Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification of CpCR, Expanding Its Substrate Scope and Kinetic Studies
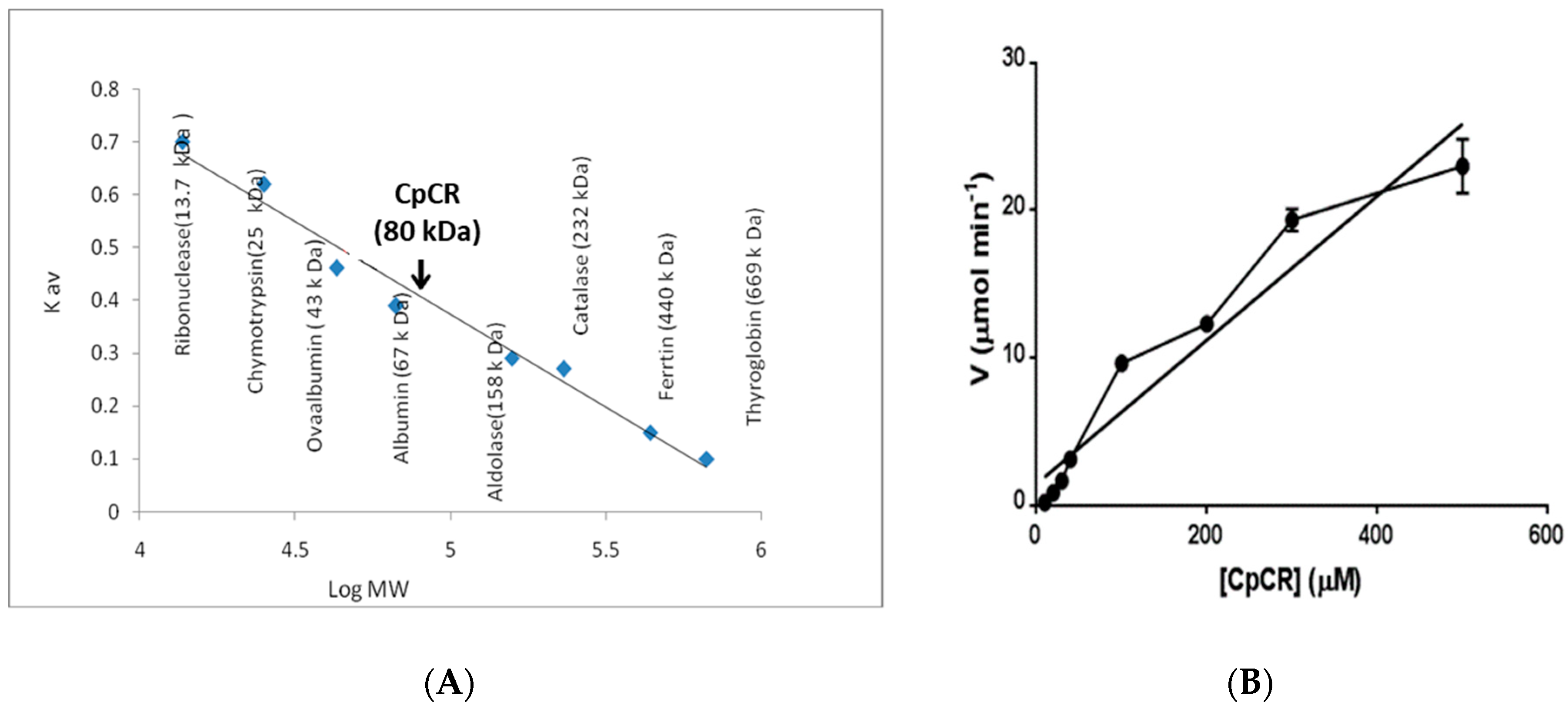
2.2. Oligomeric State of CpCR
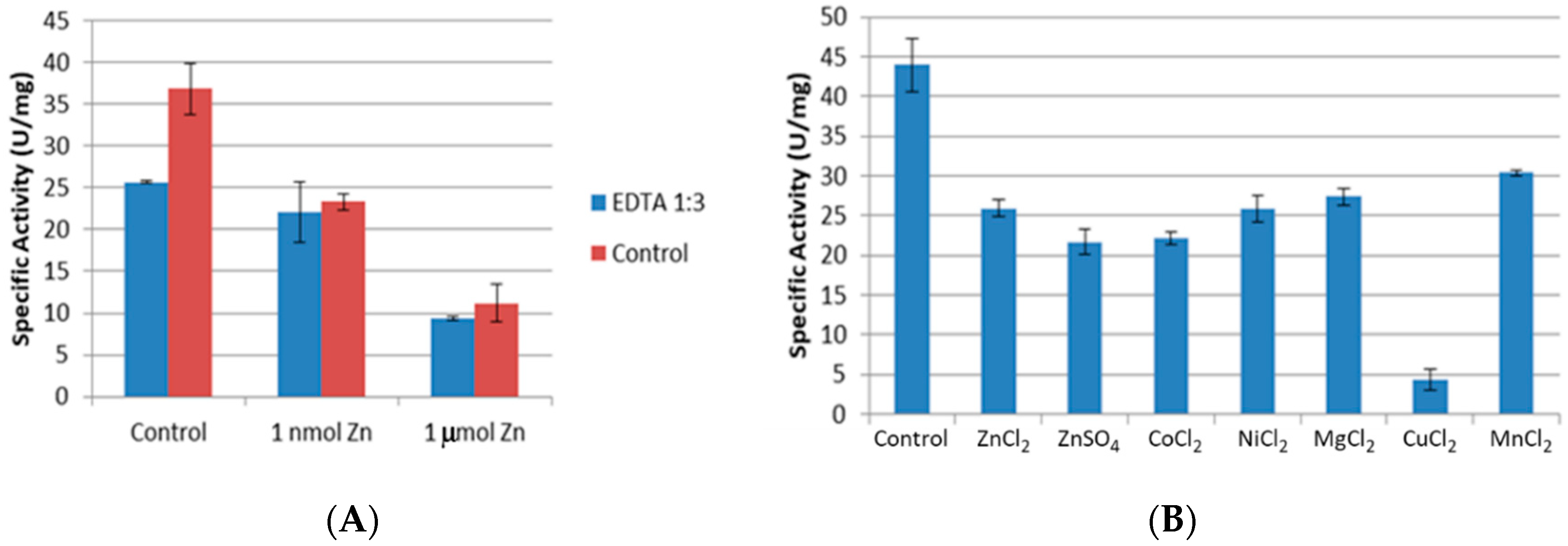
2.3. Effect of Chelating Agent EDTA and Divalent Metal Salts on Activity of CpCR
2.3.1. Effect of Time on Chelation
2.3.2. Removal of EDTA
2.3.3. Inhibition of CpCR by Divalent Metal Salts
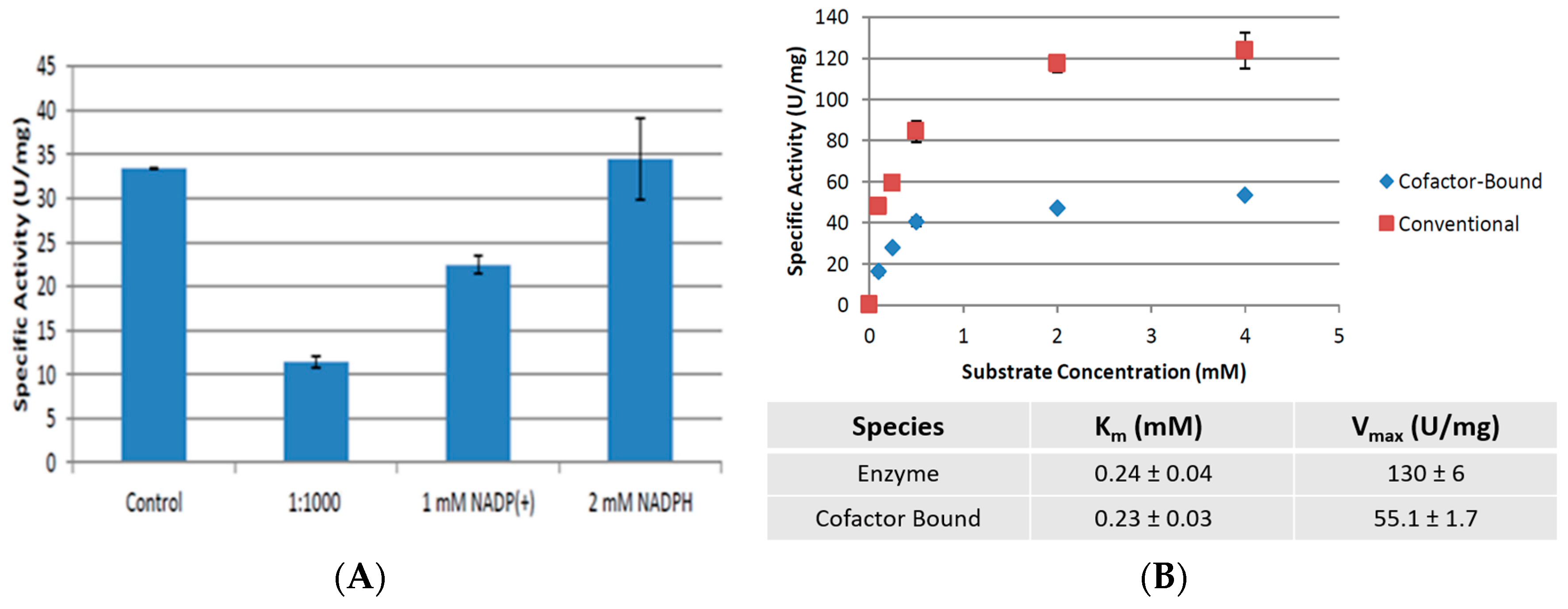
2.4. Cofactor Pre-Treatment Prevents CpCR Activity Loss
3. Materials and Methods
3.1. Chemicals and Media
3.2. Enzyme Expression and Purification
3.3. Specific Activity, Substrate Scope and Kinetic Studies of CpCR
3.4. Oligomeric State of CpCR
3.5. Treatment of CpCR with EDTA, Divalent Metal Salts
3.6. CpCR—Cofactor Binding Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Persson, B.; Hedlund, J.; Jörnvall, H. Medium- and short-chain dehydrogenase/reductase gene and protein families: The MDR superfamily. Cell. Mol. Life Sci. 2008, 65, 3879–3894. [Google Scholar] [CrossRef] [PubMed]
- Jörnvall, H.; Hedlund, J.; Bergman, T.; Oppermann, U.; Persson, B. Biochemical and Biophysical Research Communications Superfamilies SDR and MDR: From early ancestry to present forms. Emergence of three lines, a Zn-metalloenzyme, and distinct variabilities. Biochem. Biophys. Res. Commun. 2010, 396, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Ananthathamula, R.; Karanam, V.K.; Doble, M.; Chadha, A. Understanding substrate specificity and enantioselectivity of carbonyl reductase from Candida parapsilosis ATCC 7330 (CpCR): Experimental and modeling studies. Mol. Catal. 2018, 460, 40–45. [Google Scholar] [CrossRef]
- Chadha, A.; Venkataraman, S.; Preetha, R.; Padhi, S.K. Candida parapsilosis: A versatile biocatalyst for organic oxidation-reduction reactions. Bioorg. Chem. 2016, 68, 187–213. [Google Scholar] [CrossRef] [PubMed]
- Kvassman, J.; Pettersson, G. Kinetic Transients in the Reduction of Aldehydes Catalysed by Liver Alcohol Dehydrogenase. Eur. J. Biochem. 1976, 69, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Deetz, J.S.; Luehr, C.A.; Vallee, B.L. Human Liver Alcohol Dehydrogenase Isozymes: Reduction of Aldehydes and Ketones. Biochemistry 1984, 23, 6822–6828. [Google Scholar] [CrossRef]
