Industrial Application of 2-Oxoglutarate-Dependent Oxygenases
Abstract
:1. Introduction
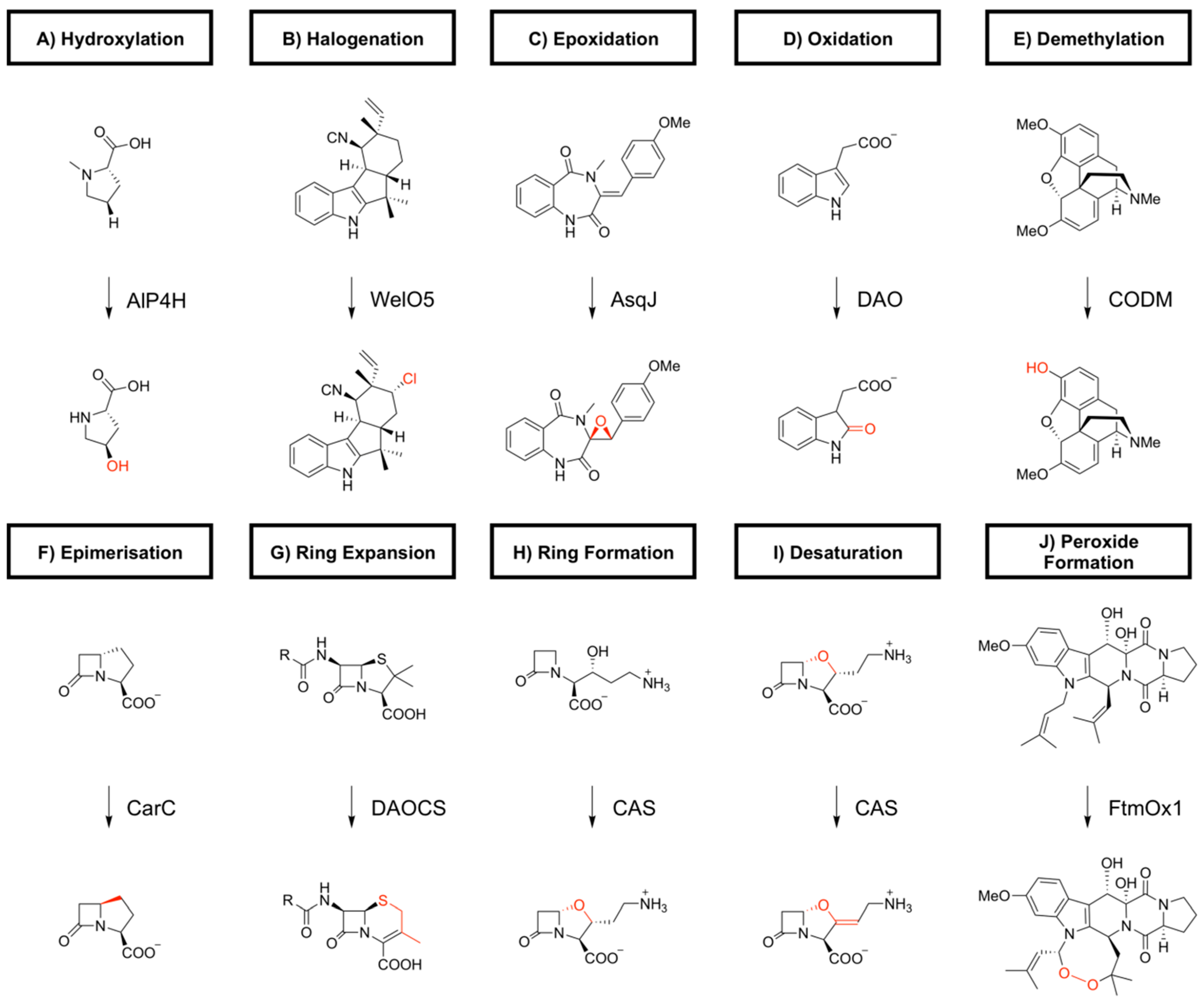
2. Biocatalytic Applications of 2-oxoglutarate-Dependent Oxygenases
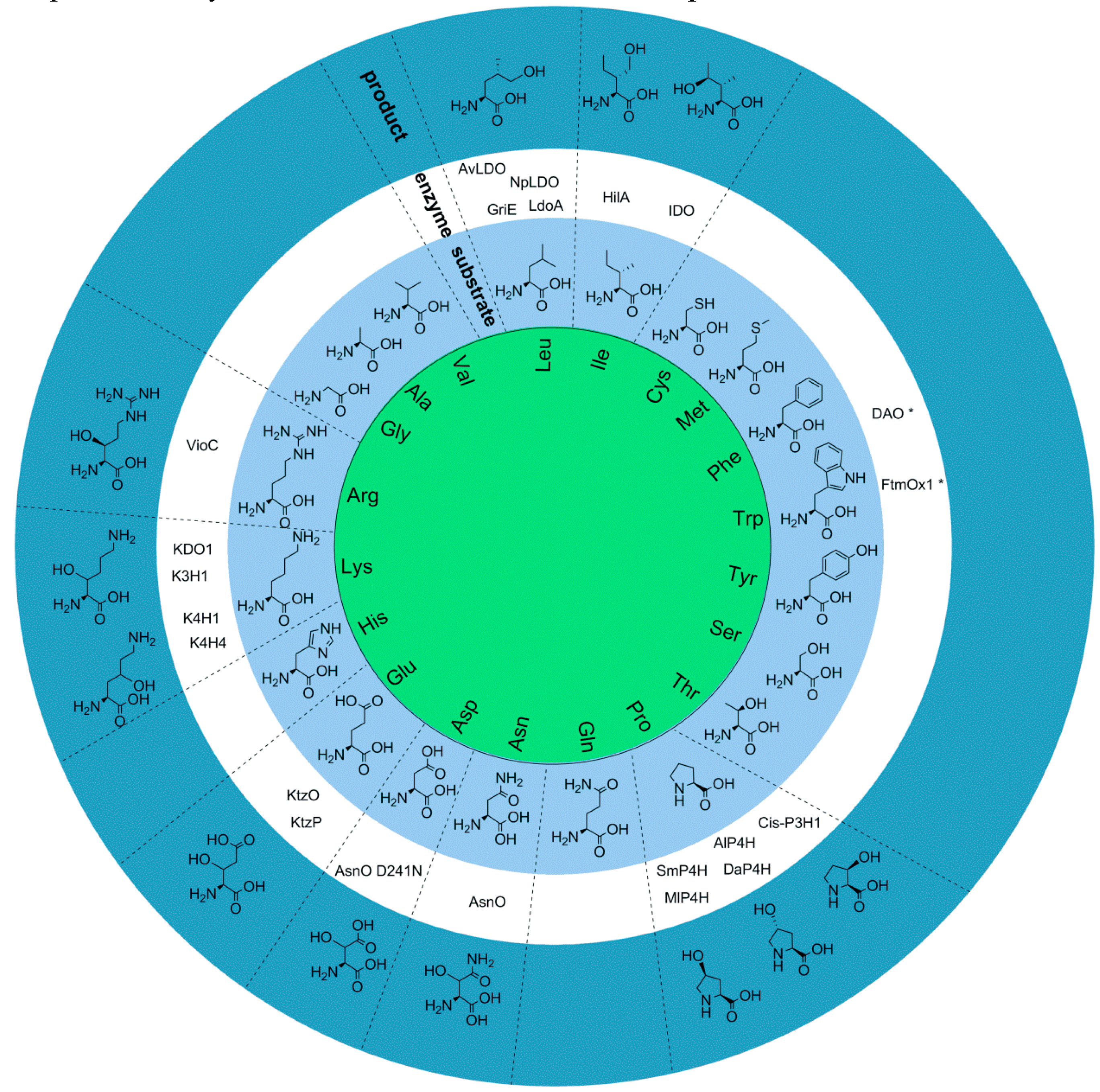
2.1. Aliphatic Amino Acid Oxygenases
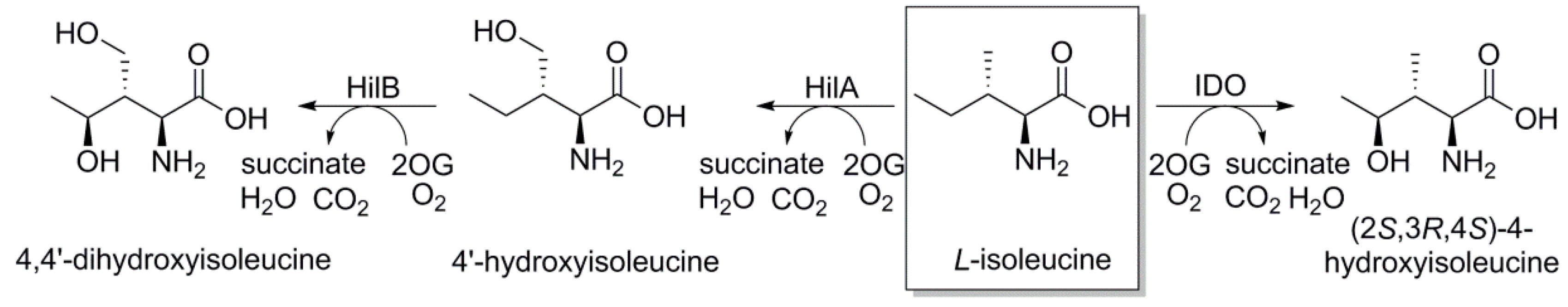
2.1.1. L-Isoleucine 4-hydroxylase or L-Isoleucine Dihydroxygenase (IDO)
2.1.2. L-Isoleucine-4′-dioxygenase
2.1.3. L-Leucine 5-hydroxylase or L-leucine Dioxygenase from Cyanobacteria
2.1.4. L-Leucine 5-hydroxylase from Streptomyces
2.1.5. N-Succinyl L-leucine 3-hydroxylase
2.2. Dioxygenase for Auxinoxidation and Fumitremorgin Oxygenases
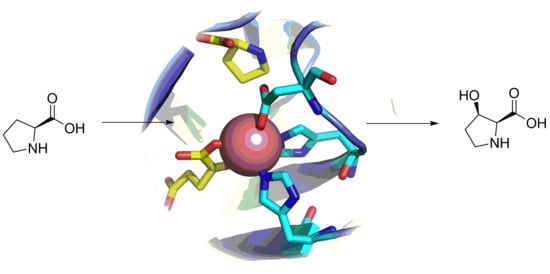
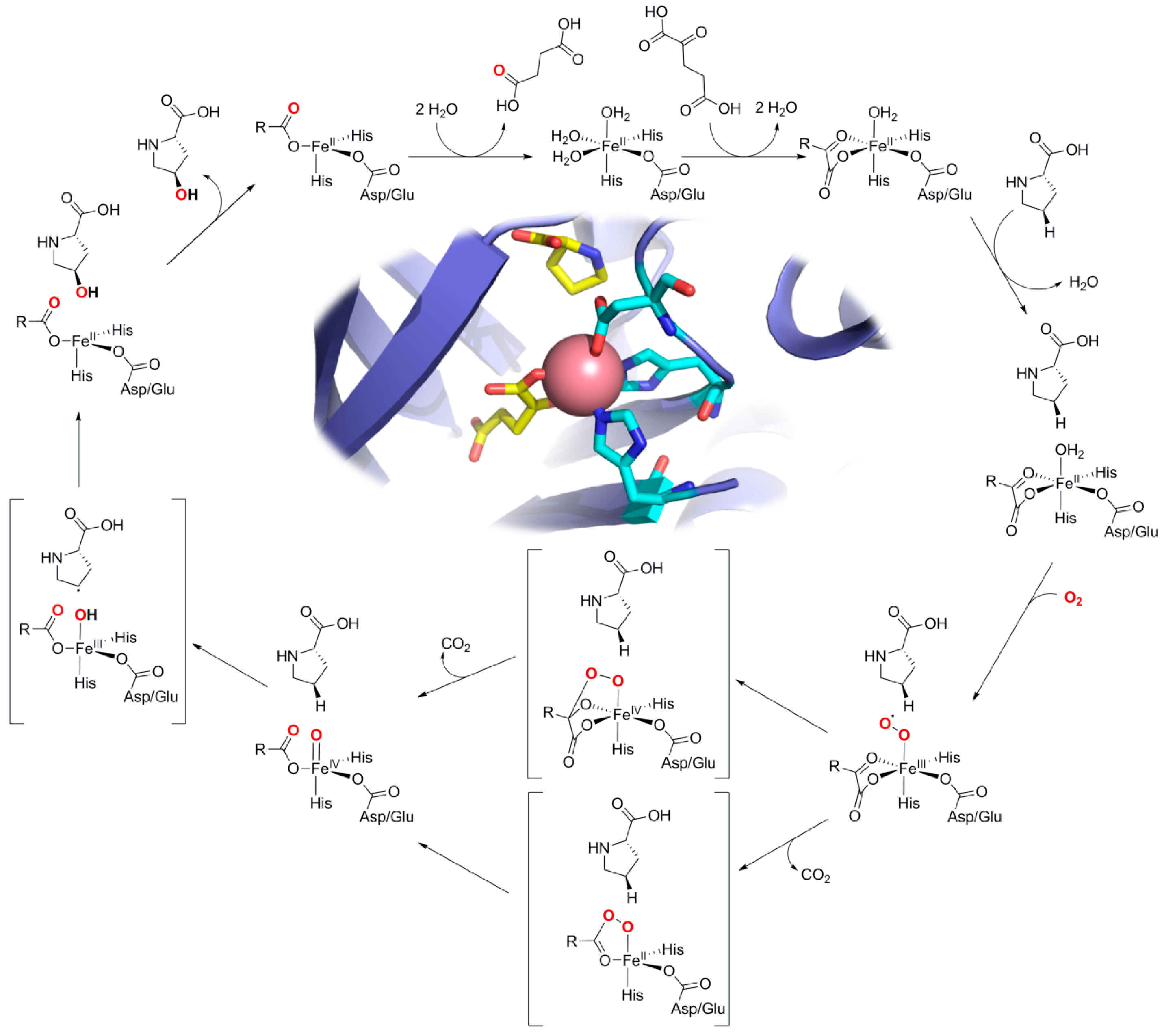
2.3. L-proline Hydroxylases
2.4. L-Hydroxyasparagine Oxygenases
2.5. L-Arginine Oxygenase
2.6. L-Lysine Hydroxylase
2.7. L-Glutamate Hydroxylase
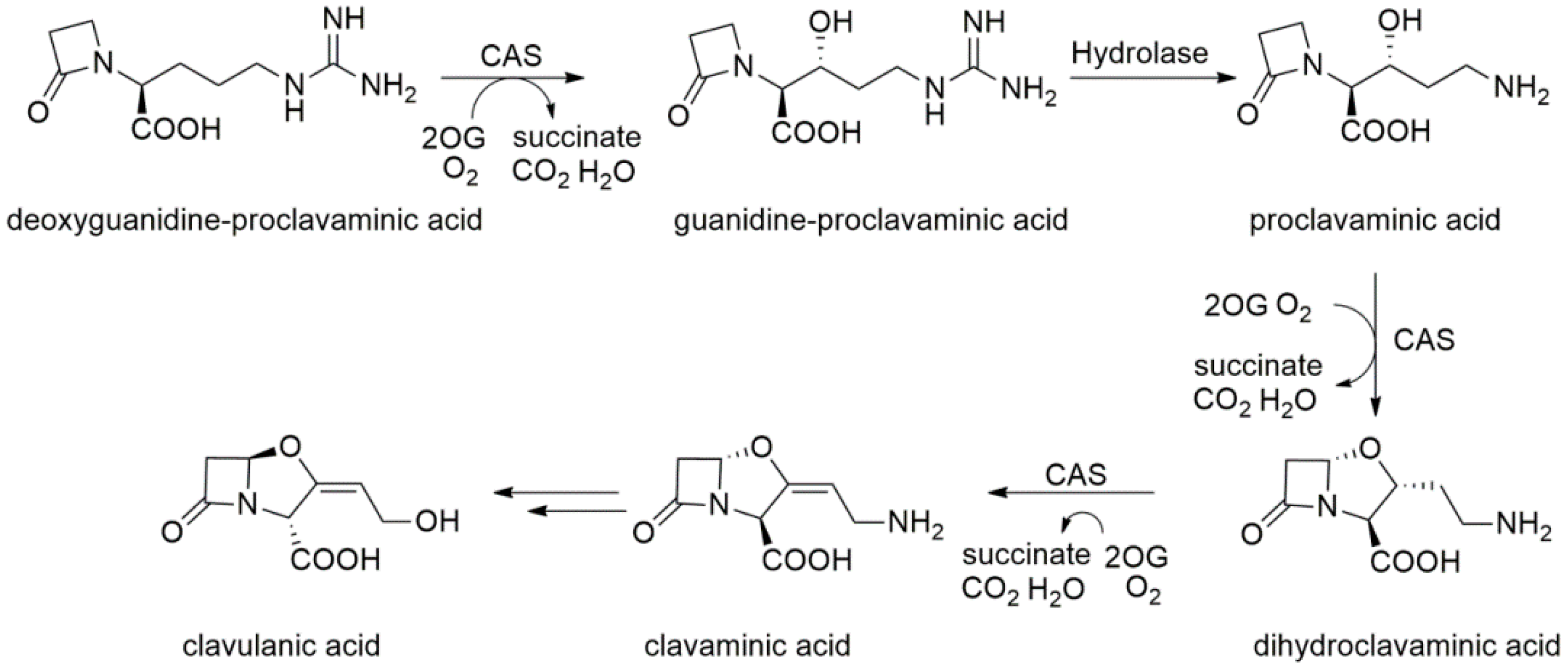
2.8. Clavaminate Synthase
2.9. Deacetoxycephalosporin C Synthase (DAOCS) and Deacetylcephalosporin C Synthase (DACS)
3. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Turner, N.J.; Humphreys, L. Biocatalysis in Organic Synthesis: The Retrosynthesis Approach; Royal Society of Chemistry: London, UK, 2018. [Google Scholar]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef] [PubMed]
- Croteau, R.; Kutchan, T.M.; Lewis, N.G. Natural products (secondary metabolites). In Biochemistry & Molecular Biology of Plants; Buchanan, B., Gruissem, W., Jones, R., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; pp. 1250–1318. [Google Scholar]
