Site-Specific Addressing of Particles and Coatings via Enzyme-Mediated Destabilization
Abstract
1. Introduction
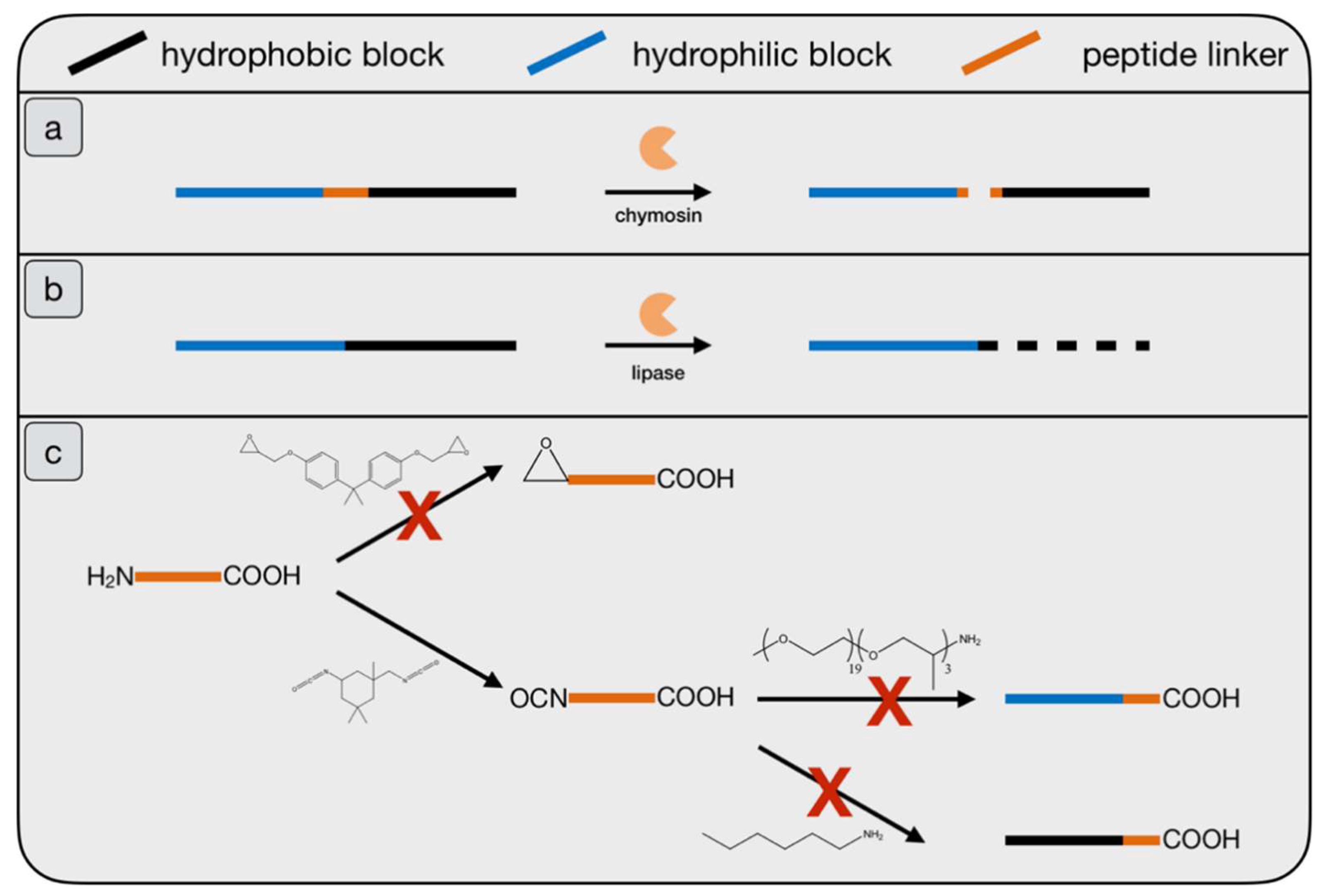
1.1. Enzyme Mediated Addressing
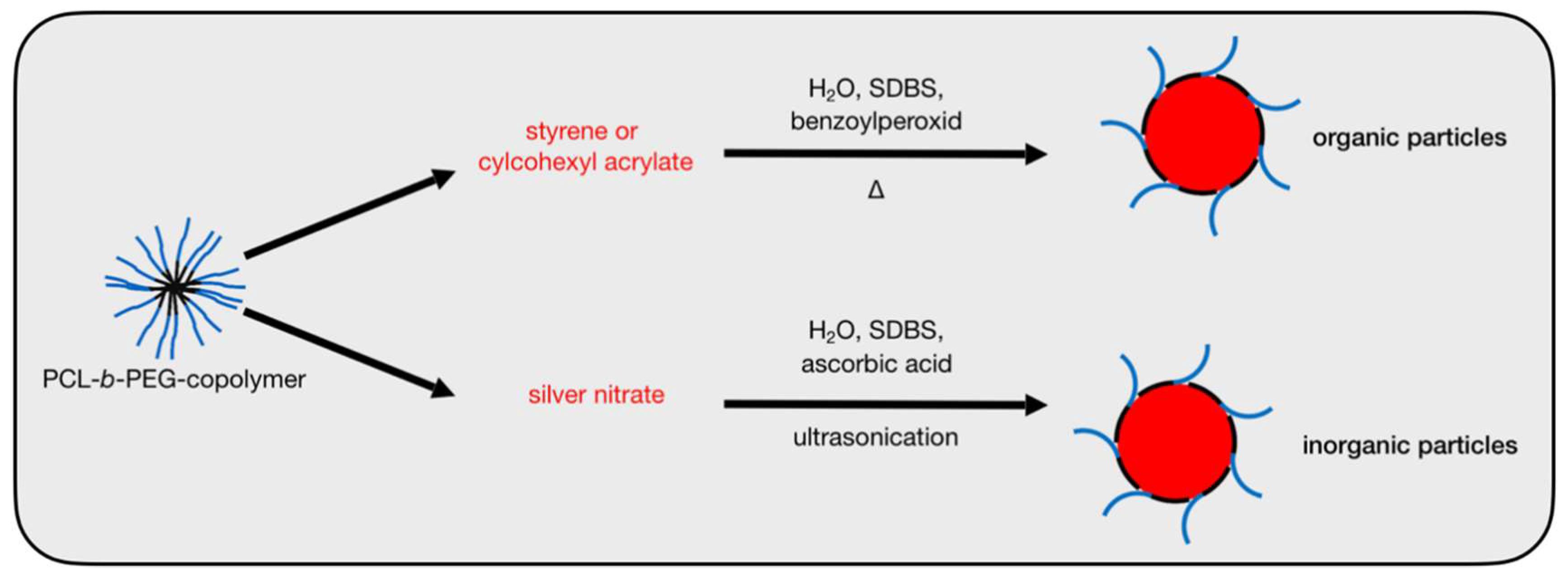
1.2. Biomimetic Hybrid Particles
