Synthesis of Ribavirin, Tecadenoson, and Cladribine by Enzymatic Transglycosylation
Abstract
1. Introduction
2. Results and Discussion
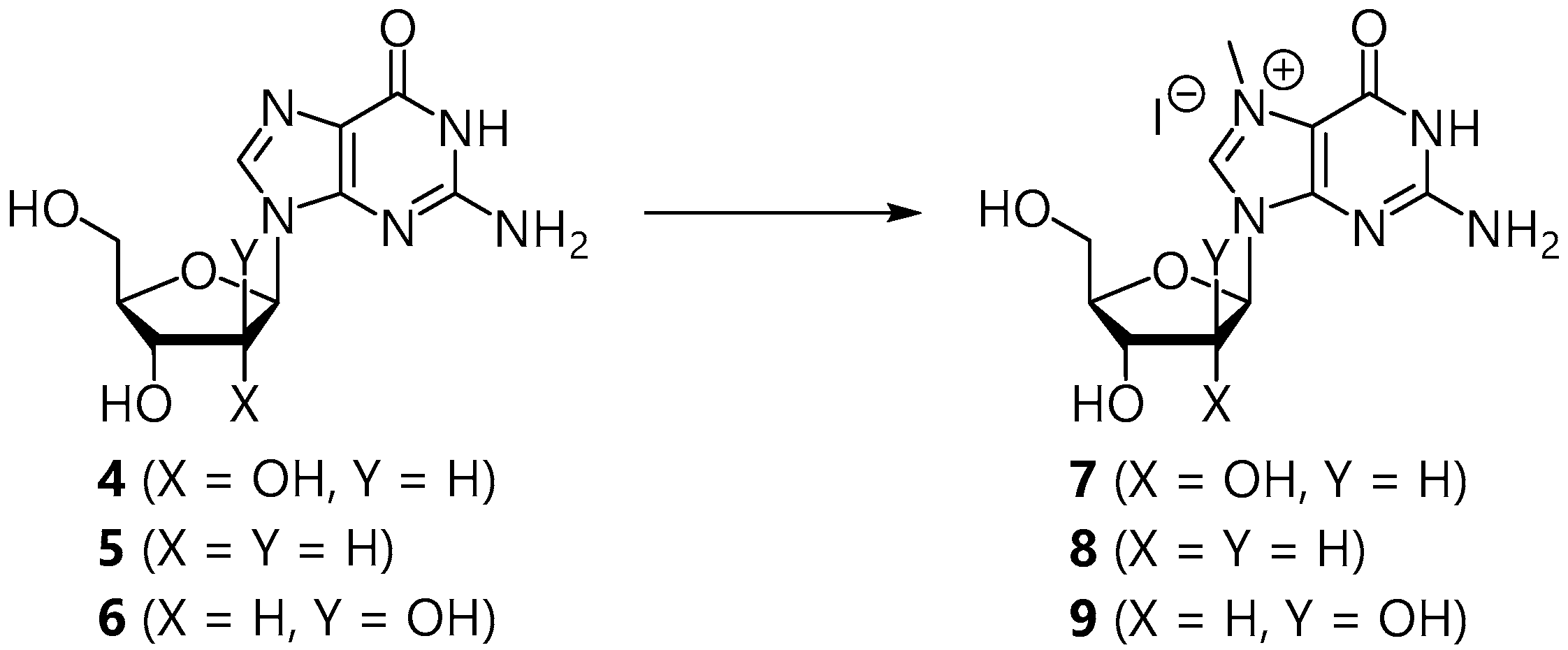
2.1. Synthesis of the Sugar Donors
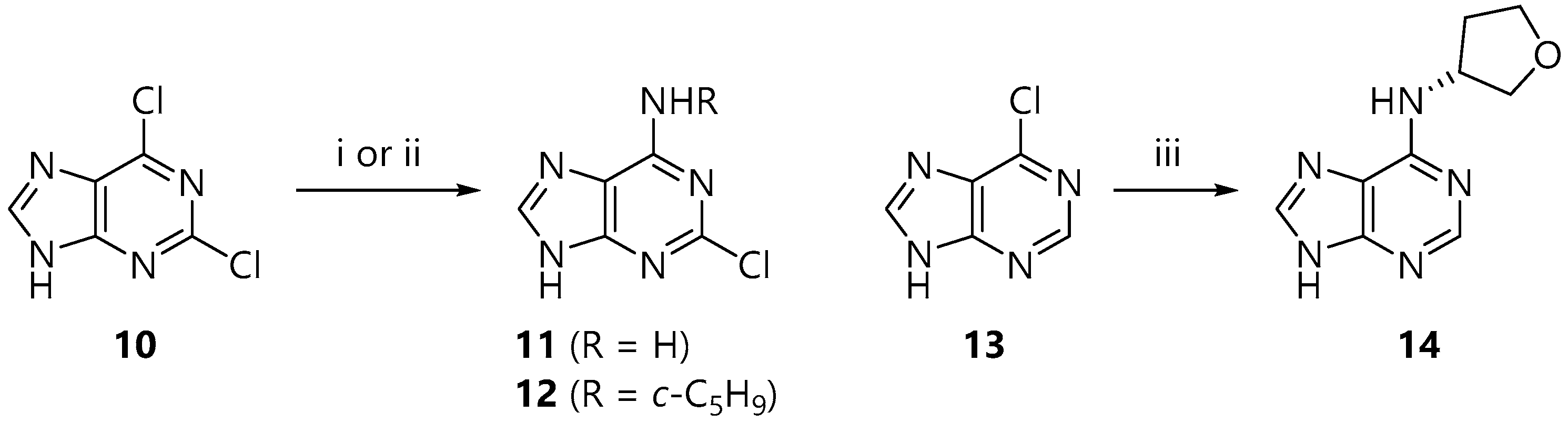
2.2. Synthesis of the Base Acceptors
2.3. “One-Pot, One-Enzyme” Transglycosylations
3. Materials and Methods
3.1. General
3.1.1. Chemicals
3.1.2. Methods
3.2. Chemical Synthesis of Sugar Donors
3.3. Chemical Synthesis of Base Acceptors and Tecadenoson
3.4. Enzymatic Synthesis of Nucleoside Analogues: General Procedure of Transglycosylation Reactions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McConachie, S.M.; Wilhelm, S.M.; Kale-Pradhan, P.B. New direct-acting antivirals in hepatitis C therapy: A review of sofosbuvir, ledipasvir, daclatasvir, simeprevir, paritaprevir, ombitasvir and dasabuvir. Expert Rev. Clin. Pharmacol. 2016, 9, 287–302. [Google Scholar] [CrossRef]
- Vorbrueggen, H.; Ruh-Pohlenz, C. Synthesis of nucleosides. Org. React. 2000, 55. [Google Scholar] [CrossRef]
- Mikhailopulo, I.; Miroshnikov, A.I. Biologically important nucleosides: Modern trends in biotechnology and application. Mendeleev Commun. 2011, 21, 57–68. [Google Scholar] [CrossRef]
- Ubiali, D.; Serra, C.D.; Serra, I.; Morelli, C.F.; Terreni, M.; Albertini, A.M.; Manitto, P.; Speranza, G. Production, characterization and synthetic application of a purine nucleoside phosphorylase from Aeromonas hydrophila. Adv. Synth. Catal. 2012, 354, 96–104. [Google Scholar] [CrossRef]
- Serra, I.; Ubiali, D.; Piškur, J.; Christoffersen, S.; Lewkowicz, E.S.; Iribarren, A.M.; Albertini, A.M.; Terreni, M. Developing a collection of immobilized nucleoside phosphorylases for the preparation of nucleoside analogues: Enzymatic synthesis of arabinosyladenine and 2′,3′-dideoxyinosine. ChemPlusChem 2013, 78, 157–165. [Google Scholar] [CrossRef]
- Serra, I.; Daly, S.; Alcantara, A.R.; Bianchi, D.; Terreni, M.; Ubiali, D. Redesigning the synthesis of Vidarabine via a multienzymatic reaction catalyzed by immobilized nucleoside phosphorylases. RSC Adv. 2015, 5, 23569–23577. [Google Scholar] [CrossRef]
- Ubiali, D.; Morelli, C.F.; Rabuffetti, M.; Cattaneo, G.; Serra, I.; Bavaro, T.; Albertini, A.; Speranza, G. Substrate specificity of a purine nucleoside phosphorylase from Aeromonas hydrophila toward 6-substituted purines and its use as a biocatalyst in the synthesis of the corresponding ribonucleosides. Curr. Org. Chem. 2015, 19, 2220–2225. [Google Scholar] [CrossRef]
