Inclusion Complexes of Naringenin in Dimethylated and Permethylated β-Cyclodextrins: Crystal Structures and Molecular Dynamics Studies
Abstract
1. Introduction
2. Results
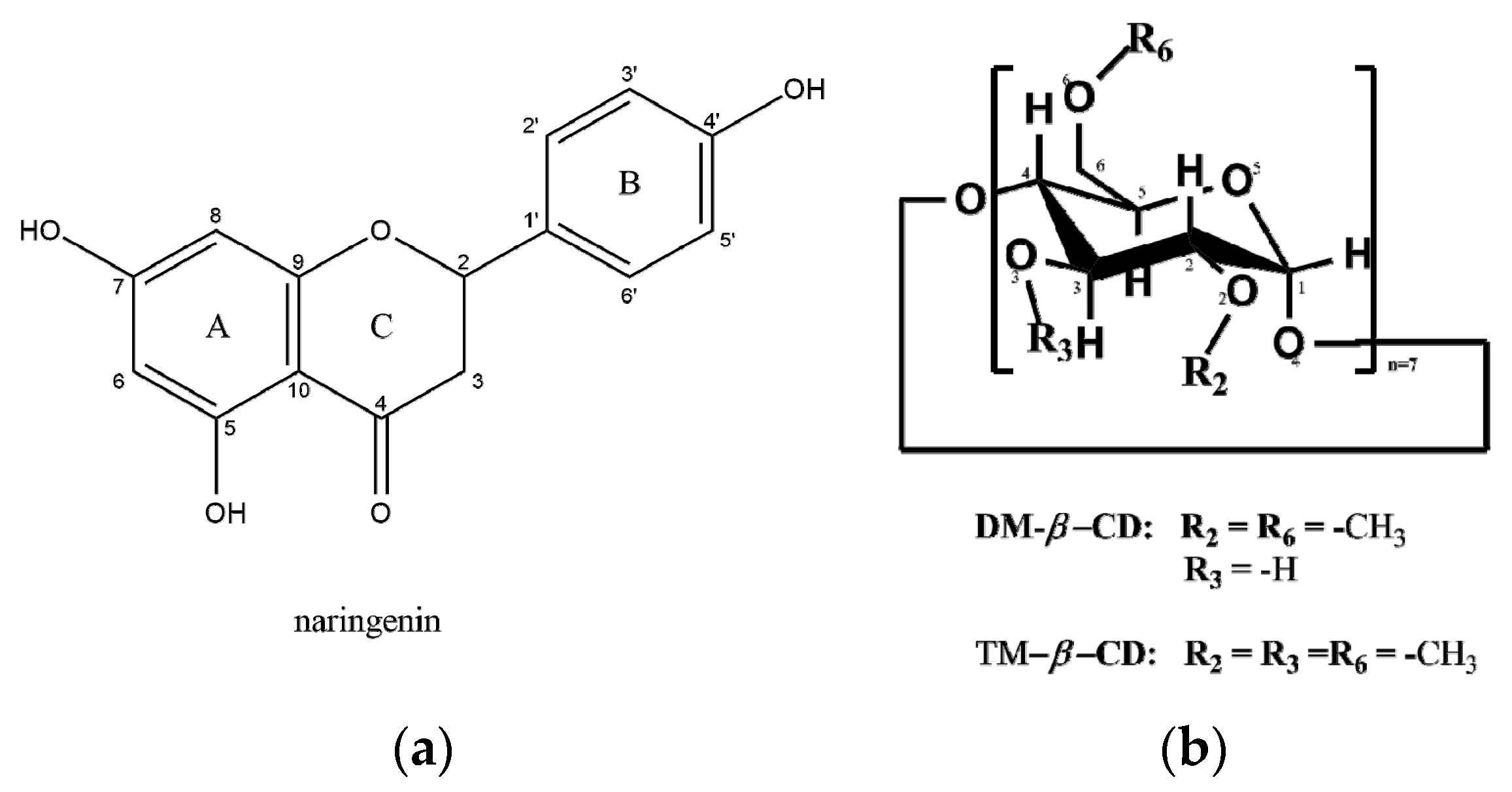
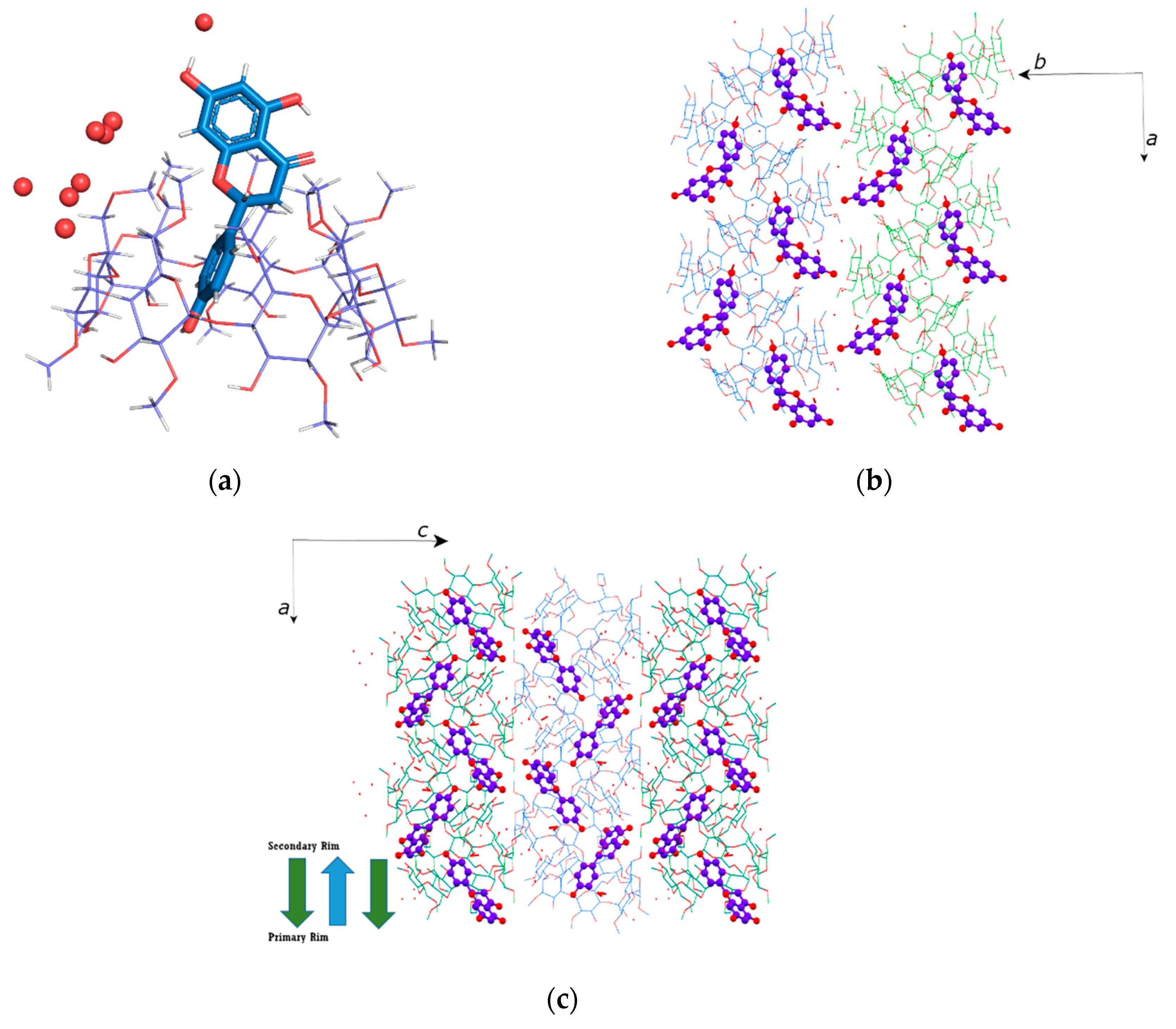
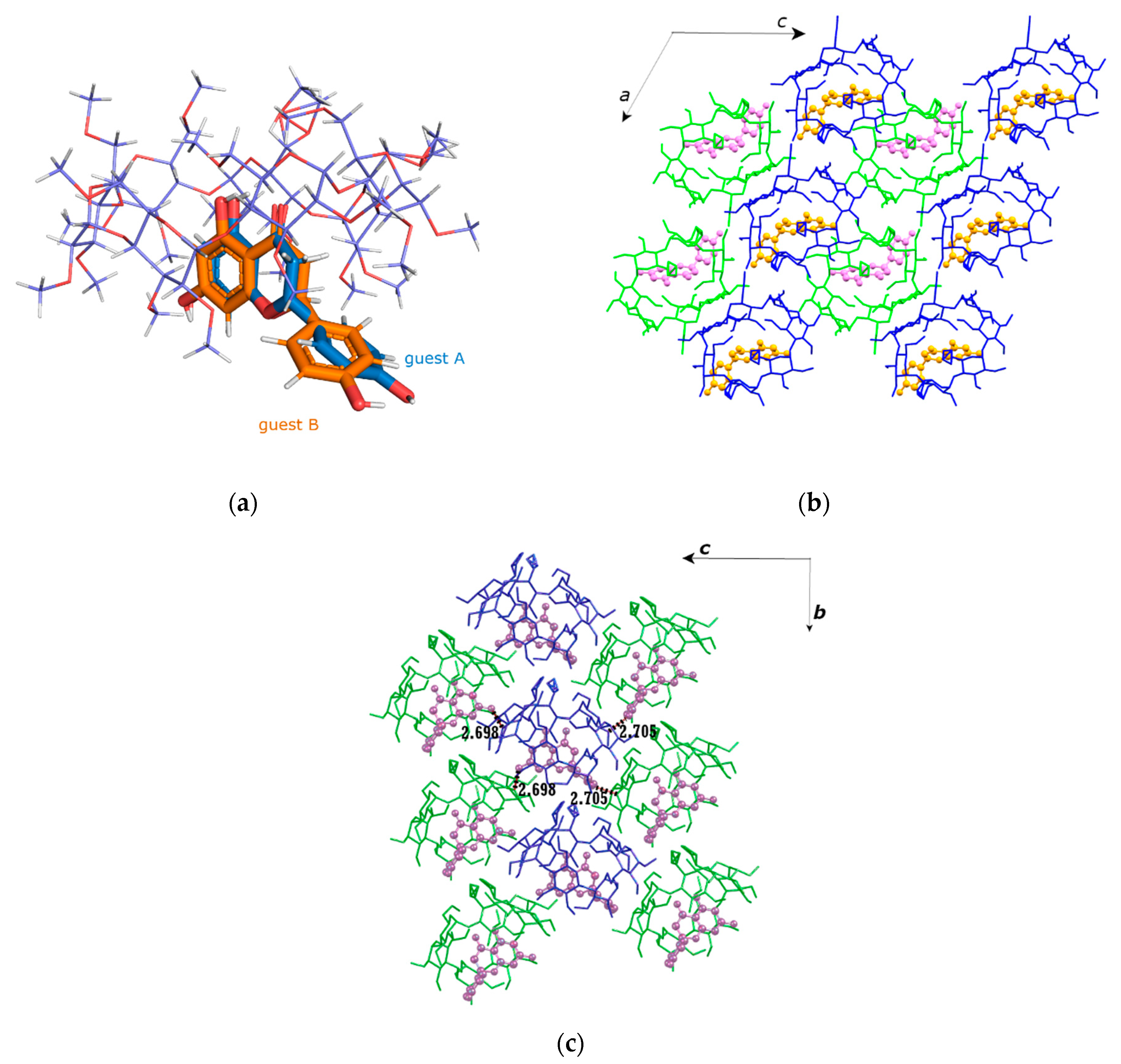
2.1. Crystal Structures Description
2.1.1. Naringenin/DM-β-CD Inclusion Complex
2.1.2. Naringenin/TM-β-CD Inclusion Complex
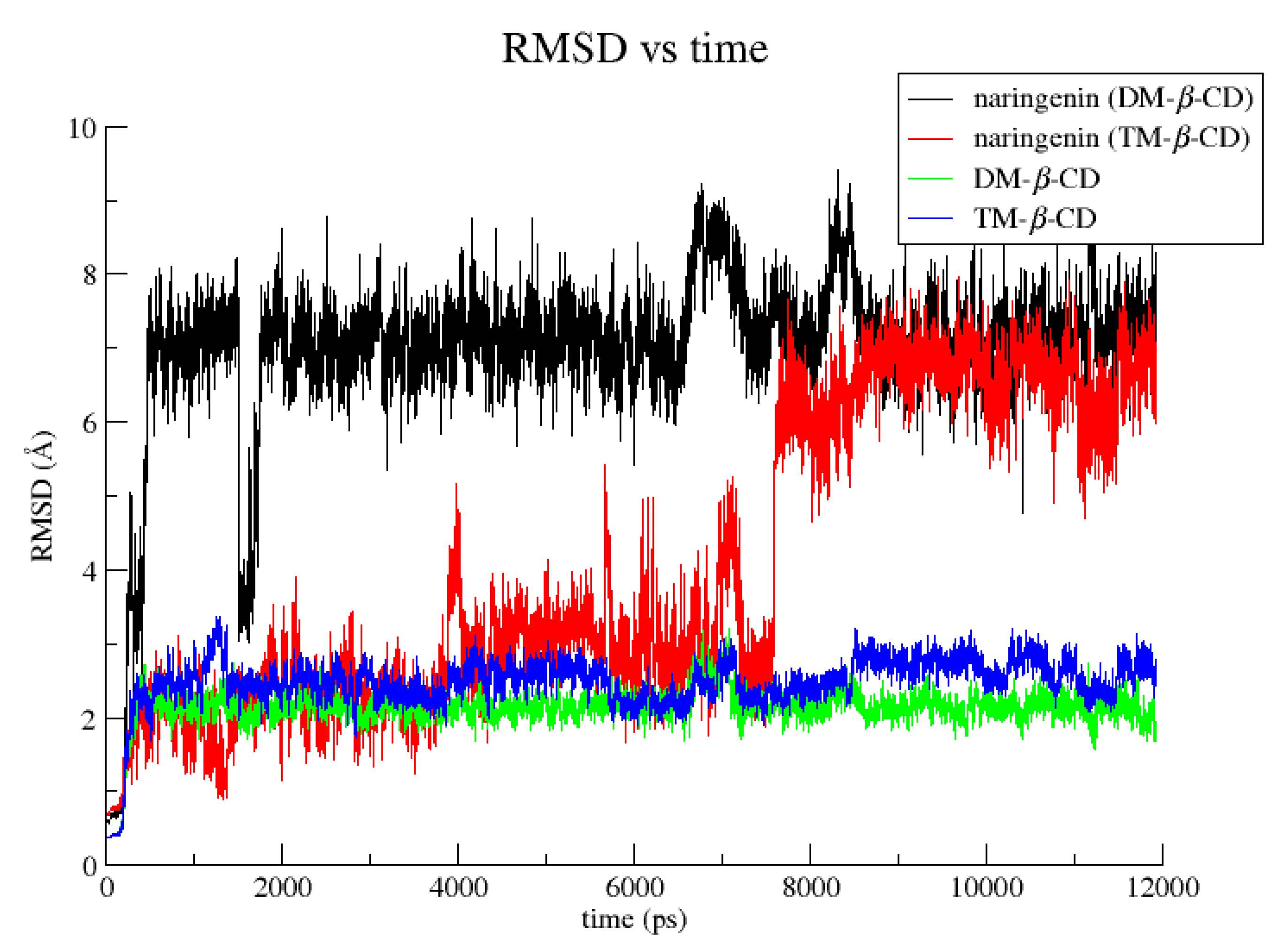
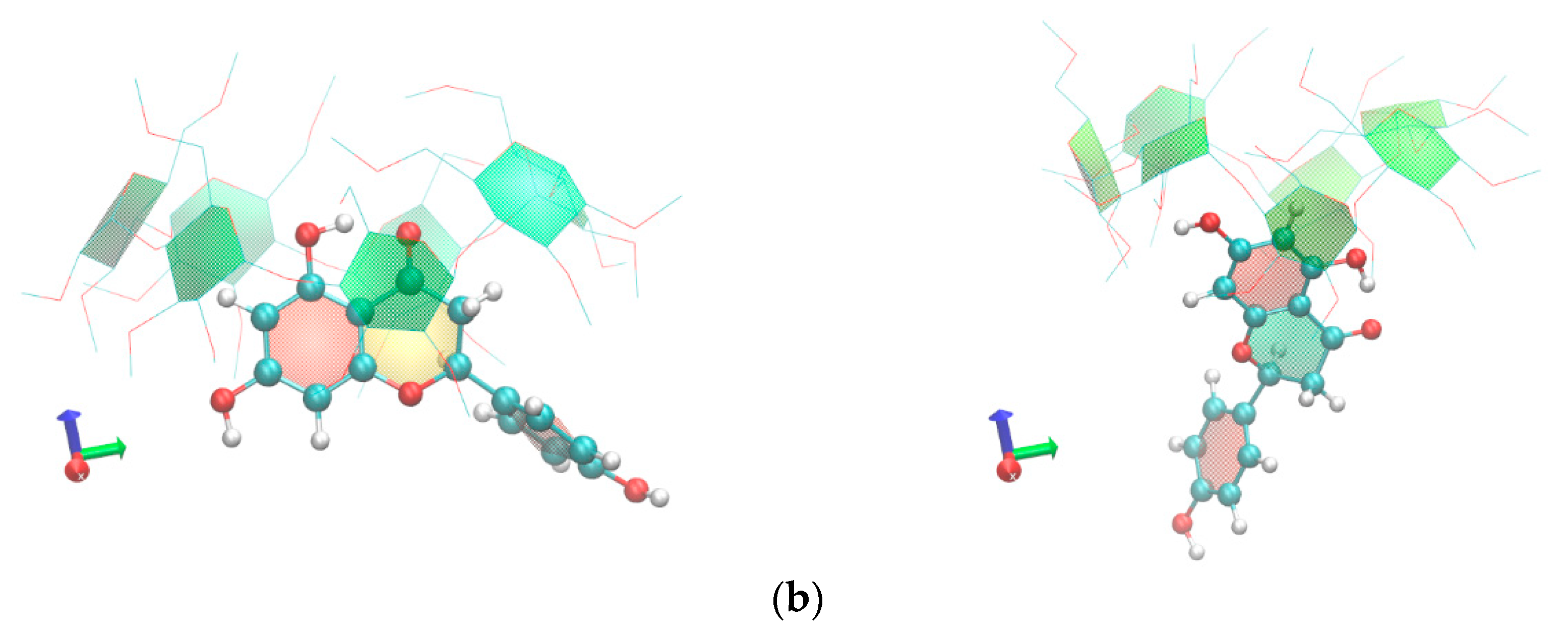
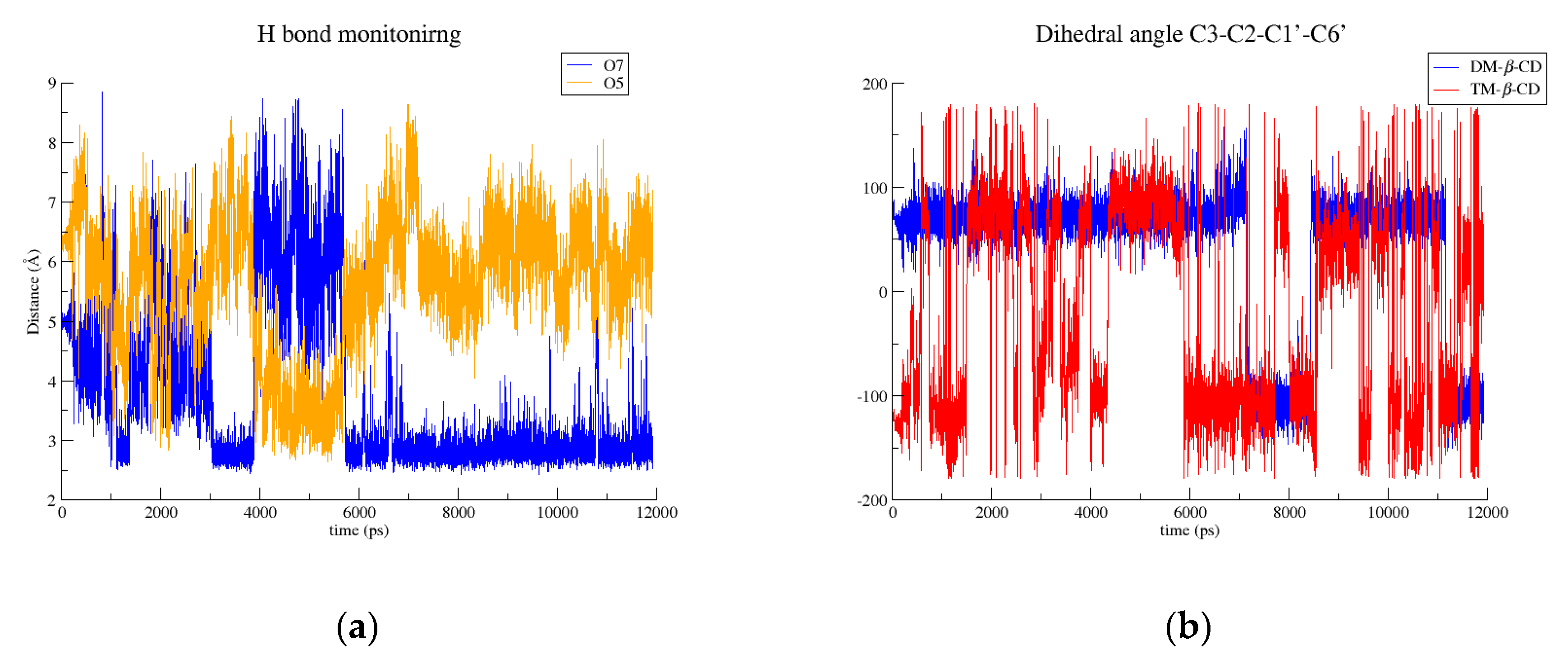
2.2. Molecular Dynamics Studies
3. Materials and Methods
3.1. X-ray Crystallography
3.1.1. Sample Preparation
3.1.2. Data Collection and Reduction
3.1.3. Crystal Structure Determination
3.1.4. Crystallographic Analysis and Coordinates Deposition
3.2. Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rao, P.V.; Kiran, S.; Rohini, P.; Bhagyasree, P. Flavonoid: A review on Naringenin. J. Pharmacogn. Phytochem. 2017, 6, 2778–2783. [Google Scholar]
