, 38, 4774–4782. [Google Scholar] [CrossRef]
- Ball, R.J.; Genge, A.R.J.; Radford, A.L.; Skelton, B.W.; Tolhurst, V.-A.; White, A.H. Complexes of late transition metals containing the mixed donor ligands 2,6-(RSCH2)2C5H3N (R = Ph, Me): Crystal structures of [{Cu(2,6-(PhSCH2)2C5H3N)(μ-X)}2] (X = Cl, Br) and [Ni(2,6-(PhSCH2)2C5H3N)Br2]. J. Chem. Soc. Dalton Trans. 2001, 2807–2812. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).