- Calleri, E.; Cattaneo, G.; Rabuffetti, M.; Serra, I.; Bavaro, T.; Massolini, G.; Speranza, G.; Ubiali, D. Flow-synthesis of nucleosides catalyzed by an immobilized purine nucleoside phosphorylase from Aeromonas hydrophila: Integrated systems of reaction control and product purification. Adv. Synth. Catal. 2015, 357, 2520–2528. [Google Scholar] [CrossRef]
- Cattaneo, G.; Rabuffetti, M.; Speranza, G.; Kupfer, T.; Peters, B.; Massolini, G.; Ubiali, D.; Calleri, E. Synthesis of adenine nucleosides by transglycosylation using two sequential nucleoside phosphorylase-based bioreactors with on-line reaction monitoring by using HPLC. ChemCatChem 2017, 9, 4614–4620. [Google Scholar] [CrossRef]
- Leyssen, P.; De Clercq, E.; Neyts, J. Molecular strategies to inhibit the replication of RNA viruses. Antivir. Res. 2008, 78, 9–25. [Google Scholar] [CrossRef]
- Broder, C. Henipavirus outbreaks to antivirals: The current status of potential therapeutics. Curr. Opin. Virol. 2012, 2, 176–187. [Google Scholar] [CrossRef]
- Lau, J.Y.N.; Tam, R.C.; Liang, T.J.; Hong, Z. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 2002, 35, 1002–1009. [Google Scholar] [CrossRef]
- Chung, R.T.; Gale, M., Jr.; Polyak, S.J.; Lemon, S.M.; Liang, T.J.; Hoofnagle, J.H. Mechanisms of action of interferon and ribavirin in chronic hepatitis C: Summary of a workshop. Hepatology 2008, 47, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Joosen, M.J.; Bueters, T.J.; van Helden, H.P. Cardiovascular effects of the adenosine A1 receptor agonist N6-cyclopentyladenosine (CPA) decisive for its therapeutic efficacy in sarin poisoning. Arch. Toxicol. 2004, 78, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Peterman, C.; Sanoski, C.A. Tecadenoson: A novel, selective A1 adenosine receptor agonist. Cardiol. Rev. 2005, 13, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Balakumar, P.; Singh, H.; Reddy, K.; Anand-Srivastava, K.; Madhu, B. Adenosine-A1 receptors activation restores the suppressed cardioprotective effects of ischemic preconditioning in hyperhomocysteinemic rat hearts. J. Cardiovasc. Pharm. 2009, 54, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Juliusson, G.; Samuelsson, H. Hairy cell leukemia: Epidemiology, pharmacokinetics of cladribine, and long-term follow-up of subcutaneous therapy. Leuk. Lymphoma 2011, 52, 46–49. [Google Scholar] [CrossRef]
- Giovannoni, G.; Comi, G.; Cook, S.; Rammohan, K.; Rieckmann, P.; Sørensen, P.S.; Vermersch, P.; Chang, P.; Hamlett, A.; Musch, B.; et al. A placebo-controlled trial of oral cladribine for relapsing multiple sclerosis. N. Eng. J. Med. 2010, 362, 416–426. [Google Scholar] [CrossRef]
