Characterisation of the First Archaeal Mannonate Dehydratase from Thermoplasma acidophilum and Its Potential Role in the Catabolism of D-Mannose
Abstract
:1. Introduction
2. Results and Discussion
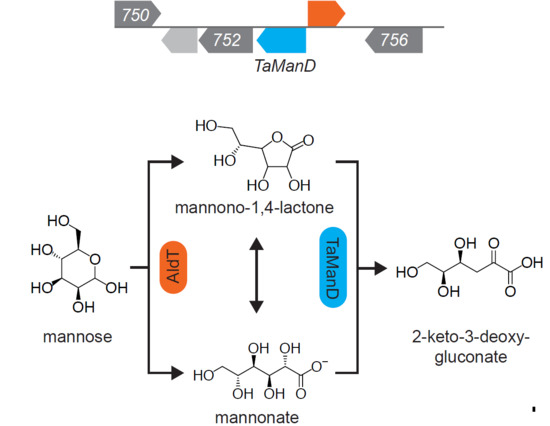
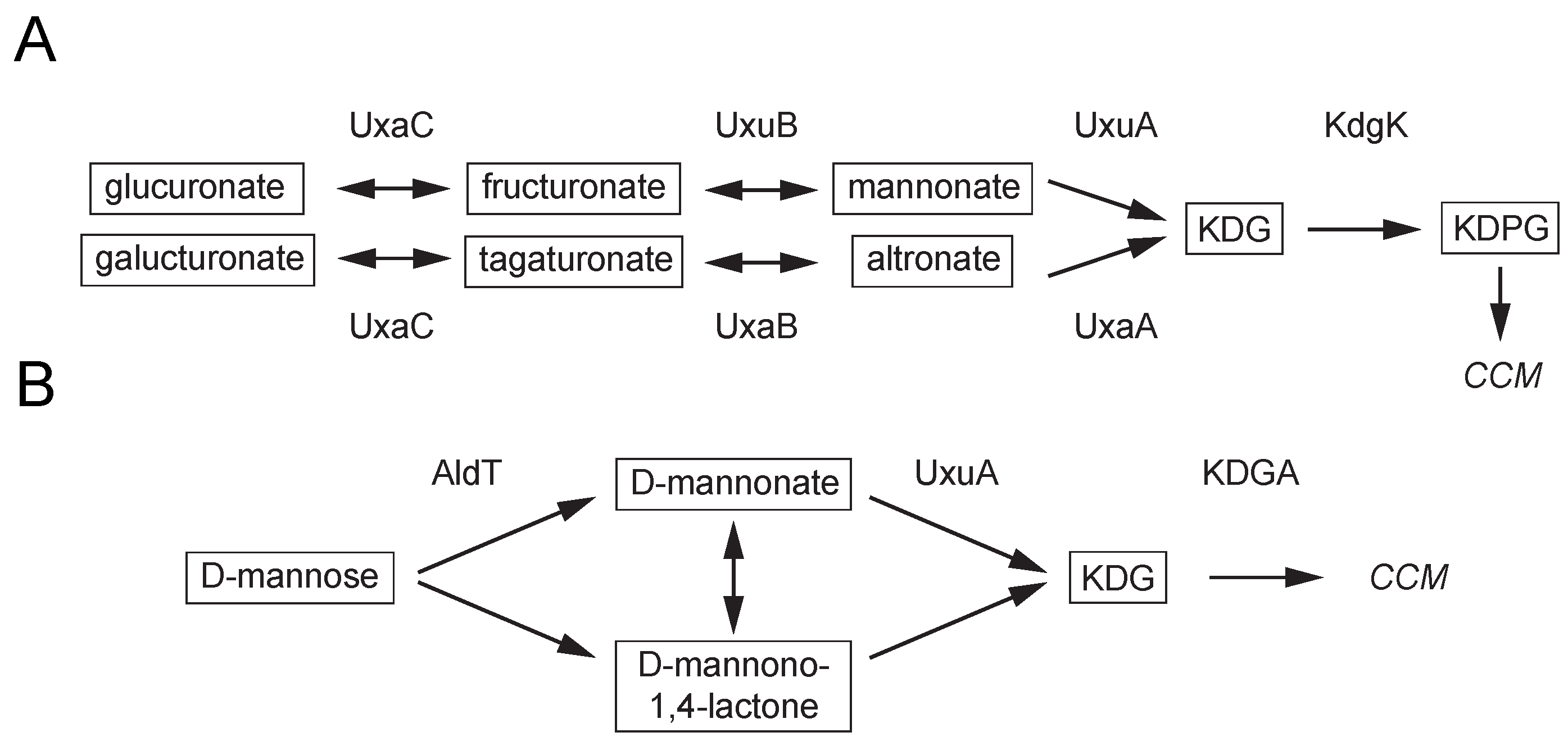
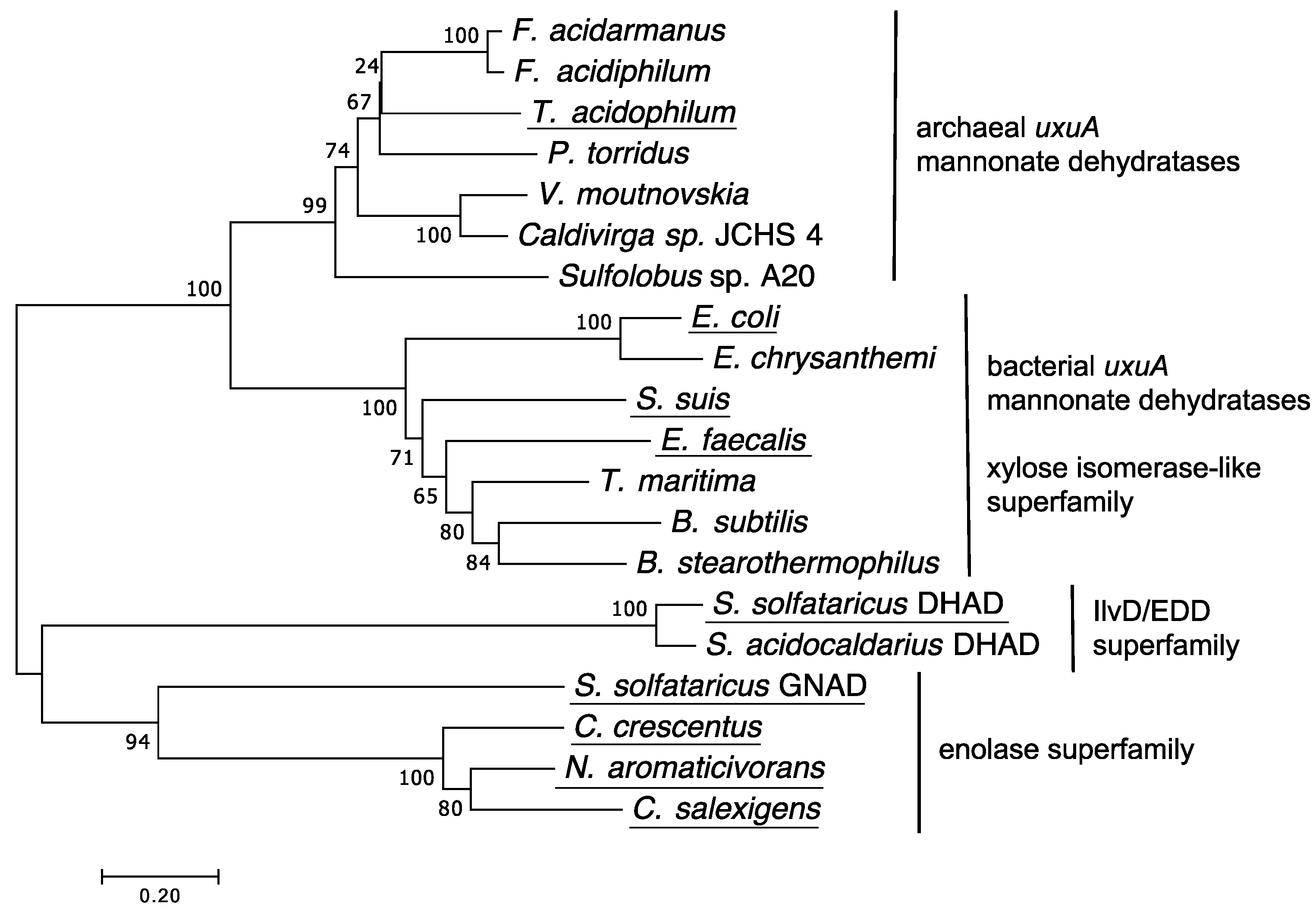
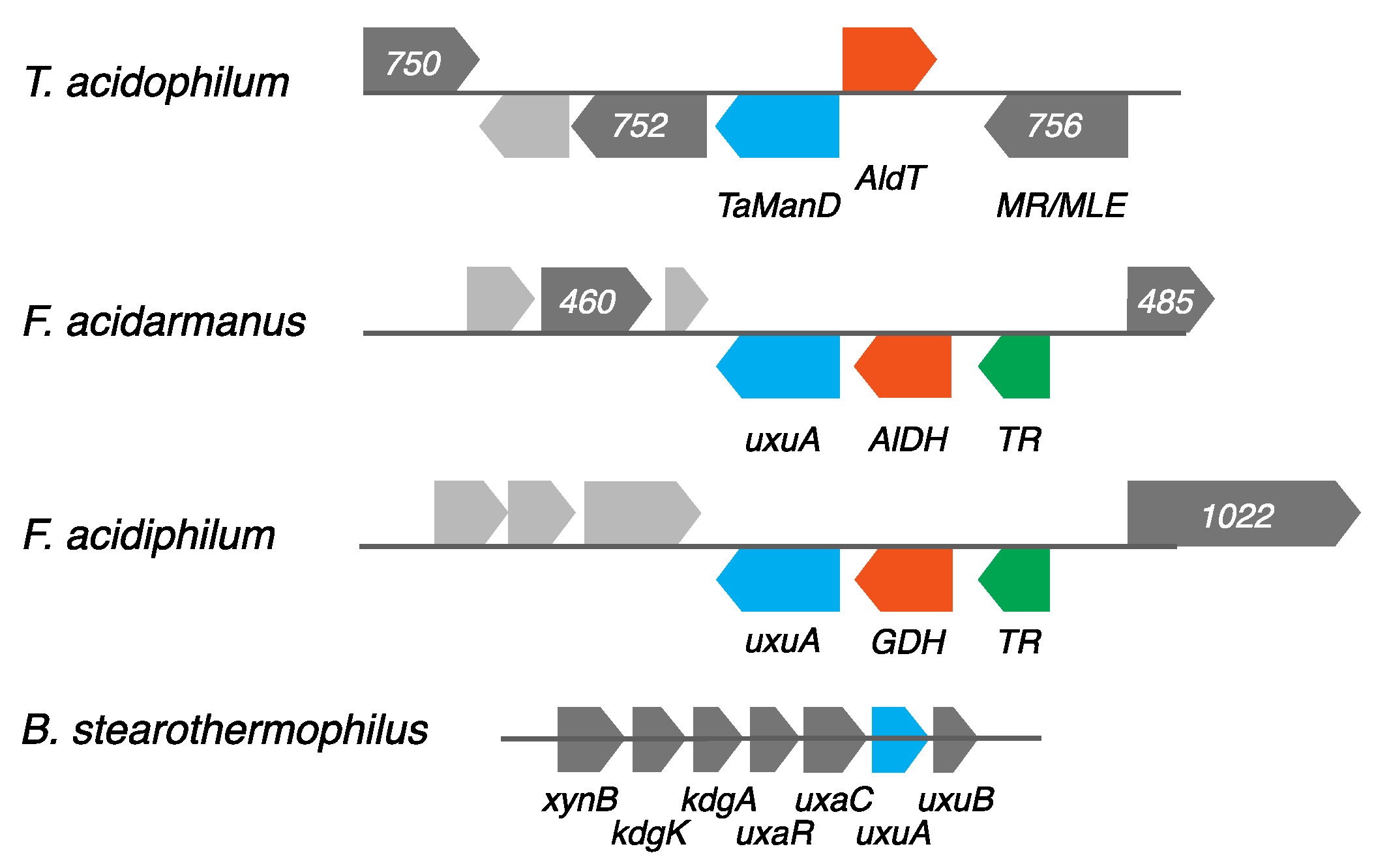
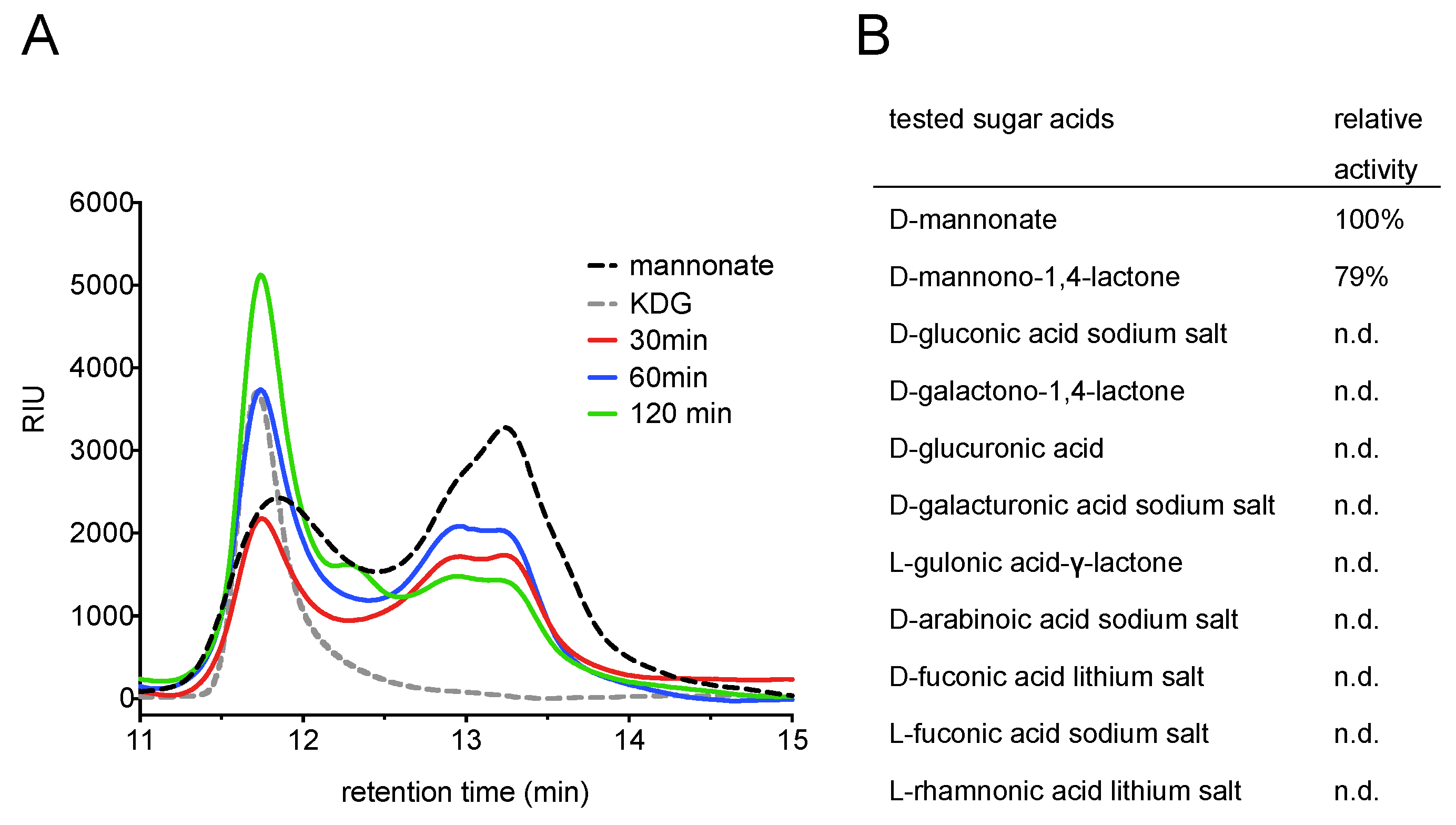
2.1. Screening for a Functional Mannonate Dehydratase in T. acidophilum
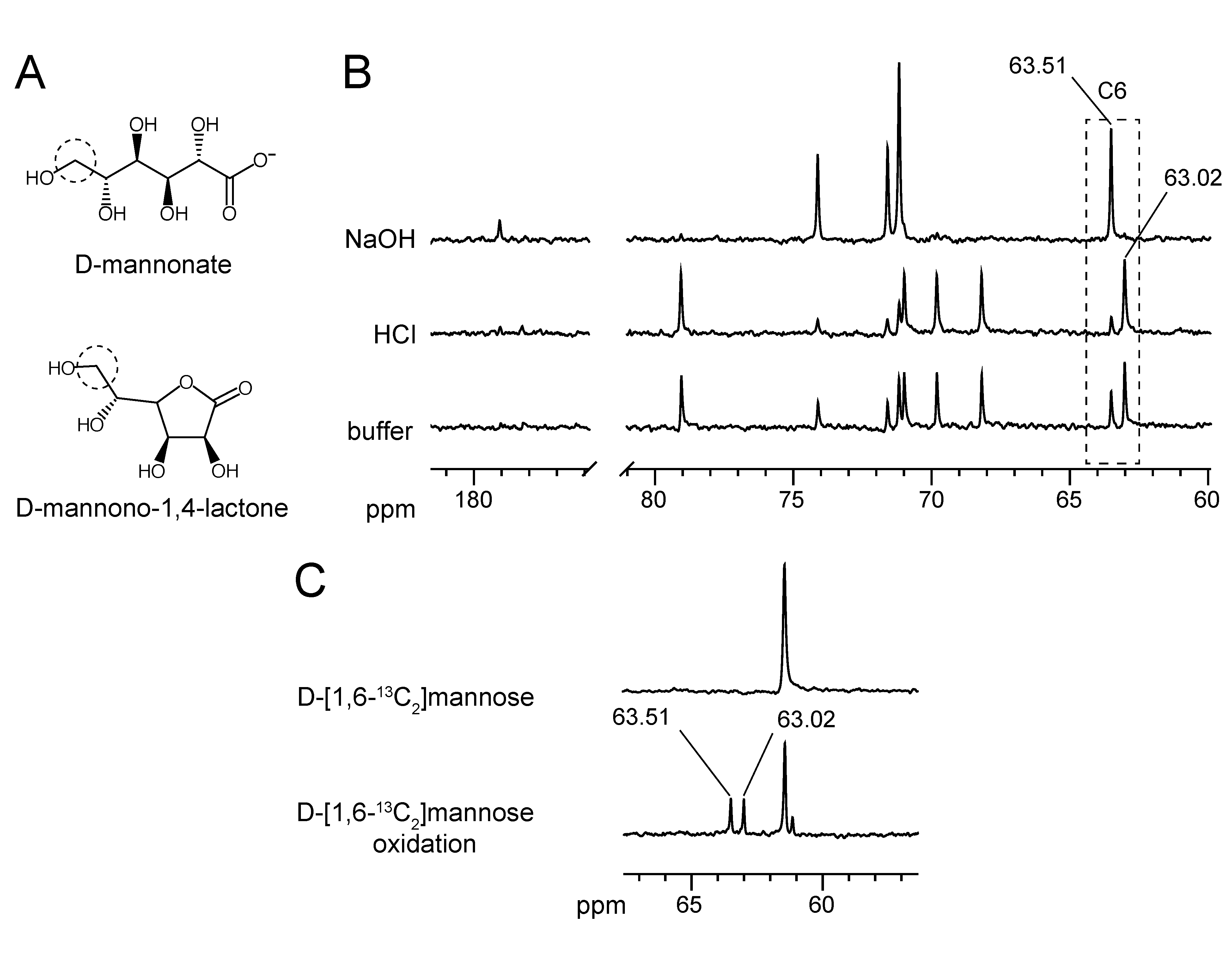
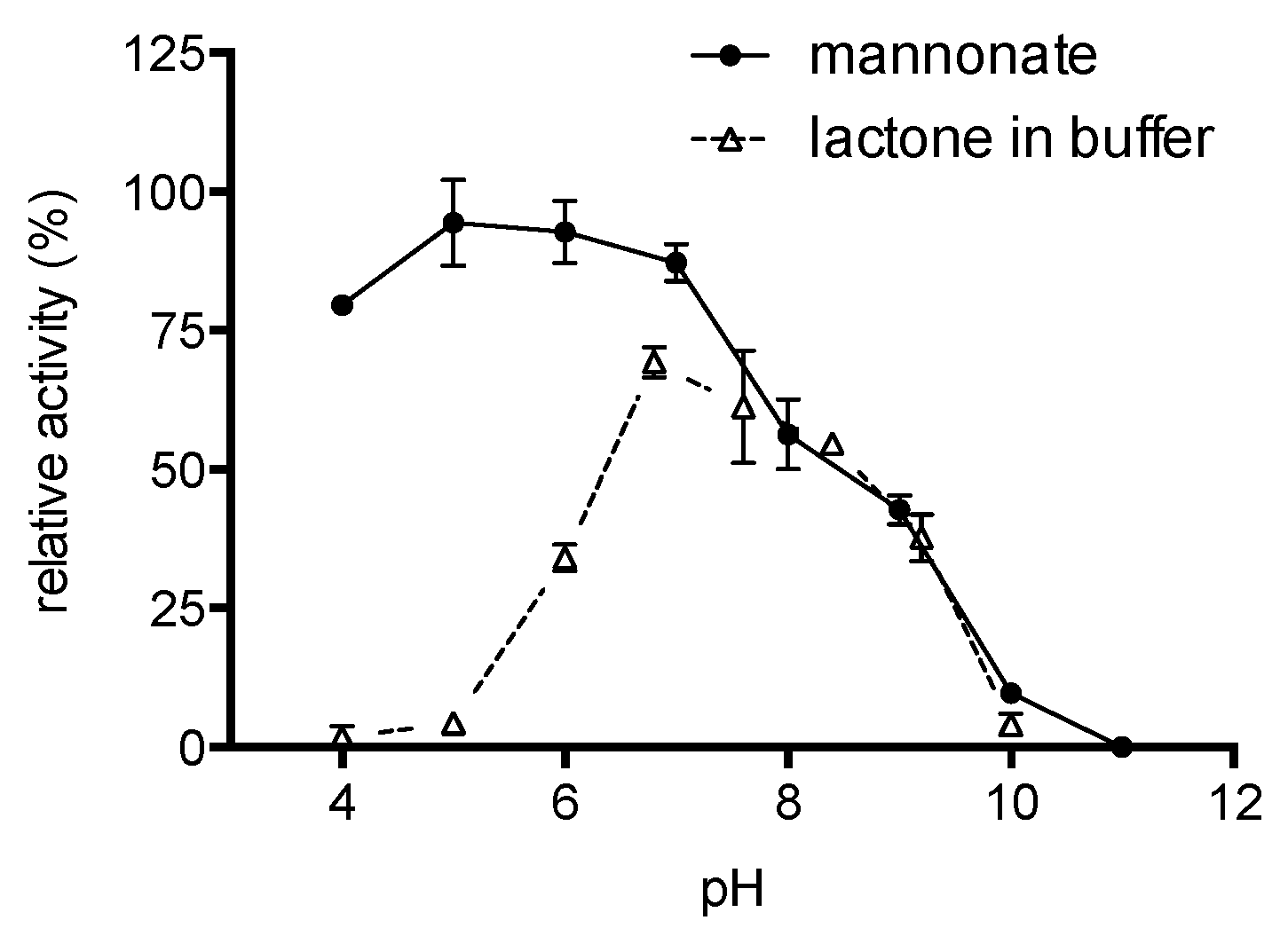
2.2. Substrate Conformation for TaManD Activity
2.3. Expression and Purification of TaManD
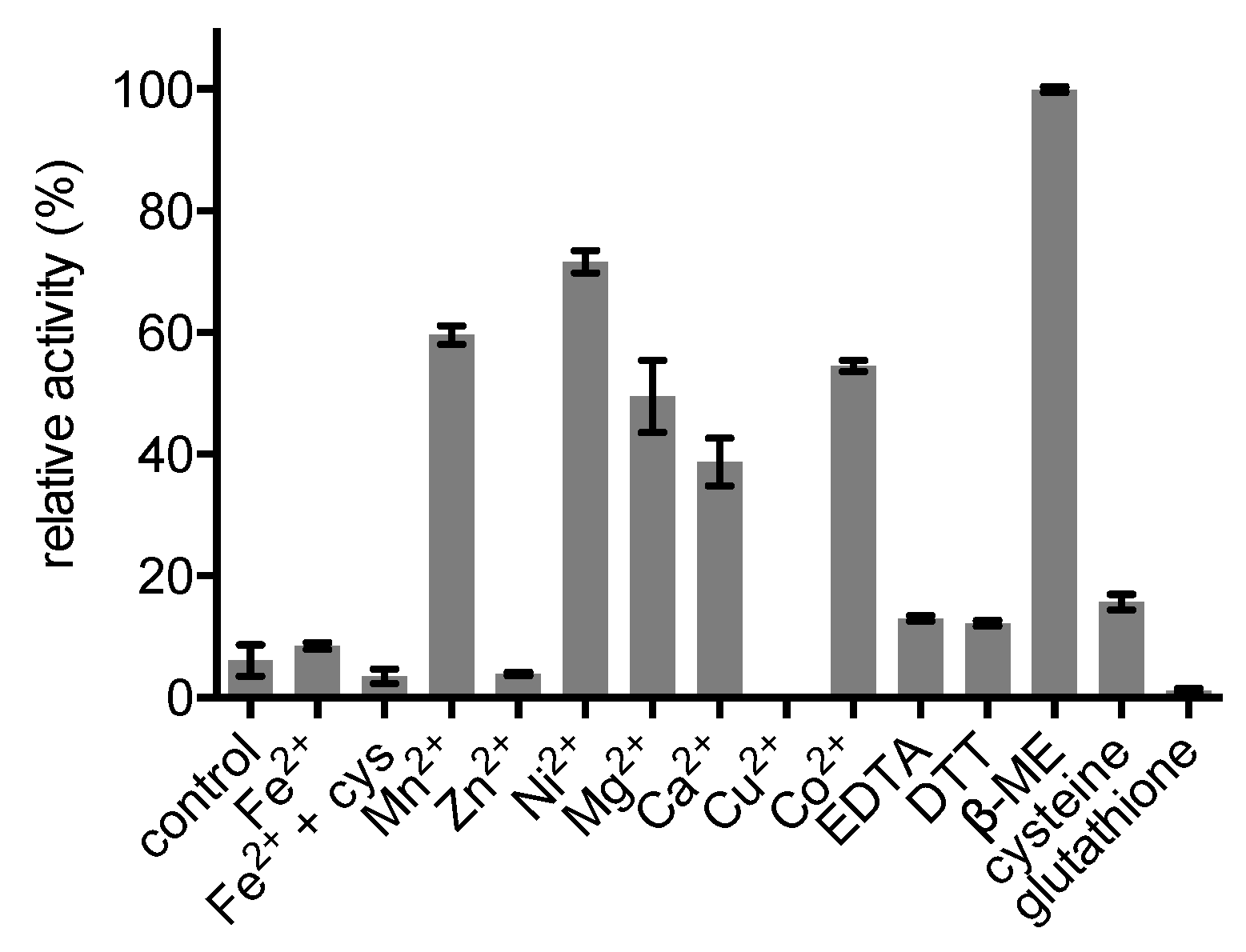
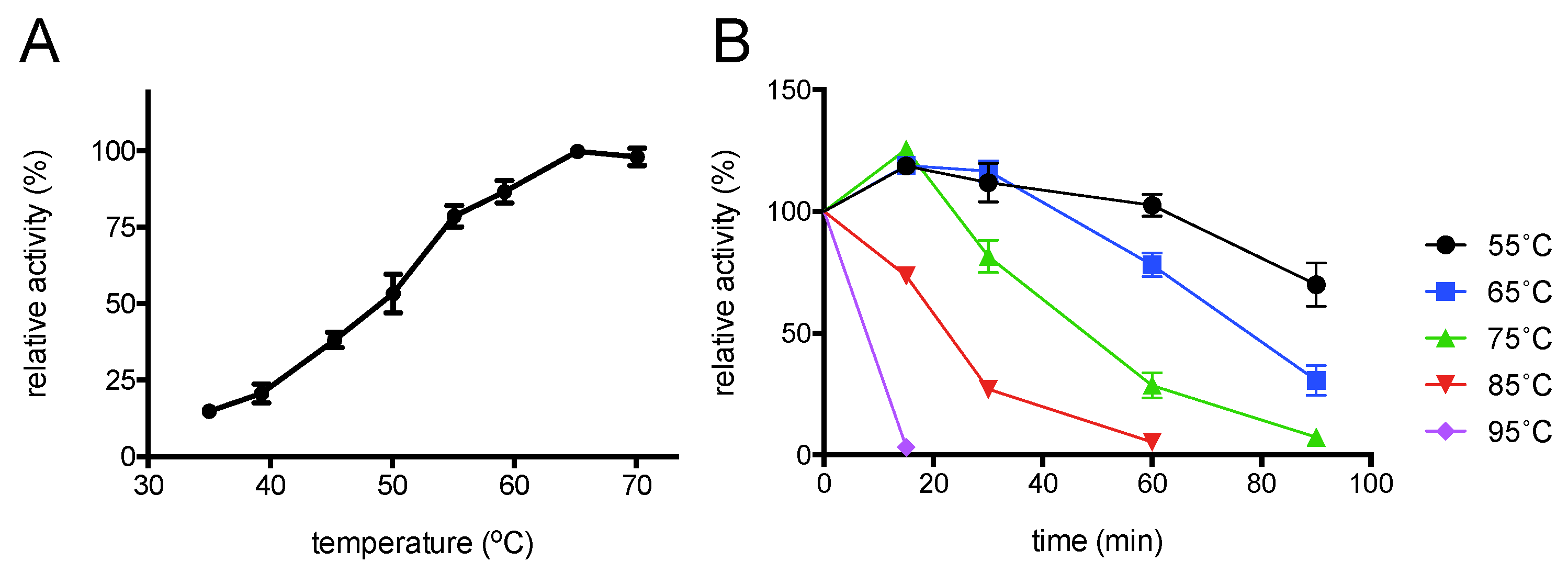
2.4. Enzyme Characterisation
2.5. Product Identification and Substrate Specificity
3. Materials and Methods
3.1. Cloning, Expression and Purification of Enzymes
3.2. Protein Analysis
3.3. Nuclear Magentic Resonance (NMR) Analysis
3.4. Enzyme Activity
3.5. High Performance Liquid Chromatography (HPLC) Analysis
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Robert-Baudouy, J.; Jimeno-Abendano, J.; Stoeber, F. D-Mannonate and D-altronate dehydratases of Escherichia coli K12. Methods Enzymol. 1982, 90, 288–294. [Google Scholar] [PubMed]