- Patel, K.; Gadewar, M.; Tahilyani, V.; Patel, D.K. A review on pharmacological and analytical aspects of diosgenin: A concise report. Nat. Prod. Bioprospecting 2012, 2, 46–52. [Google Scholar] [CrossRef]
- Zeng, W.; Jin, L.; Zhang, F.; Zhang, C.; Liang, W. Naringenin as a potential immunomodulator in therapeutics. Pharmacol. Res. 2018, 135, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Zaidun, N.H.; Thent, Z.C.; Latiff, A.A. Combating oxidative stress disorders with citrus flavonoid: Naringenin. Life Sci. 2018, 208, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Fokou, P.V.T.; Sharifi-Rad, M.; Zucca, P.; Pezzani, R.; Martins, N.; Sharifi-Rad, J. The Therapeutic Potential of Naringenin: A Review of Clinical Trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef]
- Karim, N.; Jia, Z.; Zheng, X.; Cui, S.; Chen, W. A recent review of citrus flavanone naringenin on metabolic diseases and its potential sources for high yield-production. Trends Food Sci. Technol. 2018, 79, 35–54. [Google Scholar] [CrossRef]
- Sangpheak, W.; Kicuntod, J.; Schuster, R.; Rungrotmongkol, T.; Wolschann, P.; Kungwan, N.; Viernstein, H.; Mueller, M.; Pongsawasdi, P. Physical properties and biological activities of hesperetin and naringenin in complex with methylated β-cyclodextrin. Beilstein J. Org. Chem. 2015, 11, 2763–2773. [Google Scholar] [CrossRef]
- Shulman, M.; Cohen, M.; Soto-Gutierrez, A.; Yagi, H.; Wang, H.; Goldwasser, J.; Lee-Parsons, C.W.; Benny-Ratsaby, O.; Yarmush, M.L.; Nahmias, Y. Enhancement of naringenin bioavailability by complexation with hydroxypropyl-beta-cyclodextrin. PLoS ONE 2011, 6, e18033. [Google Scholar] [CrossRef]
- Valle, E.M.M.D. Cyclodextrins and their uses: A review. Process. Biochem. 2004, 39, 1033–1046. [Google Scholar] [CrossRef]
- Astray, G.; Gonzalez-Barreiro, C.; Mejuto, J.C.; Rial-Otero, R.; Simal-Gándara, J. A review on the use of cyclodextrins in foods. Food Hydrocoll. 2009, 23, 1631–1640. [Google Scholar] [CrossRef]