- Dong, J.; Fernandez-Fueyo, E.; Hollmann, F.; Paul, C.E.; Pesic, M.; Schmidt, S.; Wang, Y.; Younes, S.; Zhang, W. Biocatalytic oxidation reactions: A chemist’s perspective. Angew. Chem. Int. Ed. 2018, 57, 9238–9261. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, F.T.; Bureik, M.; Maurer, H.H. Biotechnological synthesis of drug metabolites using human cytochrome P450 isozymes heterologously expressed in fission yeast. Bioanalysis 2009, 1, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Schroer, K.; Kittelmann, M.; Lütz, S. Recombinant human cytochrome P450 monooxygenases for drug metabolite synthesis. Biotechnol. Bioeng. 2010, 106, 699–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Filsinger Interrante, M.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2015, 349, 1095–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, K.J.; Hans, M.; Meijrink, B.; van Scheppingen, W.B.; Vollebregt, A.; Tee, K.L.; van der Laan, J.M.; Leys, D.; Munro, A.W.; van den Berg, M.A. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2015, 112, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buller, R.; Hecht, K.; Mirata, M.A.; Meyer, H.-P. An appreciation of biocatalysis in the Swiss manufacturing environment. In Biocatalysis: An Industrial Perspective; The Royal Society of Chemistry: Cambridge, UK, 2018; pp. 1–43. [Google Scholar]
- Durairaj, P.; Hur, J.-S.; Yun, H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb. Cell Fact. 2016, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Kolev, J.N.; O’Dwyer, K.M.; Jordan, C.T.; Fasan, R. Discovery of potent parthenolide-based antileukemic agents enabled by late-stage P450-mediated C—H functionalization. ACS Chem. Biol. 2014, 9, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Negretti, S.; Narayan, A.R.H.; Chiou, K.C.; Kells, P.M.; Stachowski, J.L.; Hansen, D.A.; Podust, L.M.; Montgomery, J.; Sherman, D.H. Directing group-controlled regioselectivity in an enzymatic C–H bond oxygenation. J. Am. Chem. Soc. 2014, 136, 4901–4904. [Google Scholar] [CrossRef] [PubMed]
- Loskot, S.A.; Romney, D.K.; Arnold, F.H.; Stoltz, B.M. Enantioselective total synthesis of nigelladine A via late-stage C–H oxidation enabled by an engineered P450 enzyme. J. Am. Chem. Soc. 2017, 139, 10196–10199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.K.; Huang, X.; Arnold, F.H. Selective CH bond functionalization with engineered heme proteins: New tools to generate complexity. Curr. Opin. Chem. Biol. 2019, 49, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, R.; Urlacher, V.B. Cytochromes P450 as promising catalysts for biotechnological application: Chances and limitations. Appl. Microbiol. Biotechnol. 2014, 98, 6185–6203. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, E.; Köhler, V.; Flitsch, S.L.; Turner, N.J. Cytochromes P450 as useful biocatalysts: Addressing the limitations. Chem. Commun. 2011, 47, 2490–2501. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M.; Ullrich, R. Oxidations catalyzed by fungal peroxygenases. Curr. Opin. Chem. Biol. 2014, 19, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lan, D.; Durrani, R.; Hollman, F. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Herr, C.Q.; Hausinger, R.P. Amazing diversity in biochemical roles of Fe(II)/2-oxoglutarate oxygenases. Trends Biochem. Sci. 2018, 43, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Schofield, C.; Hausinger, R. 2-Oxoglutarate-Dependent Oxygenases; The Royal Society of Chemistry: Cambridge, UK, 2015. [Google Scholar]