- Shulami, S.; Gat, O.; Sonenshein, A.L.; Shoham, Y. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 1999, 181, 3695–3704. [Google Scholar] [PubMed]
- Mekjian, K.R.; Bryan, E.M.; Beall, B.W.; Moran, C.P. Regulation of hexuronate utilization in Bacillus subtilis. J. Bacteriol. 1999, 181, 426–433. [Google Scholar] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Robert-Baudouy, J. Hexuronate catabolism in Erwinia Chrysanthemi. J. Bacteriol. 1987, 169, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Mandrand-Berthelot, M.-A.; Condemine, G.; Hugouvieux-Cotte-Pattat, N. Catabolism of hexuronides, hexuronates, aldonates, and aldarates. EcoSal Plus 2004, 1, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Reis, D.; Vian, B.; Roland, J.C. Cellulose-glucuronoxylans and plant cell wall structure. Micron 1994, 25, 171–187. [Google Scholar] [CrossRef]
- Lawford, H.G.; Rousseau, J.D. Fermentation of biomass-derived glucuronic acid by pet expressing recombinants of E. coli B. Appl. Biochem. Biotechnol. 1997, 63–65, 221–241. [Google Scholar] [CrossRef]
- Peekhaus, N.; Conway, T. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 1998, 180, 3495–3502. [Google Scholar] [PubMed]
