Deep Neural Networks for Analysis of Microscopy Images—Synthetic Data Generation and Adaptive Sampling
Abstract
1. Introduction
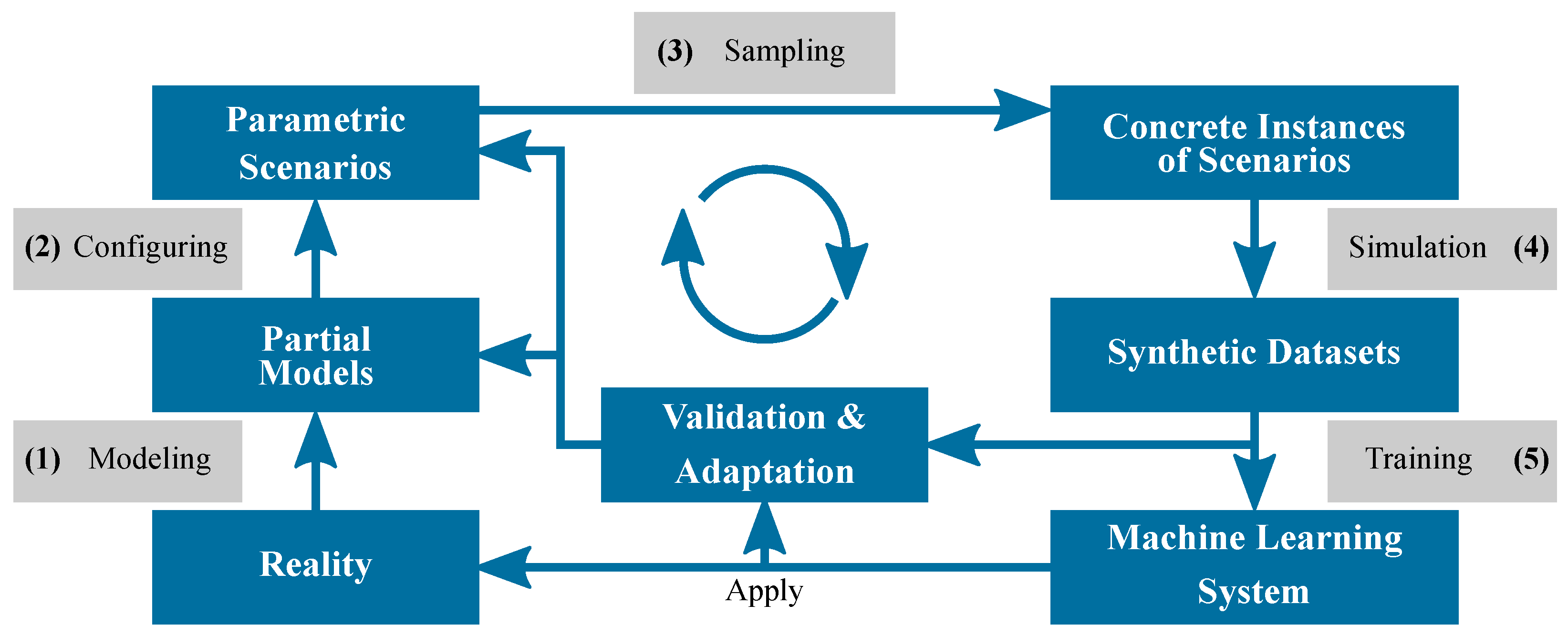
2. Materials and Methods
2.1. Synthetic Training Data
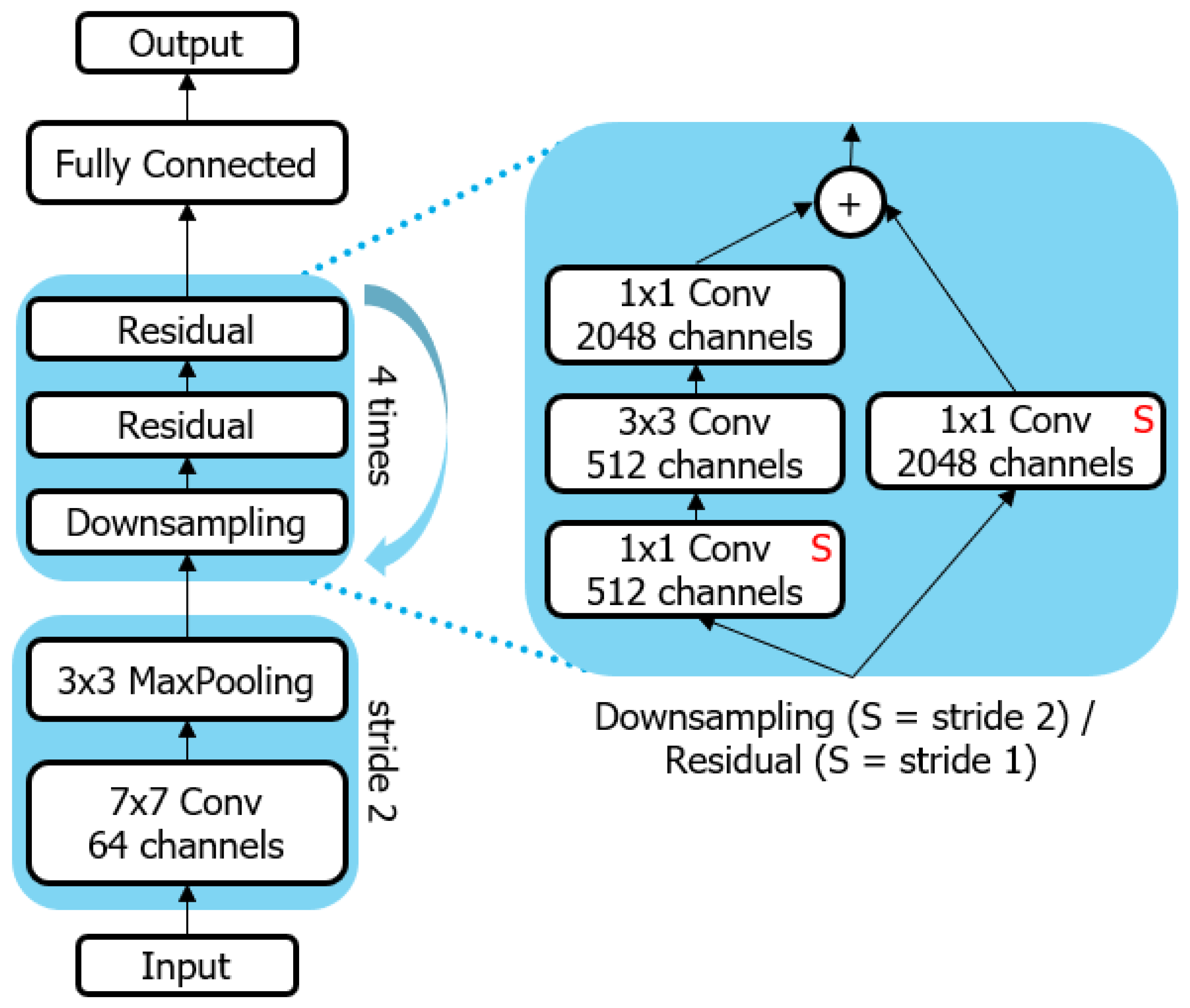
2.2. Machine Learning Setup
3. Results
3.1. Surface Crack Detection during Optical Screening
3.1.1. Synthetic Data Generation for Training
3.1.2. Training the Model
3.1.3. Impact of Synthetic Data
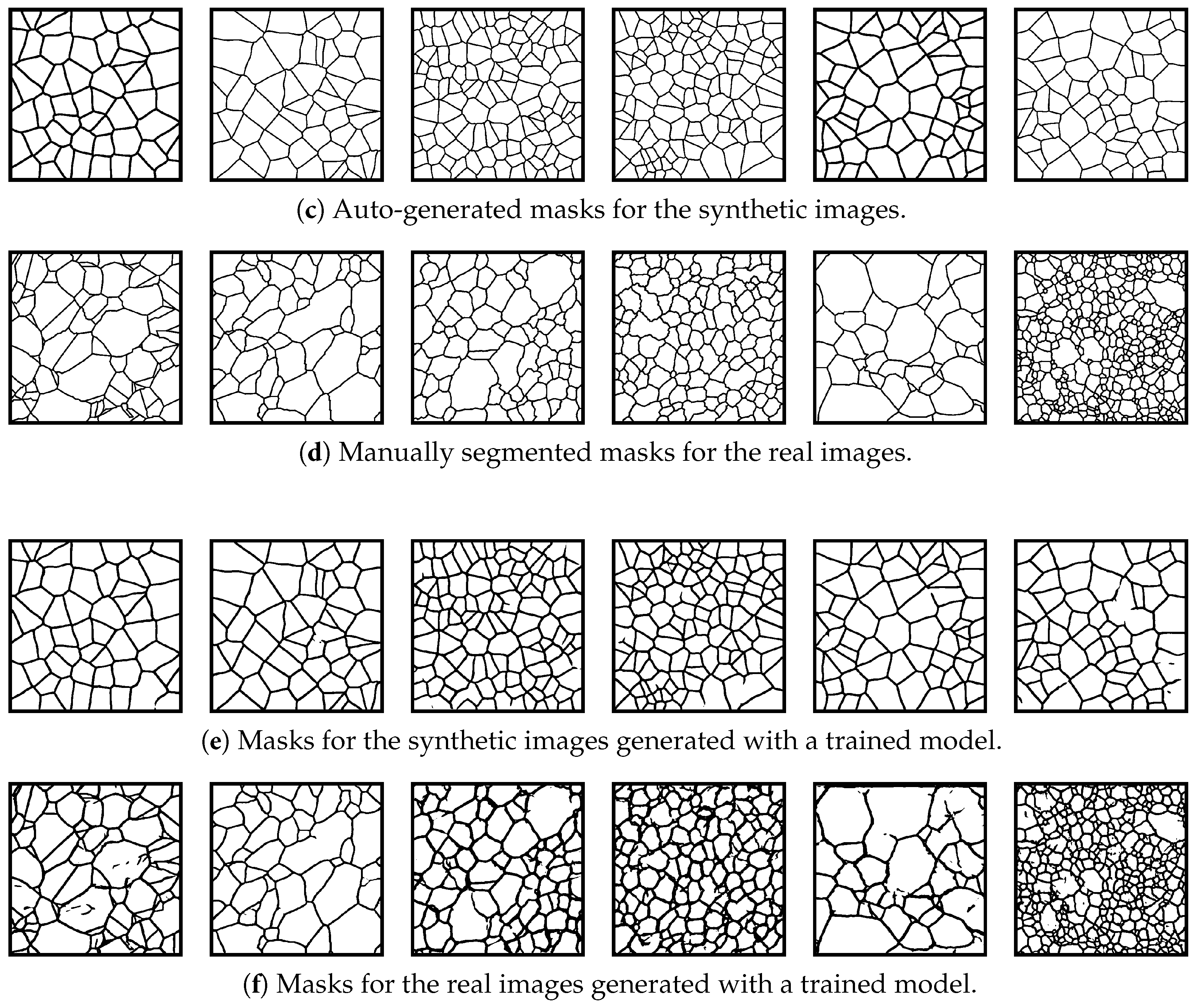
3.2. Grain Boundary Segmentation
3.2.1. Synthetic Data Generation for Training
3.2.2. Training the Model
3.2.3. Evaluation of Synthetic Model
3.3. Adaptation Data Generation and Fine-Tuning
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Roberts, G.; Haile, S.Y.; Sainju, R.; Edwards, D.J.; Hutchinson, B.; Zhu, Y. Deep Learning for Semantic Segmentation of Defects in Advanced STEM Images of Steels. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Masubuchi, S.; Watanabe, E.; Seo, Y.; Okazaki, S.; Sasagawa, T.; Watanabe, K.; Taniguchi, T.; Machida, T. Deep-learning-based image segmentation integrated with optical microscopy for automatically searching for two-dimensional materials. NPJ 2D Mater. App. 2020, 4. [Google Scholar] [CrossRef]
- Dong, X.; Li, H.; Jiang, Z.; Grünleitner, T.; Güler, I.; Dong, J.; Wang, K.; Köhler, M.H.; Jakobi, M.; Menze, B.H.; et al. 3D Deep Learning Enables Accurate Layer Mapping of 2D Materials. ACS Nano 2021. [Google Scholar] [CrossRef]
- Furat, O.; Wang, M.; Neumann, M.; Petrich, L.; Weber, M.; Krill, C.E.; Schmidt, V. Machine Learning Techniques for the Segmentation of Tomographic Image Data of Functional Materials. Front. Mater. 2019, 6, 145. [Google Scholar] [CrossRef]