- Liu, W.Y.; Li, H.Y.; Zhao, B.X.; Shin, D.S.; Lian, S.; Miao, J.Y. Synthesis of novel ribavirin hydrazone derivatives and anti-proliferative activity against A549 lung cancer cells. Carbohydr. Res. 2009, 344, 1270–1275. [Google Scholar] [CrossRef]
- Li, Y.S.; Zhang, J.J.; Mei, L.Q.; Tan, C.X. An improved procedure for the preparation of Ribavirin. Org. Prep. Proced. Int. 2012, 44, 387–391. [Google Scholar] [CrossRef]
- Bookser, B.C.; Raffaele, N.B. High-throughput five minute microwave accelerated glycosylation approach to the synthesis of nucleoside libraries. J. Org. Chem. 2007, 72, 173–179. [Google Scholar] [CrossRef]
- Shirae, H.; Yokozeki, K.; Kubota, K. Enzymatic production of Ribavirin. Agric. Biol. Chem. 1988, 52, 295–296. [Google Scholar] [CrossRef]
- Shirae, H.; Yokozeki, K.; Uchiyama, M.; Kubota, K. Enzymatic production of Ribavirin from purine nucleosides by Brevibacterium acetylicum ATCC 954. Agric. Biol. Chem. 1988, 52, 1777–1783. [Google Scholar] [CrossRef][Green Version]
- Hennen, W.J.; Wong, C.H. A new method for the enzymic synthesis of nucleosides using purine nucleoside phosphorylase. J. Org. Chem. 1989, 54, 4692–4695. [Google Scholar] [CrossRef]
- Shirae, H.; Yokozeki, K.; Kubota, K. Adenosine phosphorolyzing enzymes from microorganisms and ribavirin production by the application of the enzyme. Agric. Biol. Chem. 1991, 55, 605–607. [Google Scholar] [CrossRef]
- Barai, V.N.; Zinchenko, A.I.; Eroshevskaya, L.A.; Kalinichenko, E.N.; Kulak, T.I.; Mikhailopulo, I.A. A universal biocatalyst for the preparation of base- and sugar-modified nucleosides via an enzymatic transglycosylation. Helv. Chim. Acta 2002, 85, 1901–1908. [Google Scholar] [CrossRef]
- Trelles, J.A.; Fernandez, M.; Lewkowicz, E.S.; Iribarren, A.M.; Sinisterra, J.V. Purine nucleoside synthesis from uridine using immobilized Enterobacter gergoviae CECT 875 whole cells. Tetrahedron Lett. 2003, 44, 2605–2609. [Google Scholar] [CrossRef]
- Konstantinova, I.D.; Leont’eva, N.A.; Galegov, G.A.; Ryzhova, O.I.; Chuvikovskii, D.V.; Antonov, K.V.; Esipov, R.S.; Taran, S.A.; Verevkina, K.N.; Feofanov, S.A.; et al. Ribavirin: Biotechnological synthesis and effect on the reproduction of Vaccinia virus. J. Bioorg. Chem. 2004, 30, 553–560. [Google Scholar] [CrossRef]
- Nobile, M.; Terreni, M.; Lewkowicz, E.; Iribarren, A.M. Aeromonas hydrophila strains as biocatalysts for transglycosylation. Biocatal. Biotransfor. 2011, 28, 395–402. [Google Scholar] [CrossRef]
- Ding, Q.B.; Ou, L.; Wei, D.Z.; Wei, X.K.; Xu, Y.M.; Zhang, C.Y. Enzymatic synthesis of nucleosides by nucleoside phosphorylase co-expressed in Escherichia coli. J. Zhejiang Univ. Sci. B 2010, 11, 880–888. [Google Scholar] [CrossRef]