- Chang, D.-E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, W.E.; Stevenson, S.J.; Anderson, A.B.; Grissom, J.E.; Laux, D.C.; Cohen, P.S.; et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gao, F.; Peng, H.; Cheng, H.; Liu, Y.; Tang, J.; Thompson, J.; Wei, G.; Zhang, J.; Du, Y.; et al. Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: Genetic and biochemical evidence for a catalytic mechanism. J. Bacteriol. 2009, 191, 5832–5837. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Tao, Y.; Zhu, Y.; Yuan, Y.; Zhang, Y.; Liu, H.; Gao, Y.; Teng, M.; Niu, L. Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium. J. Struct. Biol. 2012, 180, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Wichelecki, D.J.; Balthazor, B.M.; Chau, A.C.; Vetting, M.W.; Fedorov, A.A.; Fedorov, E.V.; Lukk, T.; Patskovsky, Y.V.; Stead, M.B.; Hillerich, B.S.; et al. Discovery of function in the enolase superfamily: D-mannonate and D-gluconate dehydratases in the D-mannonate dehydratase subgroup. Biochemistry 2014, 53, 2722–2731. [Google Scholar] [CrossRef] [PubMed]
- Wichelecki, D.J.; Alyxa, J.; Vendiola, F.; Jones, A.M.; Al-obaidi, N.; Almo, S.C.; Gerlt, J.A. Investigating the physiological roles of low-efficiency D-mannonate and D-gluconate dehydratases in the enolase superfamily: pathways for the catabolism of L-gulonate and L-idonate. Biochemistry 2014, 53, 5692–5699. [Google Scholar] [CrossRef] [PubMed]