- Marques, H.M.C. A review on cyclodextrin encapsulation of essential oils and volatiles. Flavour Fragr. J. 2010, 25, 313–326. [Google Scholar] [CrossRef]
- Suvarna, V.; Gujar, P.; Murahari, M. Complexation of phytochemicals with cyclodextrin derivatives - An insight. Biomed. Pharmacother. Biomedecine Pharmacother. 2017, 88, 1122–1144. [Google Scholar] [CrossRef] [PubMed]
- Saokham, P.; Muankaew, C.; Jansook, P.; Loftsson, T. Solubility of Cyclodextrins and Drug/Cyclodextrin Complexes. Molecules 2018, 23, 1161. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-J.; Ma, S.-X.; Zhou, S.-Y.; Chen, W.; Yuan, M.-W.; Yin, Y.-Q.; Yang, X.-D. Preparation and characterization of inclusion complexes of naringenin with beta-cyclodextrin or its derivative. Carbohydr. Polym. 2013, 98, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yu, H.; Hang, L.; Shao, Y.; Ding, S.; Yang, X. Preparation of naringenin/ beta-cyclodextrin complex and its more potent alleviative effect on choroidal neovascularization in rats. BioMed Res. Int. 2014, 2014, 623509. [Google Scholar] [CrossRef]