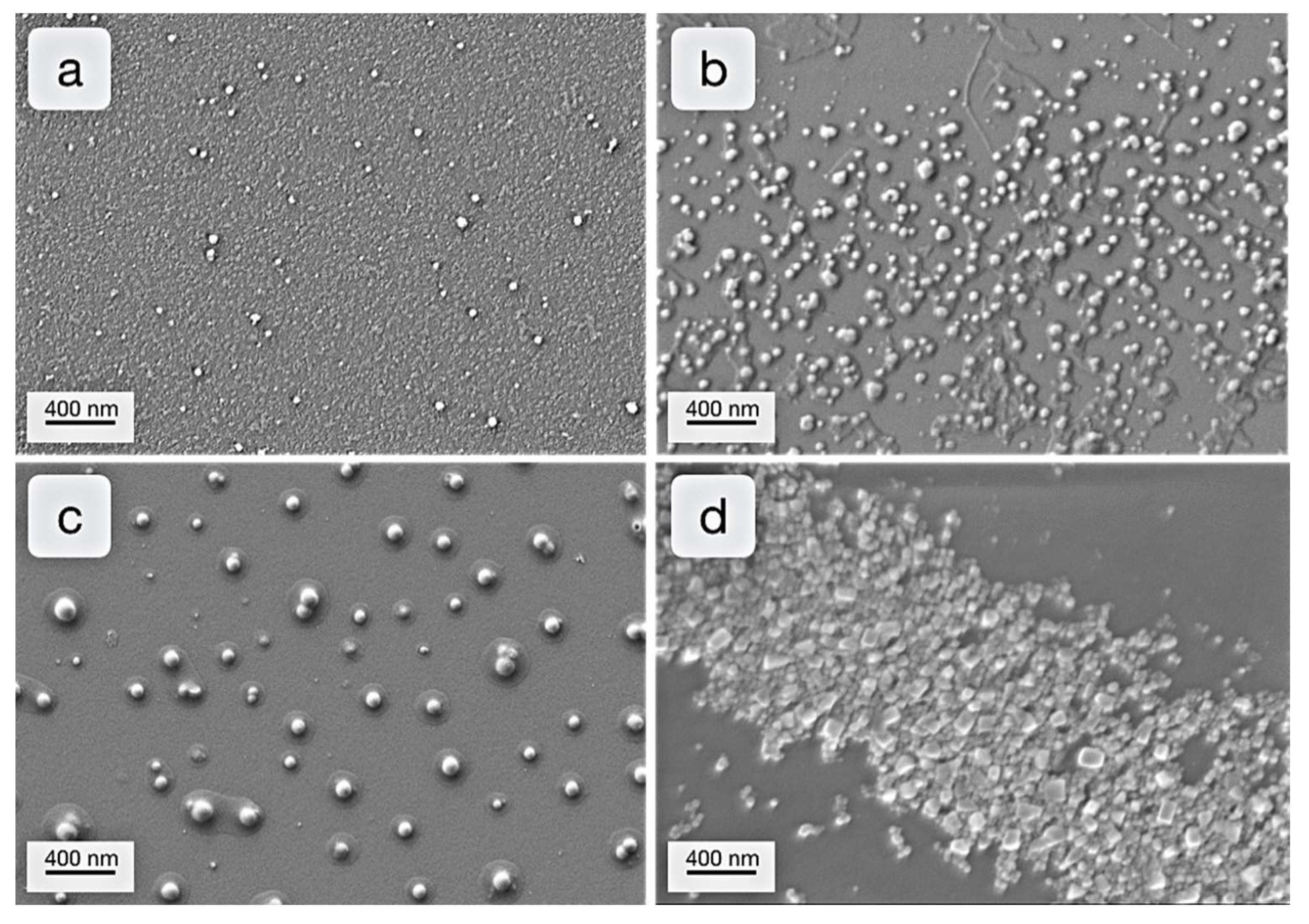
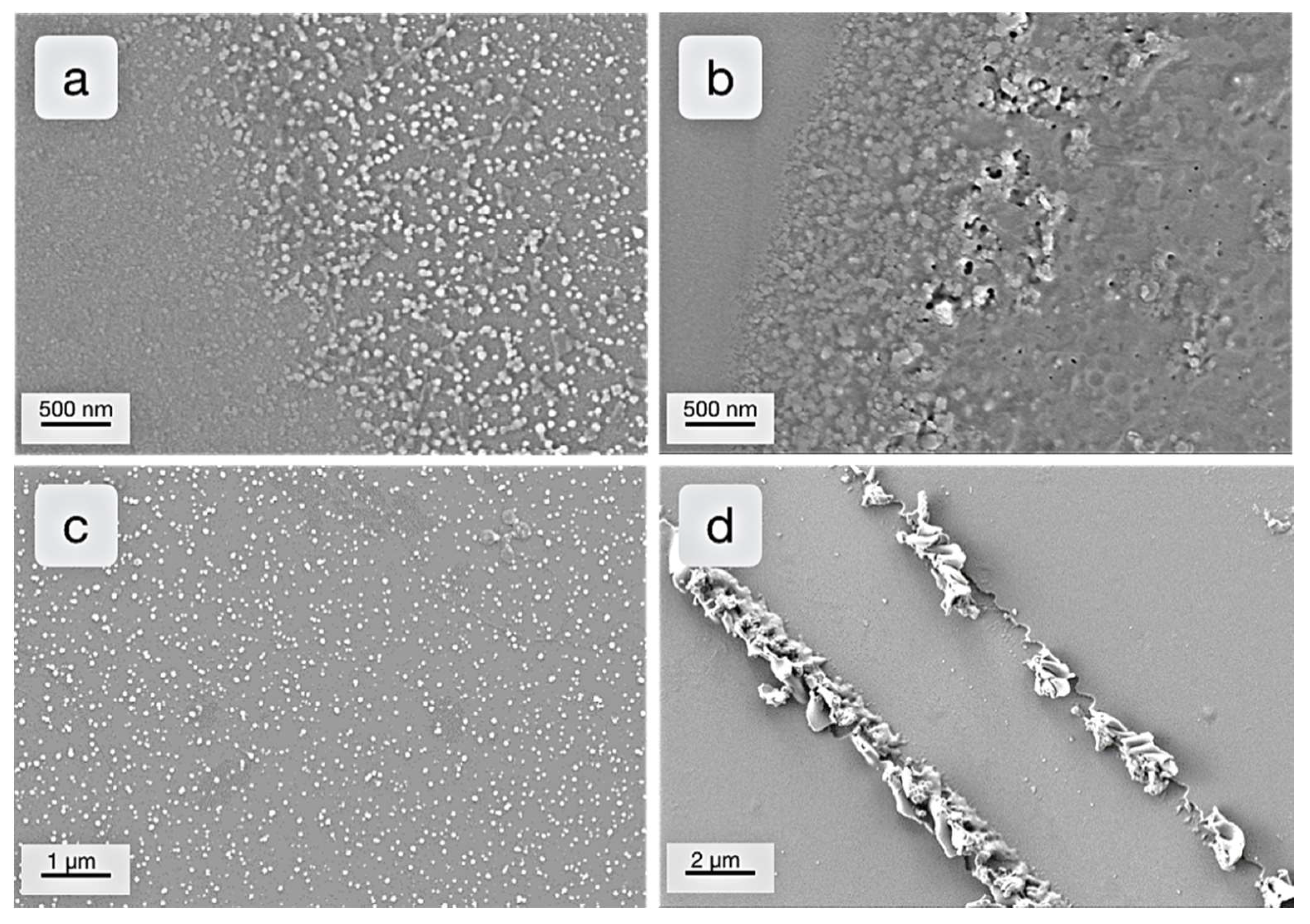
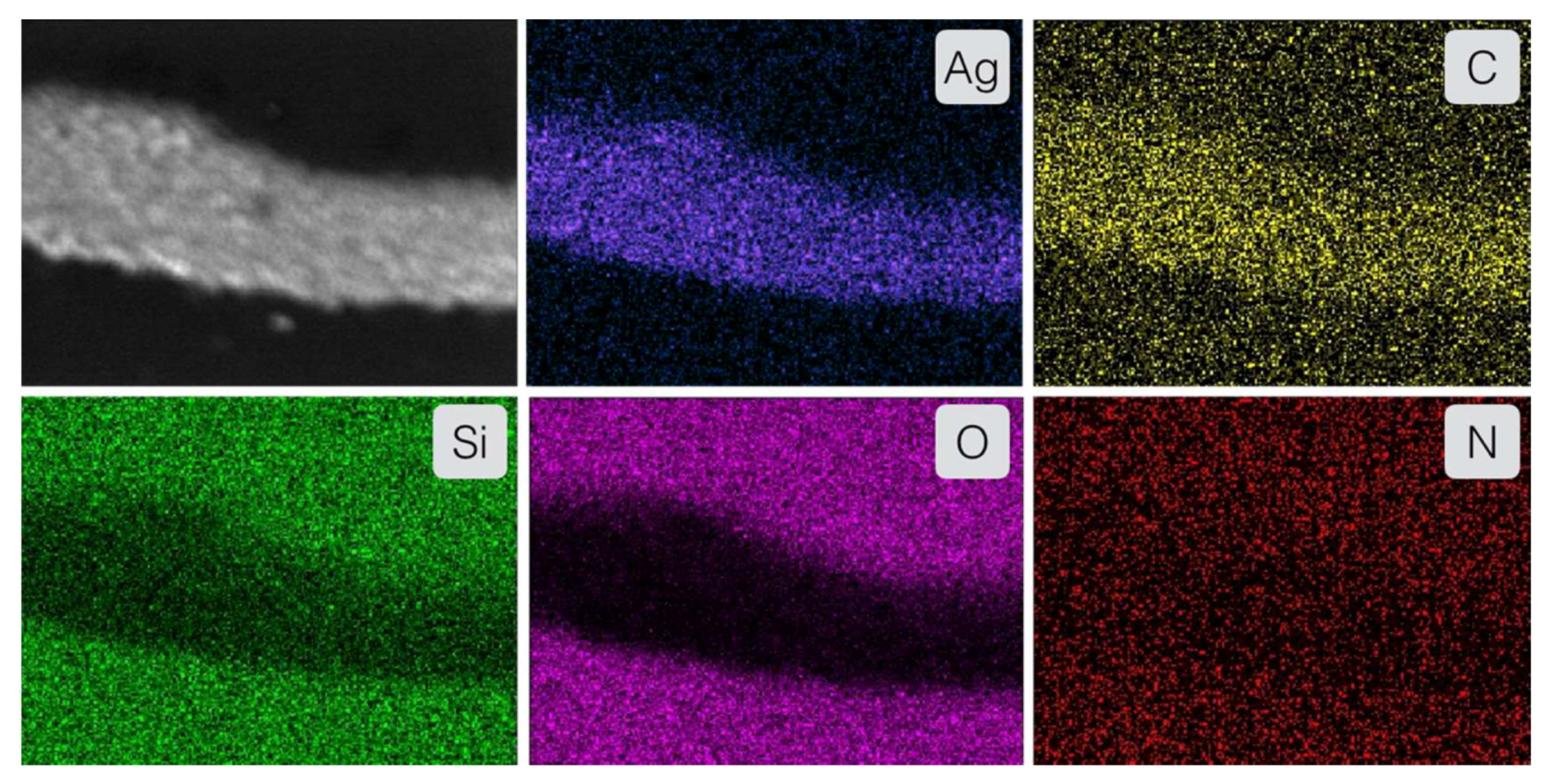
2. Results & Discussion
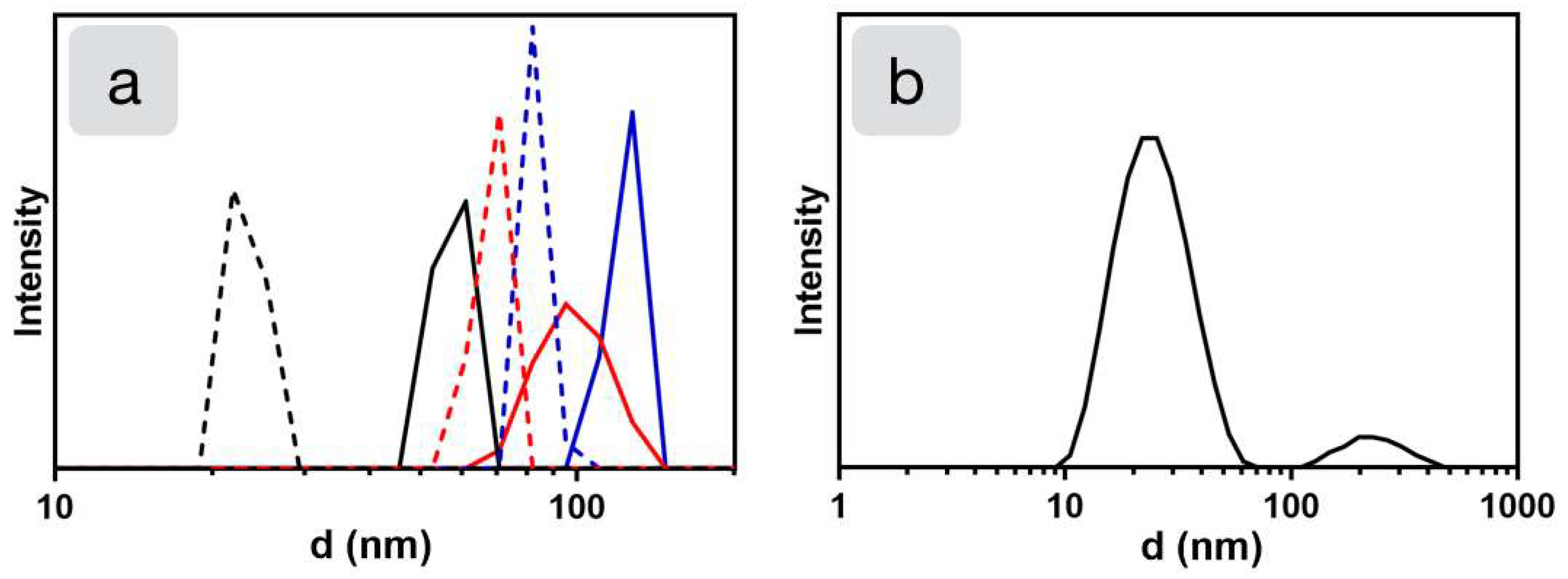
2.1. Examination of Stabilizing Block Copolymers