- Pal, S.; Park, D.H.; Plapp, B.V. Activity of yeast alcohol dehydrogenases on benzyl alcohols and benzaldehydes. Characterization of ADH1 from Saccharomyces carlsbergensis and transition state analysis. Chem. Biol. Interact. 2009, 178, 16–23. [Google Scholar] [CrossRef]
- Jelokova, J.; Karlsson, C.; Estonius, M.; Jornvall, H.; Hoog, J.O. Features of structural zinc in mammalian alcohol dehydrogenase. Site-directed mutagenesis of the zinc ligands. Eur. J. Biochem. 1994, 225, 1015–1019. [Google Scholar] [CrossRef]
- Magonet, E.; Hayen, P.; Delforge, D.; Delaive, E.; Remacle, J. Importance of the structural zinc atom for the stability of yeast alcohol dehydrogenase. Biochem. J. 1992, 287 Pt 2, 361–365. [Google Scholar] [CrossRef]
- Auld, D.S.; Bergman, T. Medium- and short-chain dehydrogenase/reductase gene and protein families: The role of zinc for alcohol dehydrogenase structure and function. Cell. Mol. Life Sci. 2008, 65, 3961–3970. [Google Scholar] [CrossRef]
- Kägi, J.H.R.; Vallee, B.L. The Role of Zinc in Alcohol Dehydrogenase. J. Biol. Chem. 1960, 235, 3188–3192. [Google Scholar]
- Baker, P.J.; Britton, K.L.; Fisher, M.; Esclapez, J.; Pire, C.; Bonete, M.J.; Ferrer, J.; Rice, D.W. Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily. Proc. Natl. Acad. Sci. USA 2009, 106, 779–784. [Google Scholar] [CrossRef] [Green Version]
- Drum, D.E.; Harrison, J.H.; Li, T.K.; Bethune, J.L.; Vallee, B.L. Structural and functional zinc in horse liver alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 1967, 57, 1434–1440. [Google Scholar] [CrossRef]
- Vallee, B.L.; Auld, D.S. Active-site zinc ligands and activated H20 of zinc enzymes. Proc. Natl. Acad. Sci. USA 1990, 87, 220–224. [Google Scholar] [CrossRef]
- Ryde, U. The coordination chemistry of the structural zinc ion in alcohol dehydrogenase studied by ab initio quantum chemical calculations. Eur. Biophys. J. 1996, 24, 213–221. [Google Scholar] [CrossRef]