- Semalty, A.; Tanwar, Y.S.; Semalty, M. Preparation and characterization of cyclodextrin inclusion complex of naringenin and critical comparison with phospholipid complexation for improving solubility and dissolution. J. Therm. Anal. Calorim. 2014, 115, 2471–2478. [Google Scholar] [CrossRef]
- Wen, J.; Liu, B.; Yuan, E.; Ma, Y.; Zhu, Y. Preparation and physicochemical properties of the complex of naringenin with hydroxypropyl-beta-cyclodextrin. Molecules 2010, 15, 4401–4407. [Google Scholar] [CrossRef]
- Sangpheak, W.; Khuntawee, W.; Wolschann, P.; Pongsawasdi, P.; Rungrotmongkol, T. Enhanced stability of a naringenin/2,6-dimethyl beta-cyclodextrin inclusion complex: Molecular dynamics and free energy calculations based on MM- and. J. Mol. Graph. Model. 2014, 50, 10–15. [Google Scholar] [CrossRef]
- Aree, T.; Saenger, W.; Leibnitz, P.; Hoier, H. Crystal structure of heptakis(2,6-di-O-methyl)-β-cyclodextrin dihydrate: A water molecule in an apolar cavity. Carbohydr. Res. 1999, 315, 199–205. [Google Scholar] [CrossRef]