- Kusche, C.; Reclik, T.; Freund, M.; Al-Samman, T.; Kerzel, U.; Korte-Kerzel, S. Large-area, high-resolution characterisation and classification of damage mechanisms in dual-phase steel using deep learning. PLoS ONE 2019, 14, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Cordts, M.; Omran, M.; Ramos, S.; Rehfeld, T.; Enzweiler, M.; Benenson, R.; Franke, U.; Roth, S.; Schiele, B. The cityscapes dataset for semantic urban scene understanding. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3213–3223. [Google Scholar]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A database and web-based tool for image annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Acuna, D.; Ling, H.; Kar, A.; Fidler, S. Efficient interactive annotation of segmentation datasets with polygon-rnn++. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 859–868. [Google Scholar]
- Xie, J.; Kiefel, M.; Sun, M.T.; Geiger, A. Semantic instance annotation of street scenes by 3d to 2d label transfer. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3688–3697. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Santurkar, S.; Tsipras, D.; Ilyas, A.; Madry, A. How does batch normalization help optimization? arXiv 2018, arXiv:1805.11604. [Google Scholar]
- Krogh, A.; Hertz, J.A. A simple weight decay can improve generalization. Adv. Neural Inf. Process. Syst. 1995, 4, 950–957. [Google Scholar]
- Wong, S.C.; Gatt, A.; Stamatescu, V.; McDonnell, M.D. Understanding data augmentation for classification: When to warp? In Proceedings of the 2016 IEEE International Conference on Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, QLD, Australia, 30 November–2 December 2016; pp. 1–6. [Google Scholar]
- Xu, Y.; Jia, R.; Mou, L.; Li, G.; Chen, Y.; Lu, Y.; Jin, Z. Improved relation classification by deep recurrent neural networks with data augmentation. In Proceedings of the COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers, Osaka, Japan, 11–16 December 2016; pp. 1461–1470. [Google Scholar]
- Vasconcelos, C.N.; Vasconcelos, B.N. Increasing deep learning melanoma classification by classical and expert knowledge based image transforms. CoRR abs/1702.07025 2017, 1. [Google Scholar]