2.2. Synthesis of Nanoparticles Stabilized by Bomimetic Micelles
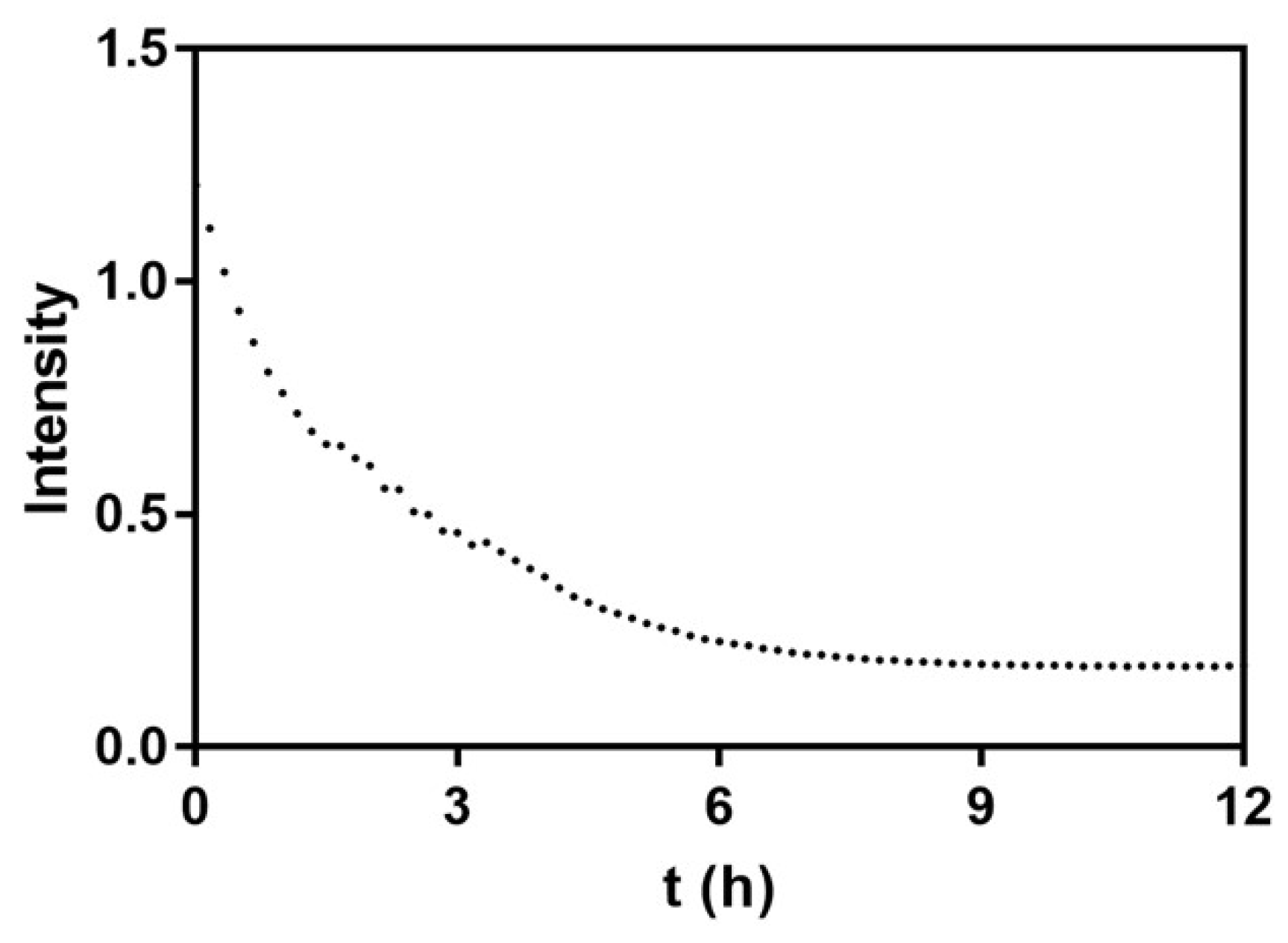
2.3. Enzyme-Medatied Addressing
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Peptide Linked Stabilizers
3.3. PEG-b-PCL Stabilised Polymer Particle Syntheses
3.4. PEG-b-PCL Stabilised Anorganic Particle Syntheses
3.5. Preparation of Supports
3.6. Adsorptive Enzyme Functionalization of Supports
3.7. Covalent Enzyme Functionalization of Supports
3.8. Preparation of the Precursor Solution
3.9. Coating of the Supports
3.10. Scanning Electron Microscopy (SEM) & Energy-Dispersive X-ray Spectroscopy (EDX)
3.11. Dynamic Light Scattering (DLS) and Zeta-Potential
3.12. UV/Vis Spectroscopy
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wanieck, K.; Fayemi, P.-E.; Maranzana, N.; Zollfrank, C.; Jacobs, S. Biomimetics and Its Tools. ICE publishing. 2017. Available online: https://sam.ensam.eu/bitstream/10985/12314/3/LCPI-BBN-WANIECK-2017.pdf (accessed on 4 March 2019).