- Bethanis, K.; Christoforides, E.; Tsorteki, F.; Fourtaka, K.; Mentzafos, D. Structural studies of the inclusion compounds of α-naphthaleneacetic acid in heptakis(2,6-di-O-methyl)-β-Cyclodextrin and heptakis(2,3,6-tri-O-methyl)-β-Cyclodextrin by X-ray crystallography and molecular dynamics. J. Incl. Phenom. Macrocycl. Chem. 2018, 92, 157–171. [Google Scholar] [CrossRef]
- Fourtaka, K.; Christoforides, E.; Mentzafos, D.; Bethanis, K. Crystal structures and molecular dynamics studies of the inclusion compounds of β-citronellol in β-cyclodextrin, heptakis(2,6-di-O-methyl)-β-cyclodextrin and heptakis(2,3,6-tri-O-methyl)-β-cyclodextrin. J. Mol. Struct. 2018, 1161, 1–8. [Google Scholar] [CrossRef]
- Steiner, T.; Saenger, W. Crystal structure of anhydrous heptakis-(2,6-di-O-methyl) cyclomaltoheptaose (dimethyl-β-cyclodextrin). Carbohydr. Res. 1995, 275, 73–82. [Google Scholar] [CrossRef]
- Caira, M.R.; Griffith, V.J.; Nassimbeni, L.R.; Oudtshoorn, B.V. X-ray structures of 1:1 complexes of (L)-menthol with β-cyclodextrin and permethylated β-cyclodextrin. Supramol. Chem. 1996, 7, 119–124. [Google Scholar] [CrossRef]
- SAINT; SAINT Version 8.34A; Bruker-AXS: Madison, WI, USA, 2013.
- Sheldrick, G.M. SADABS; SADABS Version 2012/1, 1; Bruker-AXS: Madison, WI, USA, 2012. [Google Scholar]
- Takashima, Y.; Sakamoto, K.; Oizumi, Y.; Yamaguchi, H.; Kamitori, S.; Harada, A. Complex Formation of Cyclodextrins with Various Thiophenes and their Polymerization in Water: Preparation of Poly-pseudo-rotaxanes containing Poly(thiophene)s. J. Incl. Phenom. Macrocycl. Chem. 2006, 56, 45–53. [Google Scholar] [CrossRef]
- Beurskens, P.T.; Beurskens, G.; Gelder, R.d.; Garcia-Granda, S.; Gould, R.O.; Smits, J.M.M. The DIRDIF2008 Program System; Crystallography Laboratory, University of Nijmegen: Nijmegen, The Netherlands, 2008; Available online: http://www.xtal.science.ru.nl/dirdif/software/dirdif.html (accessed on 18 December 2019).