- Hausinger, R.P. Biochemical diversity of 2-oxoglutarate-dependent oxygenases. In 2-Oxoglutarate-Dependent Oxygenases; The Royal Society of Chemistry: Cambridge, UK, 2015; pp. 1–58. [Google Scholar]
- Martinez, S.; Hausinger, R.P. Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 2015, 290, 20702–20711. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bong, Y.K.; Cabirol, F.L.; Prafulchandra, A.G.; Li, T.; Moore, J.C.; Quintanar-Audelo, M.; Hong, Y.; Collier, S.J.; Smith, D. Biocatalysts and Methods for Hydroxylation of Chemical Compounds. U.S. Patent US2017/0121744 A1, 4 May 2017. [Google Scholar]
- Hibi, M.; Ogawa, J. Characteristics and biotechnology applications of aliphatic amino acid hydroxylases belonging to the Fe(II)/alpha-ketoglutarate-dependent dioxygenase superfamily. Appl. Environ. Microbiol. 2014, 98, 3869–3876. [Google Scholar]
- Zhang, X.; Dong, L.B.; Yang, L.C.; Rudolf, J.D.; Shen, B.; Renata, H. Harnessing the biocatalytic potential of PtmO6, an α-ketoglutarate-dependent dioxygenase from platensimycin biosynthesis, for the chemoenzymatic synthesis of highly oxidized ent-kaurane diterpenes. ChemRxiv 2019. accepted. [Google Scholar] [CrossRef]
- Hüttel, W. Biocatalytic production of chemical building blocks in technical scale with α-ketoglutarate-dependent dioxygenases. Chem. Ing. Tech. 2013, 85, 809–817. [Google Scholar] [CrossRef]