- Richter, S.R.; Vineet, V.; Roth, S.; Koltun, V. Playing for data: Ground truth from computer games. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 102–118. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. arXiv 2018, arXiv:1812.04948. [Google Scholar]
- Shao, S.; Wang, P.; Yan, R. Generative adversarial networks for data augmentation in machine fault diagnosis. Comput. Ind. 2019, 106, 85–93. [Google Scholar] [CrossRef]
- Poibrenski, A.; Sprenger, J.; Müller, C. Toward a Methodology for Training with Synthetic Data on the Example of Pedestrian Detection in a Frame-by-Frame Semantic Segmentation Task. In Proceedings of the 2018 IEEE/ACM 1st International Workshop on Software Engineering for AI in Autonomous Systems (SEFAIAS), Gothenburg, Sweden, 28 May 2018; pp. 31–34. [Google Scholar]
- Tremblay, J.; Prakash, A.; Acuna, D.; Brophy, M.; Jampani, V.; Anil, C.; To, T.; Cameracci, E.; Boochoon, S.; Birchfield, S. Training deep networks with synthetic data: Bridging the reality gap by domain randomization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 969–977. [Google Scholar]
- Wang, M.; Deng, W. Deep visual domain adaptation: A survey. Neurocomputing 2018, 312, 135–153. [Google Scholar] [CrossRef]
- Sun, B.; Saenko, K. Deep coral: Correlation alignment for deep domain adaptation. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 443–450. [Google Scholar]
- Dahmen, T.; Trampert, P.; Boughorbel, F.; Sprenger, J.; Klusch, M.; Fischer, K.; Kübel, C.; Slusallek, P. Digital reality: A model-based approach to supervised learning from synthetic data. AI Perspect. 2019, 1, 2. [Google Scholar] [CrossRef]
- Su, H.; Qi, C.R.; Li, Y.; Guibas, L.J. Render for cnn: Viewpoint estimation in images using cnns trained with rendered 3d model views. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 2686–2694. [Google Scholar]