- Vincent, J.F.V. Biomimetics—A review. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2009, 223, 919–939. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Levine, M. Biomimetic Catalysis. ACS Catal. 2011, 1, 1090–1118. [Google Scholar] [CrossRef]
- Dicker, M.P.; Duckworth, P.F.; Baker, A.B.; Francois, G.; Hazzard, M.K.; Weaver, P.M. Green composites: A review of material attributes and complementary applications. Compos. Part A Appl. Sci. Manuf. 2014, 56, 280–289. [Google Scholar] [CrossRef]
- Banyard, D.A.; Bourgeois, J.M.; Widgerow, A.D.; Evans, G.R.D. Regenerative biomaterials: A review. Plast. Reconstr. Surg. 2015, 135, 1740–1748. [Google Scholar] [CrossRef]
- Dehsorkhi, A.; Hamley, I.W.; Seitsonen, J.; Ruokolainen, J. Tuning self-assembled nanostructures through enzymatic degradation of a peptide amphiphile. Langmuir 2013, 29, 6665–6672. [Google Scholar] [CrossRef] [PubMed]
- Strube, O.I.; Rüdiger, A.A.; Bremser, W. Enzymatically controlled material design with casein-from defined films to localized deposition of particles. J. Biotechnol. 2015, 201, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Ruediger, A.A.; Bremser, W.; Strube, O.I. The enzyme-mediated autodeposition of casein: Effect of enzyme immobilization on deposition of protein structures. J. Coat. Technol. Res. 2016, 13, 597–611. [Google Scholar] [CrossRef]
- Sirisha, V.L.; Jain, A.; Jain, A. Enzyme Immobilization: An Overview on Methods, Support Material, and Applications of Immobilized Enzymes. Adv. Food Nutr. Res. 2016, 79, 179–211. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ogorzalek, T.L.; Wei, S.; Zhang, X.; Yang, P.; Jasensky, J.; Brooks, C.L.; Marsh, E.N.G.; Chen, Z. Effect of immobilization site on the orientation and activity of surface-tethered enzymes. Phys. Chem. Chem. Phys. PCCP 2018, 20, 1021–1029. [Google Scholar] [CrossRef]
- Manoel, E.A.; Dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, r. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef]
- Ruediger, A.A.; Terborg, E.; Bremser, W.; Strube, O.I. Influences on the film thickness in the enzymatic autodeposition process of casein. Prog. Org. Coat. 2016, 94, 56–61. [Google Scholar] [CrossRef]
- Ruediger, A.A.; Bremser, W.; Strube, O.I. Nanoscaled Biocoatings via Enzyme Mediated Autodeposition of Casein. Macromol. Mater. Eng. 2016, 301, 1181–1190. [Google Scholar] [CrossRef]
- Dalgleish, D.G. Proteolysis and aggregation of casein micelles treated with immobilized or soluble chymosin. J. Dairy Res. 1979, 46, 653. [Google Scholar] [CrossRef]
- Rüdiger, A.A.; Brassat, K.; Lindner, J.K.N.; Bremser, W.; Strube, O.I. Easily Accessible Protein Nanostructures via Enzyme Mediated Addressing. Langmuir 2018, 34, 4264–4270. [Google Scholar] [CrossRef]
- Strube, O.I.; Büngeler, A.; Bremser, W. Site-specific in situ synthesis of eumelanin nanoparticles by an enzymatic autodeposition-like process. Biomacromolecules 2015, 16, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Strube, O.I.; Büngeler, A.; Bremser, W. Enzyme-Mediated In Situ Synthesis and Deposition of Nonaggregated Melanin Protoparticles. Macromol. Mater. Eng. 2016, 301, 801–804. [Google Scholar] [CrossRef]
- Appel, D.; Wedegärtner, D.; Lüther, M.; Strube, O.I. Enzyme-Mediated In Situ Buildup and Site-Specific Addressing of Polymeric Coatings. Macromol. Mater. Eng. 2019, 304, 1800536. [Google Scholar] [CrossRef]
- Visser, S.; van Rooijen, P.J.; Schattenkerk, C.; Kerling, K.E. Peptide substrates for chymosin (rennin). Kinetic studies with peptides of different chain length including parts of the sequence 101-112 of bovine k-casein. Biochim. Biophys. Acta 1976, 438, 265–272. [Google Scholar] [CrossRef]
- Reents, R.; Jeyaraj, D.A.; Waldmann, H. Enzymatically cleavable linker groups in polymer-supported synthesis. Drug. Discov. Today 2002, 7, 71–76. [Google Scholar] [CrossRef]
- Sabbioni, G.; Lamb, J.H.; Farmer, P.B.; Sepai, O. Reactions of 4 methylphenyl isocyanate with amino acids. Biomarkers 1997, 2, 223–232. [Google Scholar] [CrossRef]
- Glavas, L.; Olsén, P.; Odelius, K.; Albertsson, A.-C. Achieving micelle control through core crystallinity. Biomacromolecules 2013, 14, 4150–4156. [Google Scholar] [CrossRef]
- Gan, Z.; Yu, D.; Zhong, Z.; Liang, Q.; Jing, X. Enzymatic degradation of poly(ε-caprolactone)/poly(dl-lactide) blends in phosphate buffer solution. Polymer 1999, 40, 2859–2862. [Google Scholar] [CrossRef]
- Kulkarni, A.; Reiche, J.; Hartmann, J.; Kratz, K.; Lendlein, a. Selective enzymatic degradation of poly(ε-caprolactone) containing multiblock copolymers. Eur. J. Pharm. Biopharm. 2008, 68, 46–56. [Google Scholar] [CrossRef]
- Pencreac’h, G.; Leullier, M.; Baratti, J.C. Properties of free and immobilized lipase from Pseudomonas cepacia. Biotechnol. Bioeng. 1997, 56, 181–189. [Google Scholar] [CrossRef]
- Owen, S.C.; Chan, D.P.; Shoichet, M.S. Polymeric micelle stability. Nano Today 2012, 7, 53–65. [Google Scholar] [CrossRef]
- Bon, S.A.F.; Bosveld, M.; Klumperman, B.; German, A.L. Controlled Radical Polymerization in Emulsion. Macromolecules 1997, 30, 324–326. [Google Scholar] [CrossRef]
- Piirma, I. Nucleation in Monomer Droplets. In Emulsion Polymerization; Academic Press: New York, NY, USA, 1982; pp. 86–91. [Google Scholar]
- Moghimi-Rad, J.; Isfahani, T.D.; Hadi, I.; Ghalamdaran, S.; Sabbaghzadeh, J.; Sharif, M. Shape-controlled synthesis of silver particles by surfactant self-assembly under ultrasound radiation. Appl. Nanosci 2011, 1, 27–35. [Google Scholar] [CrossRef]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Mateo, C.; Fernández-Lorente, G.; Abian, O.; Fernández-Lafuente, R.; Guisán, J.M. Multifunctional Epoxy Supports: A New Tool To Improve the Covalent Immobilization of Proteins. The Promotion of Physical Adsorptions of Proteins on the Supports before Their Covalent Linkage. Biomacromolecules 2000, 1, 739–745. [Google Scholar] [CrossRef]
- Petrović, J.; Clark, R.A.; Yue, H.; Waldeck, D.H.; Bowden, E.F. Impact of surface immobilization and solution ionic strength on the formal potential of immobilized cytochrome C. Langmuir 2005, 21, 6308–6316. [Google Scholar] [CrossRef]
- Branco, R.V.; Gutarra, M.L.E.; Guisan, J.M.; Freire, D.M.G.; Almeida, R.V.; Palomo, J.M. Improving the thermostability and optimal temperature of a lipase from the hyperthermophilic archaeon Pyrococcus furiosus by covalent immobilization. BioMed. Res. Int. 2015, 2015, 250532. [Google Scholar] [CrossRef]
- Aldrich, S. Thermal Transitions of Homopolymers: Glass Transition & Melting Point. Available online: https://www.sigmaaldrich.com/technical-documents/articles/materials-science/polymer-science/thermal-transitions-of-homopolymers.html (accessed on 4 March 2019).



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wedegärtner, D.; Strube, O.I. Site-Specific Addressing of Particles and Coatings via Enzyme-Mediated Destabilization. Catalysts 2019, 9, 354. https://doi.org/10.3390/catal9040354
Wedegärtner D, Strube OI. Site-Specific Addressing of Particles and Coatings via Enzyme-Mediated Destabilization. Catalysts. 2019; 9(4):354. https://doi.org/10.3390/catal9040354
Chicago/Turabian StyleWedegärtner, David, and Oliver I. Strube. 2019. "Site-Specific Addressing of Particles and Coatings via Enzyme-Mediated Destabilization" Catalysts 9, no. 4: 354. https://doi.org/10.3390/catal9040354
APA StyleWedegärtner, D., & Strube, O. I. (2019). Site-Specific Addressing of Particles and Coatings via Enzyme-Mediated Destabilization. Catalysts, 9(4), 354. https://doi.org/10.3390/catal9040354