- Rakus, J.F.; Fedorov, A.A.; Fedorov, E.V.; Glasner, M.E.; Vick, J.E.; Babbitt, P.C.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: D-mannonate dehydratase from Novosphingobium aromaticivorans. Biochemistry 2007, 46, 12896–12908. [Google Scholar] [CrossRef] [PubMed]
- Nishiya, Y.; Tamura, N.; Tamura, T. Analysis of bacterial glucose dehydrogenase homologs from thermoacidophilic archaeon Thermoplasma acidophilum: finding and characterization of aldohexose dehydrogenase. Biosci. Biotechnol. Biochem. 2014, 68, 2451–2456. [Google Scholar] [CrossRef] [PubMed]
- Yasutake, Y.; Nishiya, Y.; Tamura, N.; Tamura, T. Structural Insights into unique substrate selectivity of Thermoplasma acidophilum D-aldohexose dehydrogenase. J. Mol. Biol. 2007, 367, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.B. Catalytic promiscuity in dihydroxy-acid dehydratase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Biochem. 2006, 139, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Ashwell, G. [21] Enzymes of glucuronic and galacturonic acid metabolism in bacteria. In Methods in enzymology; Academic Press: Cambridge, MA, USA, 1962; Volume 5, pp. 190–208. ISBN 0076-6879. [Google Scholar]