- Howard, J.; Gugger, S. Fastai: A Layered API for Deep Learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 464–472. [Google Scholar]
- Agresti, A.; Coull, B.A. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 1998, 52, 119–126. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Zabell, S.L. On Student’s 1908 Article “The Probable Error of a Mean”. J. Am. Stat. Assoc. 2008, 103, 1–7. [Google Scholar] [CrossRef]
- Criminisi, A.; Pérez, P.; Toyama, K. Region filling and object removal by exemplar-based image inpainting. IEEE Trans. Image Process. 2004, 13, 1200–1212. [Google Scholar] [CrossRef]
- Trampert, P.; Schlabach, S.; Dahmen, T.; Slusallek, P. Exemplar-Based Inpainting Based on Dictionary Learning for Sparse Scanning Electron Microscopy. Microsc. Microanal. 2018, 24, 700–701. [Google Scholar] [CrossRef]
- Smith, S.L.; Kindermans, P.; Ying, C.; Le, Q.V. Do not Decay the Learning Rate, Increase the Batch Size. In Proceedings of the 6th International Conference on Learning Representations, ICLR 2018, Conference Track Proceedings, Vancouver, BC, Canada, 30 April–3 May 2018; pp. 1–11. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Perlin, K. An image synthesizer. ACM Siggraph Comput. Graph. 1985, 19, 287–296. [Google Scholar] [CrossRef]
- Casey, D. Native-Code and Shader Implementations of Perlin Noise for Python. Available online: https://github.com/caseman/noise (accessed on 3 October 2020).
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive Growing of GANs for Improved Quality, Stability, and Variation. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018; pp. 1–26. [Google Scholar]
- Micrograph 712 by DoITPoMS is Licensed under CC-BY-NC-SA Licence. Available online: https://www.doitpoms.ac.uk/miclib/full_record.php?id=712 (accessed on 5 March 2020).
- TESCAN. Available online: https://www.tescan.com (accessed on 5 March 2020).