- Wang, J.; Sakakibara, M.; Matsuda, M.; Itoh, N. Site-directed Mutagenesis of Two Zinc- binding Centers of the NADH-dependent Phenylacetaldehyde Reductase from Styrene- assimilating Corynebacterium sp. Strain ST-10. Biosci. Biotechnol. Biochem. 2014, 63, 2216–2218. [Google Scholar] [CrossRef]
- Man, H.; Loderer, C.; Ansorge-Schumacher, M.B.; Grogan, G. Structure of NADH-dependent carbonyl reductase (CPCR2) from Candida parapsilosis provides insight into mutations that improve catalytic properties. ChemCatChem 2014, 6, 1103–1111. [Google Scholar] [CrossRef]
- Dickinson, F.M.; Berrieman, S. The reactions of 1,10-phenanthroline with yeast alcohol dehydrogenase. Biochem. J. 1977, 167, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Dołega, A. Alcohol dehydrogenase and its simple inorganic models. Coord. Chem. Rev. 2010, 254, 916–937. [Google Scholar] [CrossRef]
- Chen, F.; Wang, P.; An, Y.; Huang, J.; Xu, Y. Structural insight into the conformational change of alcohol dehydrogenase from Arabidopsis thaliana L. during coenzyme binding. Biochimie 2014, 108, 33–39. [Google Scholar] [CrossRef]
- Colonna-Cesari, F.; Perahia, D.; Karplus, M.; Eklund, H.; Brädén, C.I.; Tapia, O. Interdomain motion in liver alcohol dehydrogenase. Structural and energetic analysis of the hinge bending mode. J. Biol. Chem. 1986, 261, 15273–15280. [Google Scholar]
- Marolt, M.; Lüdeke, S. Studying NAD(P)H cofactor-binding to alcohol dehydrogenases through global analysis of circular dichroism spectra. Phys. Chem. Chem. Phys. 2019, 21, 1671–1681. [Google Scholar] [CrossRef]
- Satheesan Babu, C.; Lim, C. Efficient Binding of Flexible and Redox-Active Coenzymes by Oxidoreductases. ACS Catal. 2016, 6, 3469–3472. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.P.; Turner, N.J. Two-Enzyme Hydrogen-Borrowing Amination of Alcohols Enabled by a Cofactor-Switched Alcohol Dehydrogenase. ChemCatChem 2017, 9, 3833–3836. [Google Scholar] [CrossRef]
- Cahn, J.K.B.; Werlang, C.A.; Baumschlager, A.; Brinkmann-Chen, S.; Mayo, S.L.; Arnold, F.H. A General Tool for Engineering the NAD/NADP Cofactor Preference of Oxidoreductases. ACS Synth. Biol. 2017, 6, 326–333. [Google Scholar] [CrossRef]
- Chen, H.; Zhu, Z.; Huang, R.; Zhang, Y.H.P. Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP+ to NAD+ with Its Application to Biobatteries. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- You, C.; Huang, R.; Wei, X.; Zhu, Z.; Zhang, Y.H.P. Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology. Synth. Syst. Biotechnol. 2017, 2, 208–218. [Google Scholar] [CrossRef]
- Aggarwal, N.; Mandal, P.K.; Gautham, N.; Chadha, A. Expression, purification, crystallization and preliminary X-ray diffraction analysis of carbonyl reductase from Candida parapsilosis ATCC 7330. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 313–315. [Google Scholar] [CrossRef]
- Sudhakara, S.; Chadha, A. A carbonyl reductase from Candida parapsilosis ATCC 7330: Substrate selectivity and enantiospecificity. Org. Biomol. Chem. 2017, 15, 4165–4171. [Google Scholar] [CrossRef]
- Larsen, K.S.; Auld, D.S. Carboxypeptidase A: Mechanism of Zinc Inhibition. Biochemistry 1989, 28, 9620–9625. [Google Scholar] [CrossRef]
- Maret, W.; Yetman, C.A.; Jiang, L.J. Enzyme regulation by reversible zinc inhibition: Glycerol phosphate dehydrogenase as an example. Chem. Biol. Interact. 2001, 130, 891–901. [Google Scholar] [CrossRef]
- Ying, X.; Wang, Y.; Badiei, H.R.; Karanassios, V.; Ma, K. Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea. Arch. Microbiol. 2007, 187, 499–510. [Google Scholar] [CrossRef]
- Stiborová, M.; Leblová, S. Effect of Metals on Rape Alcohol Dehydrogenase. Biochem. Physiol. Pflanz. 2017, 174, 39–43. [Google Scholar] [CrossRef]
- Jin, L.; Szeto, K.Y.; Zhang, L.; Du, W.; Sun, H. Inhibition of alcohol dehydrogenase by bismuth. J. Inorg. Biochem. 2004, 98, 1331–1337. [Google Scholar] [CrossRef]
- Ye, Y.; Godzik, A. FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004, 32, 582–585. [Google Scholar] [CrossRef]
- Brändén, C.; Eklund, H. Coenzyme-induced Conformational Changes and Substrate Binding in Liver Alcohol Dehydrogenase. In CIBA Foundation Symposium 60—Molecular Interactions and Activity in Proteins; Porter, R., Fitzsimons, D.W., Eds.; Ciba Foundation: London, UK, 2009; pp. 63–80. [Google Scholar]
- Aggarwal, N. Cloning, Purification, Biochemical Characterisation and Crystallisation of a Carbonyl Reductase from Candida parapsilosis ATCC 7330: Towards Understanding Its Enantioselectivity at the Molecular Level. Ph.D. Thesis, Indian Institute of Technology Madras, Chennai, India, 2013. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Entry | Substrate | Specific Activity (U mg−1) 1 |
|---|---|---|
| 1 | 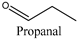 | 24.98 ± 1.06 |
| 2 |  | 19.29 ± 0.59 |
| 3 |  | 21.9 ± 0.64 |
| 4 |  | 23.06 ± 0.94 |
| 5 |  | 6.76 ± 0.63 |
| 6 | 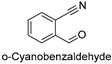 | 21.17 ± 0.73 |
| 7 | 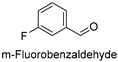 | 6.53 ± 0.82 |
| 8 | 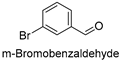 | 20.79 ± 0.76 |
| 9 | 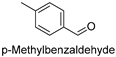 | 26.46 ± 0.92 |
| 10 |  | 21.0 ± 0.53 |
| 11 | 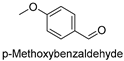 | 19.69 ± 0.97 |
| 12 | 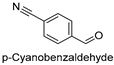 | 18.63 ± 0.72 |
| 13 | 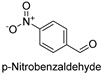 | 13.35 ± 0.69 |
| 14 | 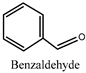 | 33.96 ± 1.02 2 |
| 15 |  | 3.8 ± 0.64 |
| Entry | Substrate | Vmax (µmol min−1 mg−1) | Km (mM) |
|---|---|---|---|
| 1 |  | 34 ± 0.9 | 0.29 ± 0.04 |
| 2 | 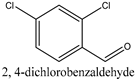 | 3.88 ± 0.56 | 7.14 ± 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karanam, V.K.; Chaudhury, D.; Chadha, A. Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc. Catalysts 2019, 9, 702. https://doi.org/10.3390/catal9090702
Karanam VK, Chaudhury D, Chadha A. Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc. Catalysts. 2019; 9(9):702. https://doi.org/10.3390/catal9090702
Chicago/Turabian StyleKaranam, Vinay Kumar, Debayan Chaudhury, and Anju Chadha. 2019. "Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc" Catalysts 9, no. 9: 702. https://doi.org/10.3390/catal9090702
APA StyleKaranam, V. K., Chaudhury, D., & Chadha, A. (2019). Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc. Catalysts, 9(9), 702. https://doi.org/10.3390/catal9090702