- Portalier, R.; Robert-Baudouy, J.; Stoeber, F. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J. Bacteriol. 1980, 143, 1095–1107. [Google Scholar] [PubMed]
- Palleroni, N.J.; Duodoroff, M. Metabolism of Carbohydrate by Pseudomonas Saccharophila III. Oxidation of Arabinose. J. Bacteriol. 1957, 74, 180–185. [Google Scholar] [PubMed]
- Watanabe, S.; Saimura, M.; Makino, K. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J. Biol. Chem. 2008, 283, 20372–20382. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef] [PubMed]
- Wałaszek, Z.; Horton, D. Conformational studies on aldonolactones by NMR spectroscopy. Conformations of d-glucono-, D-mannono, D-gulono-, D-galactono-1,4-lactone in solution. Carbohydr. Res. 1982, 105, 131–143. [Google Scholar] [CrossRef]
- Dreyer, J. The role of iron in the activation of mannonic and altronic acid hydratases, two Fe-requiring hydro-lyases. Eur. J. Biochem 1987, 166, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Robert-Baudouy, J.M.; Jimeno-Abendano, J.; Stoeber, F.R. Individualité des hydrolyases mannonique et altronique chez E. coli K-12. Biochimie 1975, 57, 1–8. [Google Scholar] [CrossRef]
- Robert-Baudouy, J.M.; Stoeber, F.R. Purification et propriétés de la D-mannonate hydrolyase d’Escherichia coli. Biochim. Biophys. Acta -Enzymology 1973, 309, 473–485. [Google Scholar] [CrossRef]