- Rheinheimer, W.; Bäurer, M.; Handwerker, C.A.; Blendell, J.E.; Hoffmann, M.J. Growth of single crystalline seeds into polycrystalline strontium titanate: Anisotropy of the mobility, intrinsic drag effects and kinetic shape of grain boundaries. Acta Mater. 2015, 95, 111–123. [Google Scholar] [CrossRef]
- Bhattacharyya, J.; Agnew, S.; Muralidharan, G. Texture enhancement during grain growth of magnesium alloy AZ31B. Acta Mater. 2015, 86, 80–94. [Google Scholar] [CrossRef]


| Synthetic Model | Real Model | Mixed Model | ||
|---|---|---|---|---|
| test data | ||||
| CI | ||||
| all data | ||||
| CI |
| Error | IOU | ||
|---|---|---|---|
| random data | |||
| CI | |||
| adaptive sampling | |||
| CI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trampert, P.; Rubinstein, D.; Boughorbel, F.; Schlinkmann, C.; Luschkova, M.; Slusallek, P.; Dahmen, T.; Sandfeld, S. Deep Neural Networks for Analysis of Microscopy Images—Synthetic Data Generation and Adaptive Sampling. Crystals 2021, 11, 258. https://doi.org/10.3390/cryst11030258
Trampert P, Rubinstein D, Boughorbel F, Schlinkmann C, Luschkova M, Slusallek P, Dahmen T, Sandfeld S. Deep Neural Networks for Analysis of Microscopy Images—Synthetic Data Generation and Adaptive Sampling. Crystals. 2021; 11(3):258. https://doi.org/10.3390/cryst11030258
Chicago/Turabian StyleTrampert, Patrick, Dmitri Rubinstein, Faysal Boughorbel, Christian Schlinkmann, Maria Luschkova, Philipp Slusallek, Tim Dahmen, and Stefan Sandfeld. 2021. "Deep Neural Networks for Analysis of Microscopy Images—Synthetic Data Generation and Adaptive Sampling" Crystals 11, no. 3: 258. https://doi.org/10.3390/cryst11030258
APA StyleTrampert, P., Rubinstein, D., Boughorbel, F., Schlinkmann, C., Luschkova, M., Slusallek, P., Dahmen, T., & Sandfeld, S. (2021). Deep Neural Networks for Analysis of Microscopy Images—Synthetic Data Generation and Adaptive Sampling. Crystals, 11(3), 258. https://doi.org/10.3390/cryst11030258