- De Benedetti, E.C.; Rivero, C.W.; Trelles, J.A. Development of a nanostabilized biocatalyst using an extremophilic microorganism for ribavirin biosynthesis. J. Mol. Catal. B Enzym. 2015, 121, 90–95. [Google Scholar] [CrossRef]
- Rivero, C.W.; De Benedetti, E.C.; Lozano, M.E.; Trelles, J.A. Bioproduction of ribavirin by green microbial biotransformation. Process Biochem. 2015, 50, 935–940. [Google Scholar] [CrossRef]
- Shirae, H.; Yokozeki, K.; Kubota, K. Enzymic production of Ribavirin from pyrimidine nucleosides by Enterobacter aerogenes AJ 11125. Agric. Biol. Chem. 1988, 52, 1233–1237. [Google Scholar] [CrossRef]
- Palle, V.P.; Varkhedkar, V.; Ibrahim, P.; Ahmed, H.; Li, Z.; Gao, Z.; Ozeck, M.; Wu, Y.; Zeng, D.; Wu, L.; et al. Affinity and intrinsic efficacy (IE) of 5′-carbamoyl adenosine analogues for the A1 adenosine receptor—Efforts towards the discovery of a chronic ventricular rate control agent for the treatment of atrial fibrillation (AF). Bioorg. Med. Chem. Lett. 2004, 14, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Ashton, T.D.; Aumann, K.M.; Baker, S.P.; Schiesser, C.H.; Scammells, P.J. Structure-activity relationships of adenosines with heterocyclic N6-substituents. Bioorg. Med. Chem. Lett. 2007, 17, 6779–6784. [Google Scholar] [CrossRef]
- Petrelli, R.; Scortichini, M.; Belardo, C.; Boccella, S.; Luongo, L.; Capone, F.; Kachler, S.; Vita, P.; Del Bello, F.; Maione, S.; et al. Structure-based design, synthesis, and in vivo antinociceptive effects of selective A1 adenosine receptor agonists. J. Med. Chem. 2018, 61, 305–318. [Google Scholar] [CrossRef]
- Robins, M.J.; Robins, R.K. Purine nucleosides. XI. The synthesis of 2′-deoxy-9-α- and -β-d-ribofuranosylpurines and the correlation of their anomeric structure with proton magnetic resonance spectra. J. Am. Chem. Soc. 1965, 87, 4934–4940. [Google Scholar] [CrossRef]
- Christensen, L.F.; Broom, A.D.; Robins, M.J.; Bloch, A. Synthesis and biological activity of selected 2,6-disubstituted-(2-deoxy-α-and-β-d-erythro-pentofuranosyl) purines. J. Med. Chem. 1972, 15, 735–739. [Google Scholar] [CrossRef]
- Kazimierczuk, Z.; Cottam, H.B.; Revankar, G.R.; Robins, R.K. Synthesis of 2′-deoxytubercidin, 2′-deoxyadenosine, and related 2′-deoxynucleosides via a novel direct stereospecific sodium salt glycosylation procedure. J. Am. Chem. Soc. 1984, 106, 6379–6382. [Google Scholar] [CrossRef]
- Xia, R.; Chen, L.S. Efficient synthesis of Cladribine via the metal-free dioxygenation. Nucleosides Nucleotides Nucleic Acids 2015, 34, 729–735. [Google Scholar] [CrossRef]