- Darland, G.; Brock, T.D.; Samsonoff, W.; Conti, S.F. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science 1970, 170, 1416–1418. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, S.B. Identification and characterization of Thermoplasma acidophilum 2-keto-3-deoxy-D-gluconate kinase: A new class of sugar kinases. Biotechnol. Bioprocess Eng. 2005, 10, 535–539. [Google Scholar] [CrossRef]
- Kim, S.M.; Paek, K.H.; Lee, S.B. Characterization of NADP+-specific L-rhamnose dehydrogenase from the thermoacidophilic Archaeon Thermoplasma acidophilum. Extremophiles 2012, 16, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Reher, M.; Schönheit, P. Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner-Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett. 2006, 580, 1198–1204. [Google Scholar] [PubMed]
- Yew, W.S.; Fedorov, A.A.; Fedorov, E.V.; Rakus, J.F.; Pierce, R.W.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry 2006, 45, 14582–14597. [Google Scholar] [CrossRef] [PubMed]
- Carsten, J.M.; Schmidt, A.; Sieber, V. Characterization of recombinantly expressed dihydroxy-acid dehydratase from Sulfobus solfataricus—A key enzyme for the conversion of carbohydrates into chemicals. J. Biotechnol. 2015, 211, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 49, ISBN 978-1-936113-42-2. [Google Scholar]
- Atack, J.M.; Srikhanta, Y.N.; Fox, K.L.; Jurcisek, J.A.; Brockman, K.L.; Clark, T.A.; Boitano, M.; Power, P.M.; Jen, F.E.C.; McEwan, A.G.; et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lamble, H.J.; Milburn, C.C.; Taylor, G.L.; Hough, D.W.; Danson, M.J. Gluconate dehydratase from the promiscuous Entner-Doudoroff pathway in Sulfolobus solfataricus. FEBS Lett. 2004, 576, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, C.L.; Connaris, H.; Danson, M.J.; Reeve, C.D.; Hough, D.W. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem. J. 1999, 343, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Skoza, L.; Mohos, S. Stable thiobarbituric acid chromophore with dimethyl sulphoxide. Application to sialic acid assay in analytical de-O-acetylation. Biochem. J. 1976, 159, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Britton, H.T.S.; Robinson, R.A. CXCVIII.—Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1931, 1456–1462. [Google Scholar] [CrossRef]
- Bräsen, C.; Esser, D.; Rauch, B.; Siebers, B. Carbohydrate metabolism in archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 2014, 78, 89–175. [Google Scholar] [CrossRef] [PubMed]
| Substrate | Km (mM) | Vmax (U/mg) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| D-mannonate | 5.37 ± 0.90 | 2.39 ± 0.11 | 1.64 | 0.30 |
| D-mannono-1,4-lactone in buffer | 4.90 ± 0.53 | 1.90 ± 0.06 | 1.33 | 0.26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopp, D.; Willows, R.; Sunna, A. Characterisation of the First Archaeal Mannonate Dehydratase from Thermoplasma acidophilum and Its Potential Role in the Catabolism of D-Mannose. Catalysts 2019, 9, 234. https://doi.org/10.3390/catal9030234
Kopp D, Willows R, Sunna A. Characterisation of the First Archaeal Mannonate Dehydratase from Thermoplasma acidophilum and Its Potential Role in the Catabolism of D-Mannose. Catalysts. 2019; 9(3):234. https://doi.org/10.3390/catal9030234
Chicago/Turabian StyleKopp, Dominik, Robert Willows, and Anwar Sunna. 2019. "Characterisation of the First Archaeal Mannonate Dehydratase from Thermoplasma acidophilum and Its Potential Role in the Catabolism of D-Mannose" Catalysts 9, no. 3: 234. https://doi.org/10.3390/catal9030234
APA StyleKopp, D., Willows, R., & Sunna, A. (2019). Characterisation of the First Archaeal Mannonate Dehydratase from Thermoplasma acidophilum and Its Potential Role in the Catabolism of D-Mannose. Catalysts, 9(3), 234. https://doi.org/10.3390/catal9030234