- Wang, X.-C.; Liu, J.; Zhao, J.; Ni, X.-M.; Zheng, P.; Guo, X.; Sun, C.-M.; Sun, J.-B.; Ma, Y.-H. Efficient production of trans-4-hydroxy-L-proline from glucose using a new trans-proline 4-hydroxylase in Escherichia coli. J. Biosci. Bioeng. 2018, 126, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Hillwig, M.L.; Liu, X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol. 2014, 10, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, N.; Tanaka, H.; Koyama, F.; Noguchi, H.; Wang, C.C.; Hotta, K.; Watanabe, K. Non-heme dioxygenase catalyzes atypical oxidations of 6,7-bicyclic systems to form the 6,6-quinolone core of viridicatin-type fungal alkaloids. Angew. Chem. Int. Ed. 2014, 53, 12880–12884. [Google Scholar] [CrossRef] [PubMed]
- Brauer, A.; Beck, P.; Hintermann, L.; Groll, M. Structure of the dioxygenase AsqJ: Mechanistic insights into a one-pot multistep quinolone antibiotic biosynthesis. Angew. Chem. Int. Ed. Engl. 2016, 55, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [PubMed]
- Hagel, J.M.; Facchini, P.J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 2010, 6, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Stapon, A.; Li, R.; Townsend, C.A. Carbapenem biosynthesis: Confirmation of stereochemical assignments and the role of CarC in the ring stereoinversion process from L-proline. J. Am. Chem. Soc. 2003, 125, 8486–8493. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Guo, Y.; Wang, C.; Butch, S.E.; Rosenzweig, A.C.; Boal, A.K.; Krebs, C.; Bollinger, J.M., Jr. Mechanism of the C5 stereoinversion reaction in the biosynthesis of carbapenem antibiotics. Science 2014, 343, 1140–1144. [Google Scholar] [CrossRef] [PubMed]
- Rabe, P.; Kamps, J.J.A.G.; Schofield, C.J.; Lohans, C.T. Roles of 2-oxoglutarate oxygenases and isopenicillin N synthase in β-lactam biosynthesis. Nat. Prod. Rep. 2018, 35, 735–756. [Google Scholar] [CrossRef] [PubMed]