- Xu, S.; Yao, P.; Chen, G.; Wang, H. A new synthesis of 2-chloro-2′-deoxyadenosine (Cladribine), CdA. Nucleosides Nucleotides Nucleic Acids 2011, 30, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Matyasovsky, J.; Perlikova, P.; Malnuit, V.; Pohl, R.; Hocek, M. 2-Substituted dATP derivatives as building blocks for polymerase-catalyzed synthesis of DNA modified in the minor groove. Angew. Chem. Int. Ed. 2016, 55, 15856–15859. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, A.; Shinozaki, M.; Suzuki, M.; Watanabe, K.; Miyasaka, T. A convenient method for the selective acylation of guanine nucleosides. Synthesis 1986, 1986, 385–386. [Google Scholar] [CrossRef]
- McGuinness, B.F.; Nakanishi, K.; Lipman, R.; Tomasz, M. Synthesis of guanine derivatives substituted in the O6 position by Mitomycin C. Tetrahedron Lett. 1988, 29, 4673–4676. [Google Scholar] [CrossRef]
- Zhong, M.; Nowak, I.; Robins, M.J. Regiospecific and highly stereoselective coupling of 6-(substituted-imidazol-1-yl) purines with 2-deoxy-3,5-di-O-(p-toluoyl)-α-d-erythro-pentofuranosyl chloride. Sodium-salt glycosylation in binary solvent mixtures: Improved synthesis of Cladribine. J. Org. Chem. 2006, 71, 7773–7779. [Google Scholar] [CrossRef]
- Satishkumar, S.; Vuram, P.K.; Relangi, S.S.; Gurram, V.; Zhou, H.; Kreitman, R.J.; MartínezMontemayor, M.M.; Yang, L.; Kaliyaperumal, M.; Sharma, S.; et al. Cladribine analogues via O6-(benzotriazolyl) derivatives of guanine nucleosides. Molecules 2015, 20, 18437–18463. [Google Scholar] [CrossRef] [PubMed]
- Hironori, K.; Araki, T. Efficient chemo-enzymatic syntheses of pharmaceutically useful unnatural 2′-deoxynucleosides. Nucleosides Nucleotides Nucleic Acids 2005, 24, 1127–1130. [Google Scholar] [CrossRef]
- Zhou, X.; Szeker, K.; Jiao, L.Y.; Oestreich, M.; Mikhailopulo, I.A.; Neubauer, P. Synthesis of 2,6-dihalogenated purine nucleosides by thermostable nucleoside phosphorylases. Adv. Synth. Catal. 2015, 357, 1237–1244. [Google Scholar] [CrossRef]
- Drenichev, M.S.; Alexeev, C.S.; Kurochkin, N.N.; Mikhailov, S.N. Use of nucleoside phosphorylases for the preparation of purine and pyrimidine 2′-deoxynucleosides. Adv. Synth. Catal. 2018, 360, 305–312. [Google Scholar] [CrossRef]
- Voegel, J.J.; Altorfer, M.M.; Benner, S.A. The donor-acceptor-acceptor purine analog: Transformation of 5-aza-7-deaza-1H-isoguanine (=4-aminoimidazo-[1,2-α]-1,3,5-triazin-2(1H)-one) to 2′-deoxy-5-aza-7-deaza-isoguanosine using purine nucleoside phosphorylase. Helv. Chim. Acta 1993, 76, 2061–2069. [Google Scholar] [CrossRef]
- Kamel, S.; Yehia, H.; Neubauer, P.; Wagner, A. Enzymatic synthesis of nucleoside analogues by nucleoside phosphorylases. In Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives, 1st ed.; Fernández-Lucas, J., Camarasa Rius, M.J., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2018; Chapter 1; pp. 1–20. [Google Scholar]
- Acosta, J.; del Arco, J.; Martinez-Pascual, S.; Clemente-Suárez, V.X.; Fernández-Lucas, J. One-pot multi-enzymatic production of purine derivatives with application in pharmaceutical and food industry. Catalysts 2018, 8, 9. [Google Scholar] [CrossRef]
- Gruen, M.; Becker, C.; Beste, A.; Siethoff, C.; Scheidig, A.J.; Goody, R.S. Synthesis of 2′-iodo- and 2′-bromo-ATP and GTP analogues as potential phasing tools for X-ray crystallography. Nucleosides Nucleotides Nucleic Acids 1999, 18, 137–151. [Google Scholar] [CrossRef]
- Borrmann, T.; Abdelrahman, A.; Volpini, R.; Lambertucci, C.; Alksnis, E.; Gorzalka, S.; Knospe, M.; Schiedel, A.C.; Cristalli, G.; Mueller, C.E. Structure-activity relationships of adenine and deazaadenine derivatives as ligands for adenine receptors, a new purinergic receptor family. J. Med. Chem. 2009, 52, 5974–5989. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.D.; Secunda, S.; Daly, J.W.; Olsson, R.A. N6-9-Disubstituted adenines: Potent, selective antagonists at the A1 adenosine receptor. J. Med. Chem. 1991, 34, 2877–2882. [Google Scholar] [CrossRef] [PubMed]


| X 2 | B-H 2 | Product | Conversion |
|---|---|---|---|
| OH | 15 | 1 (Ribavirin) | 67% |
| OH | 14 | 2 (Tecadenoson) | 49% |
| H | 11 | 3 (Cladribine) | 56% 3 |
| OH | 12 | 17 (CCPA) | 52% 3 |
| OH | 16 | 18 (Acadesine) | n.d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabuffetti, M.; Bavaro, T.; Semproli, R.; Cattaneo, G.; Massone, M.; Morelli, C.F.; Speranza, G.; Ubiali, D. Synthesis of Ribavirin, Tecadenoson, and Cladribine by Enzymatic Transglycosylation. Catalysts 2019, 9, 355. https://doi.org/10.3390/catal9040355
Rabuffetti M, Bavaro T, Semproli R, Cattaneo G, Massone M, Morelli CF, Speranza G, Ubiali D. Synthesis of Ribavirin, Tecadenoson, and Cladribine by Enzymatic Transglycosylation. Catalysts. 2019; 9(4):355. https://doi.org/10.3390/catal9040355
Chicago/Turabian StyleRabuffetti, Marco, Teodora Bavaro, Riccardo Semproli, Giulia Cattaneo, Michela Massone, Carlo F. Morelli, Giovanna Speranza, and Daniela Ubiali. 2019. "Synthesis of Ribavirin, Tecadenoson, and Cladribine by Enzymatic Transglycosylation" Catalysts 9, no. 4: 355. https://doi.org/10.3390/catal9040355
APA StyleRabuffetti, M., Bavaro, T., Semproli, R., Cattaneo, G., Massone, M., Morelli, C. F., Speranza, G., & Ubiali, D. (2019). Synthesis of Ribavirin, Tecadenoson, and Cladribine by Enzymatic Transglycosylation. Catalysts, 9(4), 355. https://doi.org/10.3390/catal9040355