- Sheldrick, G.M. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr. Sect. D 2010, 66, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Schüttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0 – new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 3, 198–210. [Google Scholar] [CrossRef]
- Cezard, C.; Trivelli, X.; Aubry, F.; Djedaini-Pilard, F.; Dupradeau, F.-Y. Molecular dynamics studies of native and substituted cyclodextrins in different media: 1. Charge derivation and force field performances. Phys. Chem. Chem. Phys. 2011, 13, 15103–15121. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Kräutler, V.; van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28, 33–38. [Google Scholar] [CrossRef]
- Miller, B.R., 3rd; McGee, T.D.J.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Wang, J.; Morin, P.; Wang, W.; Kollman, P.A. Use of MM-PBSA in Reproducing the Binding Free Energies to HIV-1 RT of TIBO Derivatives and Predicting the Binding Mode to HIV-1 RT of Efavirenz by Docking and MM-PBSA. J. Am. Chem. Soc. 2001, 123, 5221–5230. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]

| Naringenin/DM-β-CD | Naringenin/TM-β-CD | |
|---|---|---|
| Enthalpic Term | ||
| ΔEvdW | −33.50 ± 2.77 | −23.40 ± 4.63 |
| ΔEele | −8.07 ± 2.46 | −13.09 ± 3.25 |
| ΔEGB | 21.95 ± 2.11 | 24.58 ± 3.24 |
| ΔEsurf | −3.31 ± 0.16 | −2.79 ± 0.29 |
| ΔGgas | −41.58 ± 3.9 | −36.49 ± 6.02 |
| ΔGsolv | 18.64 ± 2.07 | 21.79 ± 3.18 |
| a ΔG(GB) | −22.93 ± 2.75 | −14.69 ± 3.94 |
| Entropic Term | ||
| (TΔS) | −18.19 ± 1.16 | −17.69 ± 1.71 |
| Binding Energy | ||
| b (ΔGbind) | −4.74 ± 2.98 | 3.0 ± 4.29 |
| Crystal Data | Naringenin/DM-β-CD | Naringenin/TM-β-CD |
|---|---|---|
| Chemical formula | C56H98O35·C15H12O5·4.6(H2O) | C63H112O35·C15H12O5 |
| Mr | 1687 | 1701.76 |
| Crystal system, space group | Orthorhombic, P212121 | Monoclinic, P21 |
| Temperature (K) | 100(2) | 100(2) |
| Unit cell parameters a, b, c (Å) | 15.531(1), 17.810(1), 30.149(2) | 15.758(3), 12.913(2), 22.273(3) β = 109.936 (1) (°) |
| V (Å3) | 8340.1 (10) | 4260.6 (13) |
| Z | 4 | 2 |
| Radiation type | Cu Ka | Cu Ka |
| μ (mm−1) | 0.96 | 0.90 |
| Crystal size (mm3) | 0.5 × 0.1 × 0.1 | 0.45 × 0.3 × 0.2 |
| Data collection | ||
| Diffractometer | Bruker APEX-II | Bruker APEX-II |
| Absorption correction | Multi-scan SADABS2014/5—Bruker AXS area detector scaling and absorption correction | Multi-scan SADABS2014/5 - Bruker AXS area detector scaling and absorption correction |
| Tmin, Tmax | 0.583, 0.752 | 0.496, 0.753 |
| No. of measured, independent, and observed [I > 2σ(I)] reflections | 65,321, 11,718, 10,457 | 65,916, 15,008, 14,866 |
| Rint | 0.056 | 0.046 |
| (sin θ/λ)max (Å-1) | 0.556 | 0.596 |
| Refinement | ||
| R[F2 > 2σ(F2)], wR(F2), S | 0.056, 0.127, 1.05 | 0.043, 0.110, 1.06 |
| No. of reflections | 11,718 | 15,008 |
| No. of parameters | 959 | 1050 |
| No. of restraints | 80 | 147 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.51, −0.43 | 0.38, −0.35 |
| Absolute structure | Flack x determined using 4169 quotients [(I+) − (I−)]/[(I+) + (I−)] (Parsons, Flack and Wagner, Acta Cryst. B69 (2013) 249–259). | Flack x determined using 6895 quotients [(I+) − (I−)]/[(I+) + (I−)] (Parsons, Flack and Wagner, Acta Cryst. B69 (2013) 249–259). |
| Absolute structure parameter | −0.04 (6) | 0.07 (3) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaioannou, A.; Christoforides, E.; Bethanis, K. Inclusion Complexes of Naringenin in Dimethylated and Permethylated β-Cyclodextrins: Crystal Structures and Molecular Dynamics Studies. Crystals 2020, 10, 10. https://doi.org/10.3390/cryst10010010
Papaioannou A, Christoforides E, Bethanis K. Inclusion Complexes of Naringenin in Dimethylated and Permethylated β-Cyclodextrins: Crystal Structures and Molecular Dynamics Studies. Crystals. 2020; 10(1):10. https://doi.org/10.3390/cryst10010010
Chicago/Turabian StylePapaioannou, Andreas, Elias Christoforides, and Kostas Bethanis. 2020. "Inclusion Complexes of Naringenin in Dimethylated and Permethylated β-Cyclodextrins: Crystal Structures and Molecular Dynamics Studies" Crystals 10, no. 1: 10. https://doi.org/10.3390/cryst10010010
APA StylePapaioannou, A., Christoforides, E., & Bethanis, K. (2020). Inclusion Complexes of Naringenin in Dimethylated and Permethylated β-Cyclodextrins: Crystal Structures and Molecular Dynamics Studies. Crystals, 10(1), 10. https://doi.org/10.3390/cryst10010010