- Busby, R.W.; Chang, M.D.-T.; Busby, R.C.; Wimp, J.; Townsend, C.A. Expression and purification of two isozymes of clavaminate synthase and initial characterization of the iron binding site: General error analysis in polymerase chain reaction amplification. J. Biol. Chem. 1995, 270, 4262–4269. [Google Scholar] [CrossRef] [PubMed]
- Steffan, N.; Grundmann, A.; Afiyatullov, S.; Ruan, H.; Li, S.-M. FtmOx1, a non-heme Fe(ii) and α-ketoglutarate-dependent dioxygenase, catalyses the endoperoxide formation of verruculogen in Aspergillus fumigatus. Org. Biomol. Chem. 2009, 7, 4082–4087. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Suzuki, H.; Takagi, H.; Uramoto, M.; Takahashi, S.; Osada, H. Gene disruption and biochemical characterization of verruculogen synthase of Aspergillus fumigatus. ChemBioChem 2011, 12, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Clifton, I.J.; McDonough, M.A.; Ehrismann, D.; Kershaw, N.J.; Granatino, N.; Schofield, C.J. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 2006, 100, 644–669. [Google Scholar] [CrossRef] [PubMed]
- Hegg, E.L.; Que, L. The 2-His-1-carboxylate facial triad—An emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 1997, 250, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Aik, W.; McDonough, M.A.; Thalhammer, A.; Chowdhury, R.; Schofield, C.J. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struct. Biol. 2012, 22, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Elkins, J.M.; Ryle, M.J.; Clifton, I.J.; Hotopp, J.C.D.; Lloyd, J.S.; Burzlaff, N.I.; Baldwin, J.E.; Hausinger, R.P.; Roach, P.L. X-ray crystal structure of Escherichia coli taurine/α-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry 2002, 41, 5185–5192. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, J.M.; Price, J.C.; Hoffart, L.M.; Barr, E.W.; Krebs, C. Mechanism of taurine: α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Eur. J. Inorg. Chem. 2005, 4245–4254. [Google Scholar] [CrossRef]
- Krebs, C.; Fujimori, D.G.; Walsh, C.T.; Bollinger, J.M. Non-heme Fe(IV)-oxo intermediates. Acc. Chem. Res. 2007, 40, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, E.; Breisch, M.; Julsing, M.K.; Falcioni, F.; Bühler, B.; Schmid, A. An artificial TCA cycle selects for efficient α-ketoglutarate dependent hydroxylase catalysis in engineered Escherichia coli. Biotechnol. Bioeng. 2017, 114, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, S.V.; Kodera, T.; Samsonova, N.N.; Kotlyarovа, V.А.; Rushkevich, N.Y.; Kivero, А.D.; Sokolov, P.M.; Hibi, M.; Ogawa, J.; Shimizu, S. Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine. Appl. Microbiol. Biotechnol. 2010, 88, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.-S.; Naowarojna, N.; Cheng, R.; Liu, X.; Liu, P. Recent examples of α-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Nat. Prod. Rep. 2018, 35, 792–837. [Google Scholar] [CrossRef] [PubMed]
- Latham, J.; Brandenburger, E.; Shepherd, S.A.; Menon, B.R.K.; Micklefield, J. Development of halogenase enzymes for use in synthesis. Chem. Rev. 2018, 118, 232–269. [Google Scholar] [CrossRef] [PubMed]
- Miyake, R.; Dekishima, Y. Method for Manufacturing cis-5-hydroxy-L-pipecolic Acid. U.S. