Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. FT-SSX Data Collection
2.3. Structure Determination
2.4. Accession Code
3. Results
3.1. FT-SSX Experiment
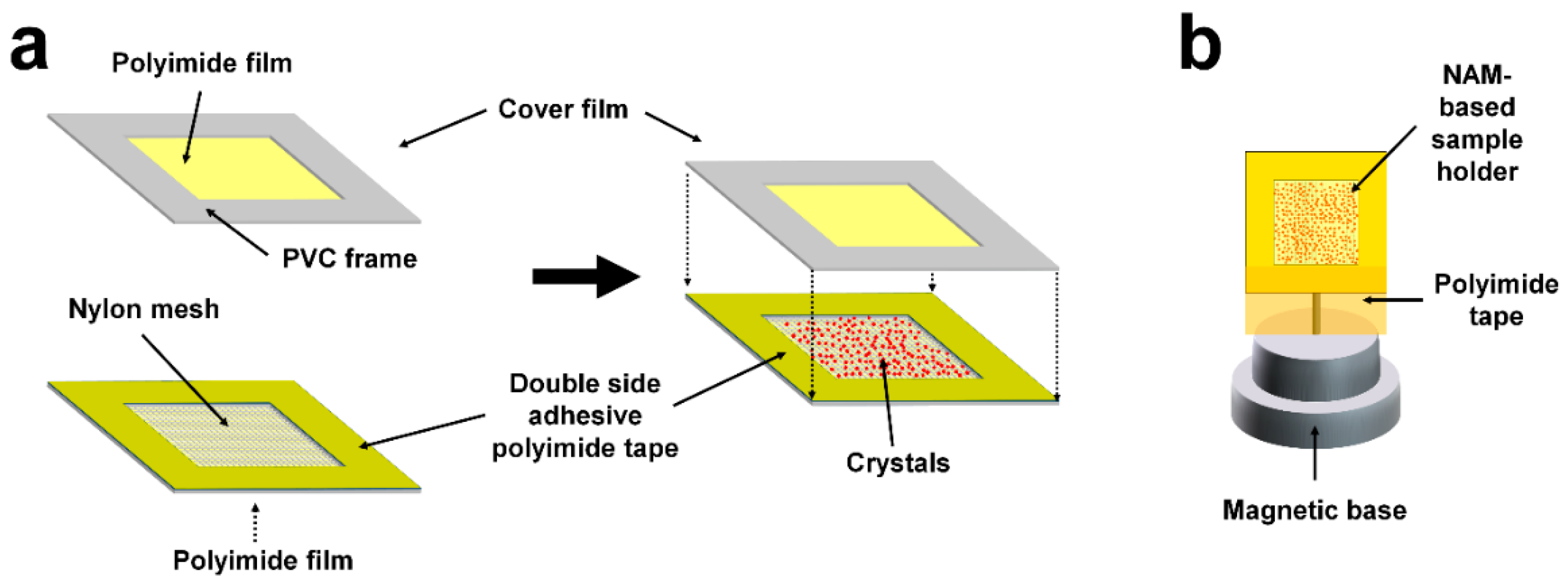
3.2. Preparation of NAM-Based Sample Holder
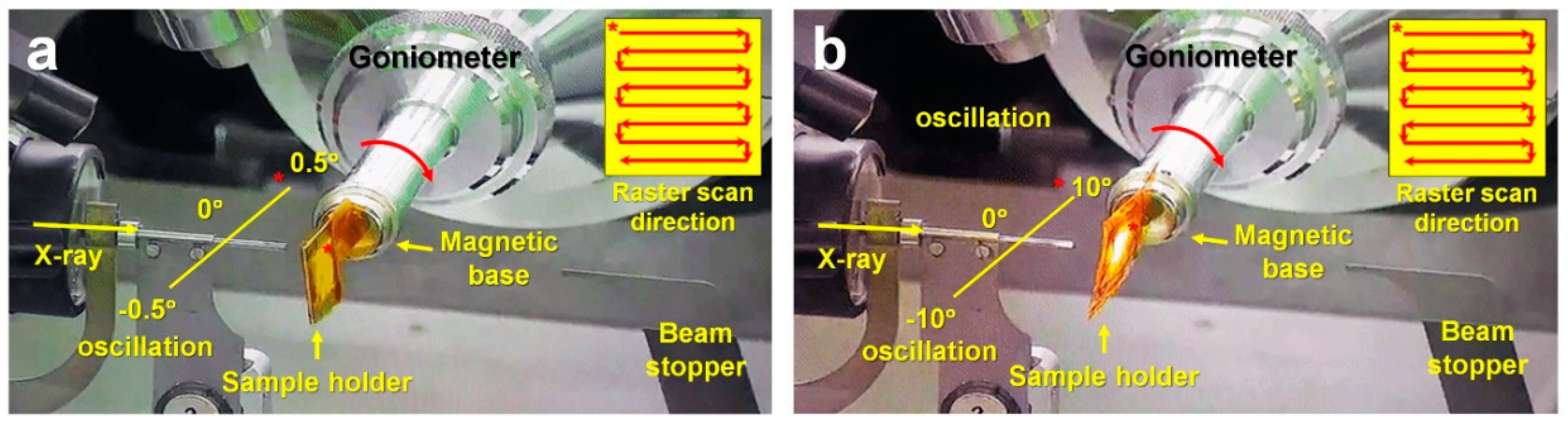
3.3. Data Collection by FT-SSX
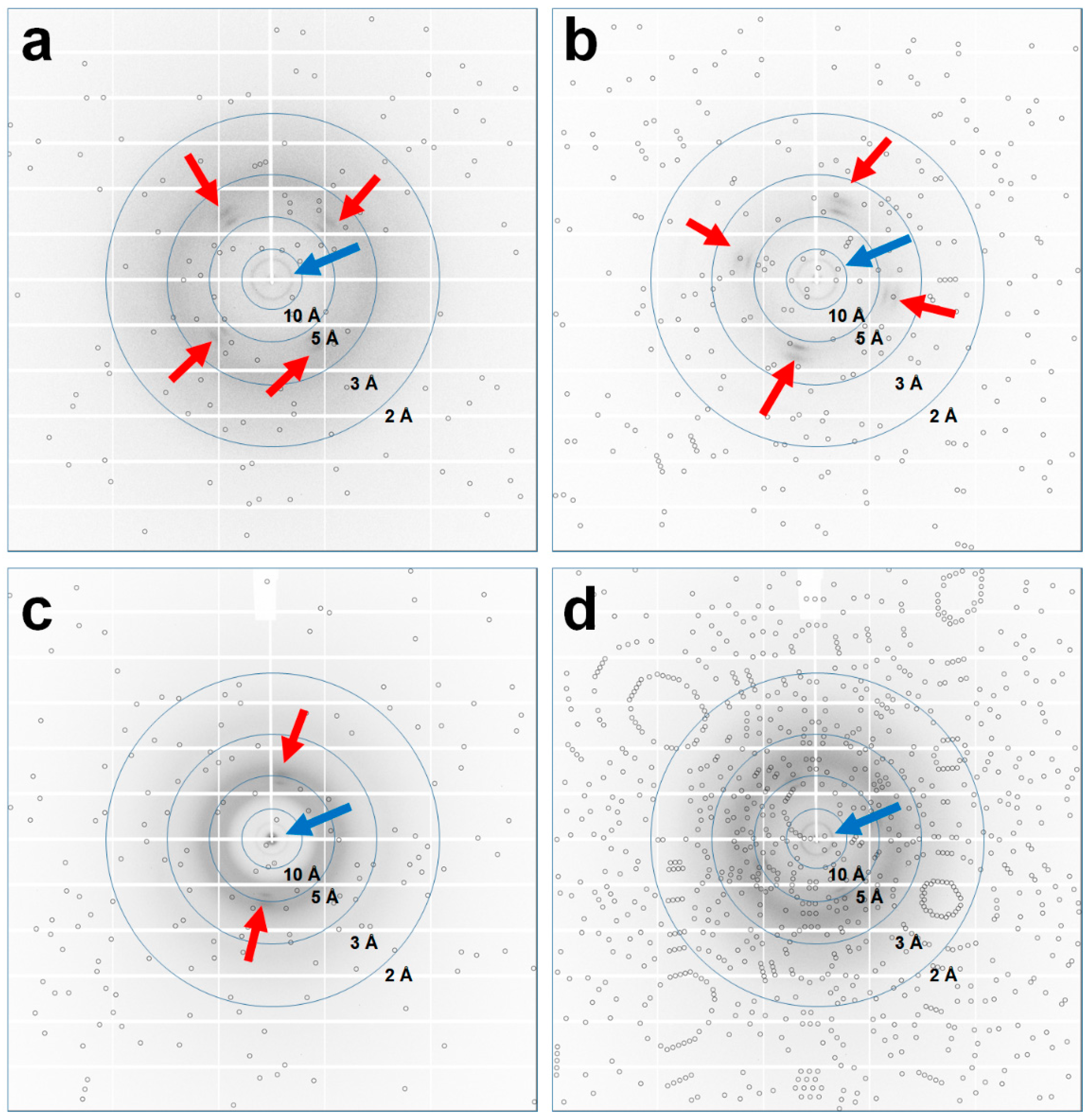
3.3.1. Data Collection from 100 ms X-ray Exposure with 0.01° Oscillation
3.3.2. Data Collection from 500 ms X-ray Exposure with 0.22° Oscillation
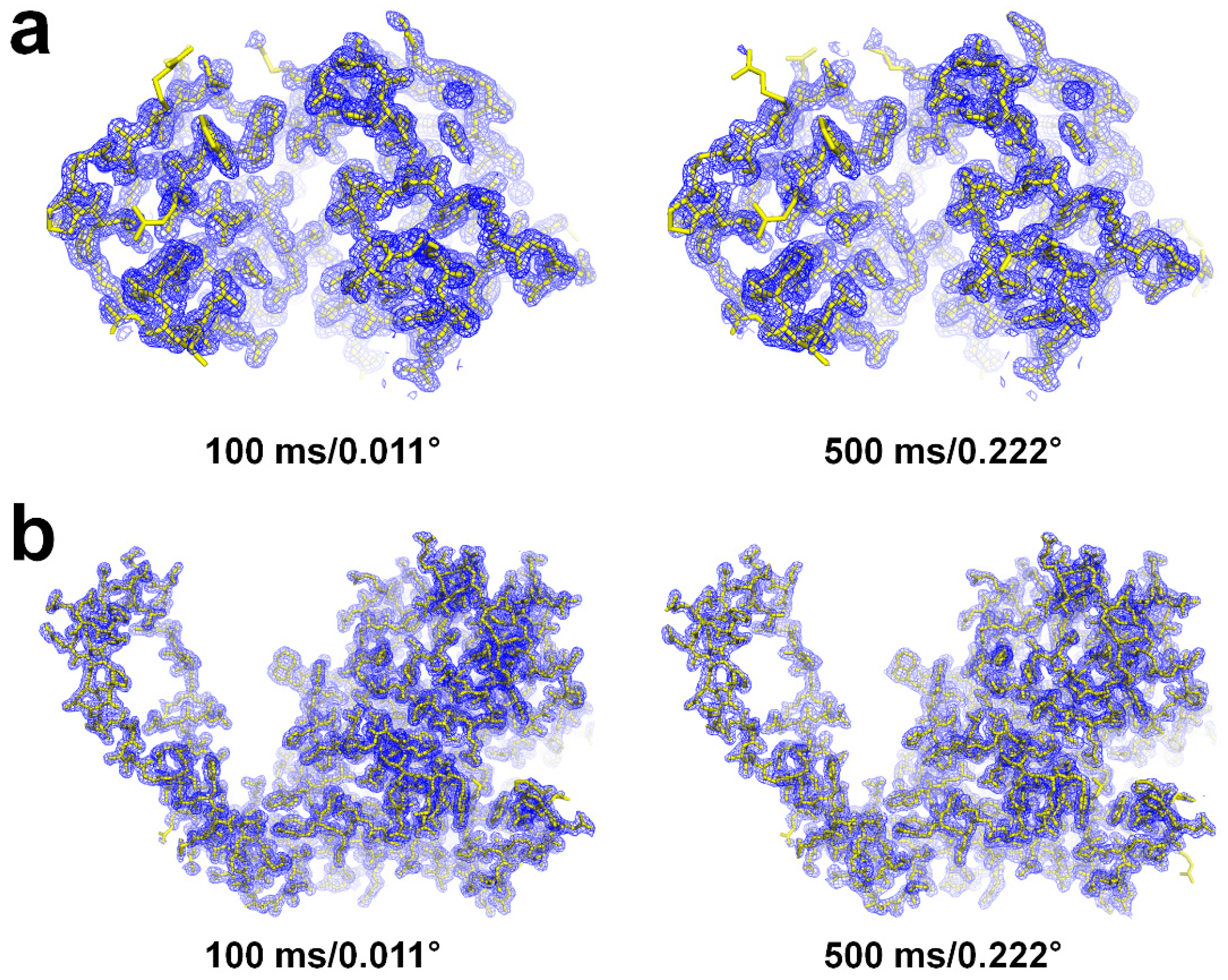
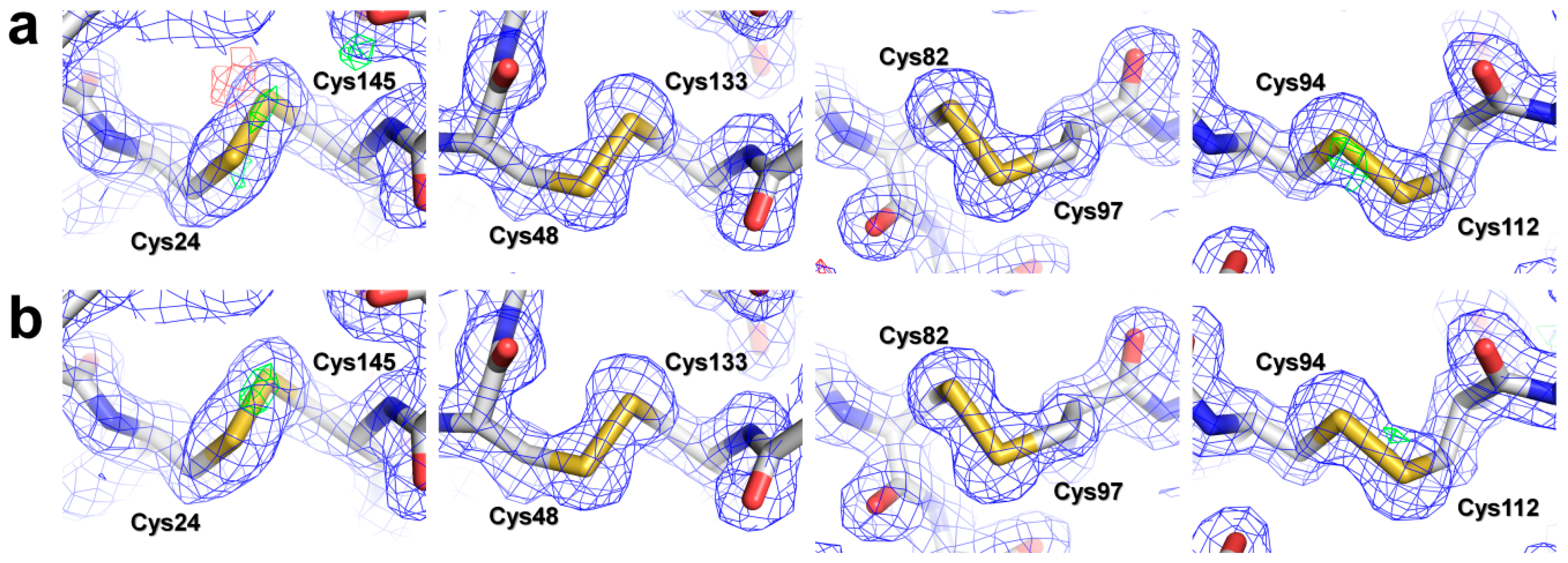
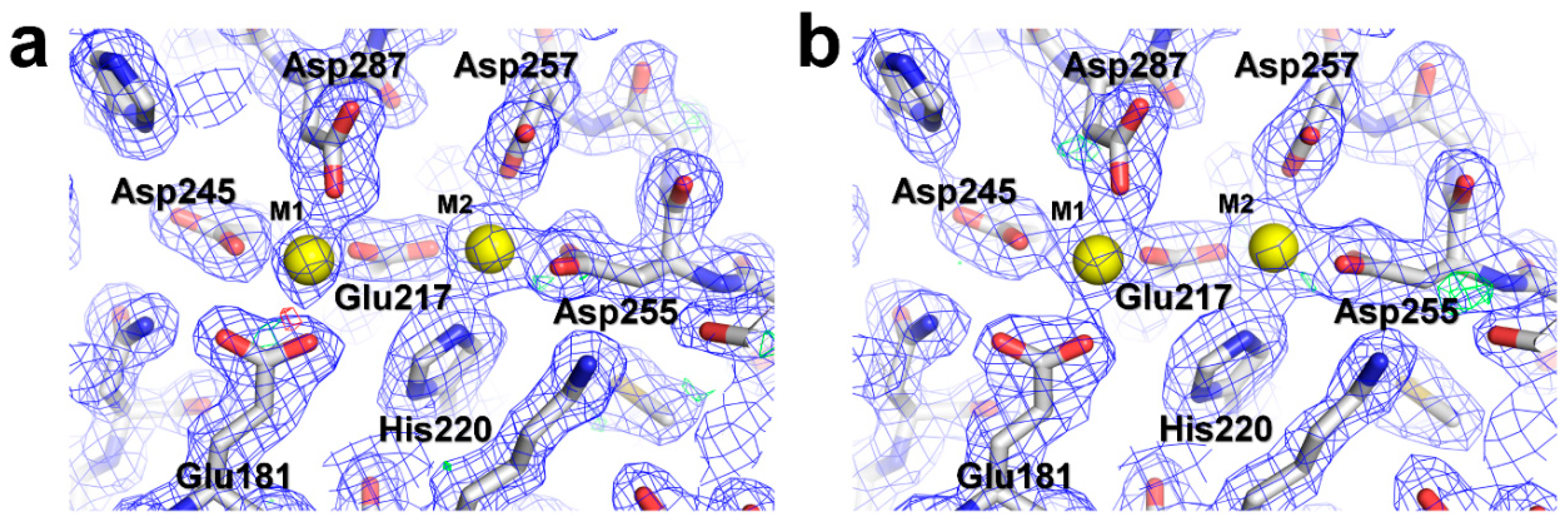
3.4. Structural Analysis of Lysozyme and Glucose Isomerase
3.5. Observation of Sample Holder after FT-SSX
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smyth, M.S. x Ray crystallography. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef]
- Müller, H.; Becker, A.K.; Palm, G.J.; Berndt, L.; Badenhorst, C.P.S.; Godehard, S.P.; Reisky, L.; Lammers, M.; Bornscheuer, U.T. Sequence-Based Prediction of Promiscuous Acyltransferase Activity in Hydrolases. Angew. Chem. Int. Ed. 2020, 59, 11607–11612. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2020, 59, 2–34. [Google Scholar] [CrossRef] [PubMed]
- Meents, A.; Gutmann, S.; Wagner, A.; Schulze-Briese, C. Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Owen, R.L.; Rudino-Pinera, E.; Garman, E.F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. USA 2006, 103, 4912–4917. [Google Scholar] [CrossRef] [PubMed]
- Weik, M.; Ravelli, R.B.G.; Kryger, G.; McSweeney, S.; Raves, M.L.; Harel, M.; Gros, P.; Silman, I.; Kroon, J.; Sussman, J.L. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. USA 2000, 97, 623–628. [Google Scholar] [CrossRef]
- Meents, A.; Dittrich, B.; Gutmann, S. A new aspect of specific radiation damage: Hydrogen abstraction from organic molecules. J. Synchrotron Radiat. 2009, 16, 183–190. [Google Scholar] [CrossRef]
- Stellato, F.; Oberthur, D.; Liang, M.; Bean, R.; Gati, C.; Yefanov, O.; Barty, A.; Burkhardt, A.; Fischer, P.; Galli, L.; et al. Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ 2014, 1, 204–212. [Google Scholar] [CrossRef]
- Weinert, T.; Olieric, N.; Cheng, R.; Brunle, S.; James, D.; Ozerov, D.; Gashi, D.; Vera, L.; Marsh, M.; Jaeger, K.; et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017, 8, 542. [Google Scholar] [CrossRef]
- Schlichting, I. Serial femtosecond crystallography: The first five years. IUCrJ 2015, 2, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Olmos, J.L.; Pandey, S.; Martin-Garcia, J.M.; Calvey, G.; Katz, A.; Knoska, J.; Kupitz, C.; Hunter, M.S.; Liang, M.; Oberthuer, D.; et al. Enzyme intermediates captured “on the fly” by mix-and-inject serial crystallography. BMC Biol. 2018, 16, 59. [Google Scholar] [CrossRef] [PubMed]
- Tenboer, J.; Basu, S.; Zatsepin, N.; Pande, K.; Milathianaki, D.; Frank, M.; Hunter, M.; Boutet, S.; Williams, G.J.; Koglin, J.E.; et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science 2014, 346, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, P.; Schulz, E.C.; Agthe, M.; Horrell, S.; Bourenkov, G.; von Stetten, D.; Leimkohl, J.-P.; Schikora, H.; Schneider, T.R.; Pearson, A.R.; et al. Liquid application method for time-resolved analyses by serial synchrotron crystallography. Nat. Methods 2019, 16, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M. Reaction Initiation in Enzyme Crystals by Diffusion of Substrate. Crystals 2020, 10, 116. [Google Scholar] [CrossRef]
- Fraser, J.S.; van den Bedem, H.; Samelson, A.J.; Lang, P.T.; Holton, J.M.; Echols, N.; Alber, T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA 2011, 108, 16247–16252. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.Y.; Park, J.; Kim, S.; Kim, S.; Rah, S.; Lim, J.; Nam, K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick-Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018, 25, 289–292. [Google Scholar] [CrossRef]
- Kang, H.S.; Min, C.K.; Heo, H.; Kim, C.; Yang, H.; Kim, G.; Nam, I.; Baek, S.Y.; Choi, H.J.; Mun, G.; et al. Hard X-ray free-electron laser with femtosecond-scale timing jitter. Nat. Photonics 2017, 11, 708–713. [Google Scholar] [CrossRef]
- Gati, C.; Bourenkov, G.; Klinge, M.; Rehders, D.; Stellato, F.; Oberthur, D.; Yefanov, O.; Sommer, B.P.; Mogk, S.; Duszenko, M.; et al. Serial crystallography on in vivo grown microcrystals using synchrotron radiation. IUCrJ 2014, 1, 87–94. [Google Scholar] [CrossRef]
- Botha, S.; Nass, K.; Barends, T.R.; Kabsch, W.; Latz, B.; Dworkowski, F.; Foucar, L.; Panepucci, E.; Wang, M.; Shoeman, R.L.; et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta Crystallogr. Sect. D Boil. Crystallogr. 2015, 71 Pt 2, 387–397. [Google Scholar] [CrossRef]
- Owen, R.L.; Axford, D.; Sherrell, D.A.; Kuo, A.; Ernst, O.P.; Schulz, E.C.; Miller, R.J.D.; Mueller-Werkmeister, H.M. Low-dose fixed-target serial synchrotron crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.; Skopintsev, P.; James, D.; Dworkowski, F.; Panepucci, E.; Kekilli, D.; Furrer, A.; Brunle, S.; Mous, S.; Ozerov, D.; et al. Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography. Science 2019, 365, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Martiel, I.; Muller-Werkmeister, H.M.; Cohen, A.E. Strategies for sample delivery for femtosecond crystallography. Acta Crystallogr. Sect. D Boil. Crystallogr. 2019, 75, 160–177. [Google Scholar] [CrossRef] [PubMed]
- DePonte, D.P.; Weierstall, U.; Schmidt, K.; Warner, J.; Starodub, D.; Spence, J.C.H.; Doak, R.B. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D Appl. Phys. 2008, 41, 195505. [Google Scholar] [CrossRef]
- Park, S.Y.; Nam, K.H. Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography. J. Synchrotron Radiat. 2019, 26, 1815–1819. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, M.; Mizohata, E.; Nango, E.; Suzuki, M.; Tanaka, T.; Masudala, T.; Tanaka, R.; Shimamura, T.; Tanaka, Y.; Suno, C.; et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods 2015, 12, 61–63. [Google Scholar] [CrossRef]
- Weierstall, U.; James, D.; Wang, C.; White, T.A.; Wang, D.J.; Liu, W.; Spence, J.C.H.; Doak, R.B.; Nelson, G.; Fromme, P.; et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014, 5, 3309. [Google Scholar] [CrossRef]
- Conrad, C.E.; Basu, S.; James, D.; Wang, D.; Schaffer, A.; Roy-Chowdhury, S.; Zatsepin, N.A.; Aquila, A.; Coe, J.; Gati, C.; et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ 2015, 2, 421–430. [Google Scholar] [CrossRef]
- Park, J.; Park, S.; Kim, J.; Park, G.; Cho, Y.; Nam, K.H. Polyacrylamide injection matrix for serial femtosecond crystallography. Sci. Rep. 2019, 9, 2525. [Google Scholar] [CrossRef]
- Nam, K.H. Sample Delivery Media for Serial Crystallography. Int. J. Mol. Sci. 2019, 20, 1094. [Google Scholar] [CrossRef]
- Nam, K.H. Shortening injection matrix for serial crystallography. Sci. Rep. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Polysaccharide-Based Injection Matrix for Serial Crystallography. Int. J. Mol. Sci. 2020, 21, 3332. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Lard Injection Matrix for Serial Crystallography. Int. J. Mol. Sci. 2020, 21, 5977. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Stable sample delivery in viscous media via a capillary for serial crystallography. J. Appl. Crystallogr. 2020, 53, 45–50. [Google Scholar] [CrossRef]
- Hunter, M.S.; Segelke, B.; Messerschmidt, M.; Williams, G.J.; Zatsepin, N.A.; Barty, A.; Benner, W.H.; Carlson, D.B.; Coleman, M.; Graf, A.; et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2014, 4, 6026. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Baek, S.; Park, J.; Lee, K.; Kim, J.; Lee, S.J.; Chung, W.K.; Lee, J.L.; Cho, Y.; Nam, K.H. Nylon mesh-based sample holder for fixed-target serial femtosecond crystallography. Sci. Rep. 2019, 9, 6971. [Google Scholar] [CrossRef]
- Lee, K.; Lee, D.; Baek, S.; Park, J.; Lee, S.J.; Park, S.; Chung, W.K.; Lee, J.-L.; Cho, H.-S.; Cho, Y.; et al. Viscous-medium-based crystal support in a sample holder for fixed-target serial femtosecond crystallography. J. Appl. Crystallogr. 2020, 53, 1051–1059. [Google Scholar] [CrossRef]
- Lee, D.; Park, S.; Lee, K.; Kim, J.; Park, G.; Nam, K.H.; Baek, S.; Chung, W.K.; Lee, J.-L.; Cho, Y.; et al. Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography. J. Appl. Crystallogr. 2020, 53, 477–485. [Google Scholar] [CrossRef]
- Roedig, P.; Ginn, H.M.; Pakendorf, T.; Sutton, G.; Harlos, K.; Walter, T.S.; Meyer, J.; Fischer, P.; Duman, R.; Vartiainen, I.; et al. High-speed fixed-target serial virus crystallography. Nat. Methods 2017, 14, 805–810. [Google Scholar] [CrossRef]
- Monteiro, D.C.F.; Vakili, M.; Harich, J.; Sztucki, M.; Meier, S.M.; Horrell, S.; Josts, I.; Trebbin, M. A microfluidic flow-focusing device for low sample consumption serial synchrotron crystallography experiments in liquid flow. J. Synchrotron Radiat. 2019, 26, 406–412. [Google Scholar] [CrossRef]
- Calvey, G.D.; Katz, A.M.; Pollack, L. Microfluidic Mixing Injector Holder Enables Routine Structural Enzymology Measurements with Mix-and-Inject Serial Crystallography Using X-ray Free Electron Lasers. Anal. Chem. 2019, 91, 7139–7144. [Google Scholar] [CrossRef] [PubMed]
- Beyerlein, K.R.; Dierksmeyer, D.; Mariani, V.; Kuhn, M.; Sarrou, I.; Ottaviano, A.; Awel, S.; Knoska, J.; Fuglerud, S.; Jönsson, O.; et al. Mix-and-diffuse serial synchrotron crystallography. IUCrJ 2017, 4, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Fuller, F.D.; Gul, S.; Chatterjee, R.; Burgie, E.S.; Young, I.D.; Lebrette, H.; Srinivas, V.; Brewster, A.S.; Michels-Clark, T.; Clinger, J.A.; et al. Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers. Nat. Methods 2017, 14, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Wierman, J.L.; Paré-Labrosse, O.; Sarracini, A.; Besaw, J.E.; Cook, M.J.; Oghbaey, S.; Daoud, H.; Mehrabi, P.; Kriksunov, I.; Kuo, A.; et al. Fixed-target serial oscillation crystallography at room temperature. IUCrJ 2019, 6, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Murray, T.D.; Lyubimov, A.Y.; Ogata, C.M.; Vo, H.; Uervirojnangkoorn, M.; Brunger, A.T.; Berger, J.M. A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions. Acta Crystallogr. Sect. D Boil. Crystallogr. 2015, 71, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Sui, S.; Wang, Y.; Kolewe, K.W.; Srajer, V.; Henning, R.; Schiffman, J.D.; Dimitrakopoulos, C.; Perry, S.L. Graphene-based microfluidics for serial crystallography. Lab Chip 2016, 16, 3082–3096. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanesh, N.S.P.; Reibenspies, J.H. A novel approach to micro-sample X-ray powder diffraction using nylon loops. J. Appl. Crystallogr. 2003, 36, 1480–1481. [Google Scholar] [CrossRef]
- Thorne, R.E.; Stum, Z.; Kmetko, J.; O‘Neill, K.; Gillilan, R. Microfabricated mounts for high-throughput macromolecular cryocrystallography. J. Appl. Crystallogr. 2003, 36, 1455–1460. [Google Scholar] [CrossRef]
- Park, S.Y.; Ha, S.C.; Kim, Y.G. The Protein Crystallography Beamlines at the Pohang Light Source II. Biodesign 2017, 5, 30–34. [Google Scholar]
- Barty, A.; Kirian, R.A.; Maia, F.R.; Hantke, M.; Yoon, C.H.; White, T.A.; Chapman, H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014, 47, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- White, T.A.; Mariani, V.; Brehm, W.; Yefanov, O.; Barty, A.; Beyerlein, K.R.; Chervinskii, F.; Galli, L.; Gati, C.; Nakane, T.; et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 2016, 49, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Boil. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode. Biochem. Biophys. Res. Commun. 2017, 493, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC. Available online: https://pymol.org/2/ (accessed on 9 September 2020).
- Bae, J.E.; Hwang, K.Y.; Nam, K.H. Structural analysis of substrate recognition by glucose isomerase in Mn2+ binding mode at M2 site in S. rubiginosus. Biochem. Biophys. Res. Commun. 2018, 503, 770–775. [Google Scholar] [CrossRef]

| Approach I | Approach II | |||
|---|---|---|---|---|
| Data collection | Lysozyme | GI | Lysozyme | GI |
| Energy (ev) | 12,659 | 12,659 | 12,659 | 12,659 |
| Exposure time (ms) | 100 | 100 | 500 | 500 |
| Oscillation (°) | 0.01 | 0.01 | 0.22 | 0.22 |
| Space group | P43212 | I222 | P43212 | I222 |
| Cell dimension (Å) | ||||
| a | 79.45 | 94.44 | 79.45 | 94.55 |
| b | 79.45 | 100.24 | 79.45 | 100.26 |
| c | 38.45 | 130.52 | 38.47 | 103.61 |
| Collected images | 56,700 | 48,600 | 8100 | 8100 |
| Hits images | 41,916 | 25,099 | 7628 | 2576 |
| Indexed images | 21,670 | 13,727 | 2737 | 1245 |
| Indexed pattern | 29,735 | 17,100 | 3476 | 1565 |
| Resolution (Å) | 80.0–1.50 (1.55–1.50) | 72.46–1.70 (1.76–1.70) | 80.0–1.60 (1.66–1.60) | 72.46–2.00 (2.07–2.00) |
| Unique reflections | 20,320 (1969) | 54,250 (5361) | 16,824 (1627) | 33,589 (3282) |
| Completeness (%) | 100.00 (100.00) | 100.00 (100.00) | 100.00 (100.00) | 100.00 (100.00) |
| Redundancy | 427.0 (296.0) | 241.2 (167.2) | 63.5 (43.9) | 30.8 (21.5) |
| SNR | 7.94 (2.24) | 3.36 (1.66) | 4.31 (1.72) | 2.05 (1.52) |
| CC | 0.9877 (0.5840) | 0.9246 (0.6616) | 0.9515 (0.5680) | 0.7069 (0.5605) |
| CC* | 0.9969 (0.8587) | 0.9802 (0.8924) | 0.9875 (0.8512) | 0.9101 (0.8475) |
| Rsplit (%) a | 8.97 (50.97) | 24.09 (57.80) | 18.26 (66.37) | 46.47 (66.92) |
| Wilson B factor (Å2) | 17.7 | 13.7 | 18.7 | 15.9 |
| Refinement | ||||
| Resolution (Å) | 56.18–1.50 | 72.01–1.70 | 56.18–1.60 | 72.05–2.00 |
| Rwork/Rwork b | 16.66/18.53 | 18.15/19.94 | 18.87/21.54 | 21.36/24.81 |
| R.m.s. deviations | ||||
| Bond length (Å) | 0.014 | 0.008 | 0.014 | 0.07 |
| Bond angle (°) | 1.703 | 0.959 | 2.386 | 0.993 |
| B factors (Å2) | ||||
| Protein | 25.95 | 18.35 | 27.87 | 18.32 |
| Metals | 25.78 | 15.69 | 27.92 | 12.89 |
| Water | 34.33 | 31.92 | 35.37 | 28.27 |
| Ramachandran (%) | ||||
| Favored | 99.21 | 96.61 | 99.21 | 96.61 |
| Allowed | 0.79 | 3.13 | 0.79 | 3.13 |
| Outliers | 0.00 | 0.26 | 0.00 | 0.26 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-Y.; Choi, H.; Eo, C.; Cho, Y.; Nam, K.H. Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder. Crystals 2020, 10, 803. https://doi.org/10.3390/cryst10090803
Park S-Y, Choi H, Eo C, Cho Y, Nam KH. Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder. Crystals. 2020; 10(9):803. https://doi.org/10.3390/cryst10090803
Chicago/Turabian StylePark, Suk-Youl, Hyeongju Choi, Cheolsoo Eo, Yunje Cho, and Ki Hyun Nam. 2020. "Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder" Crystals 10, no. 9: 803. https://doi.org/10.3390/cryst10090803
APA StylePark, S.-Y., Choi, H., Eo, C., Cho, Y., & Nam, K. H. (2020). Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder. Crystals, 10(9), 803. https://doi.org/10.3390/cryst10090803






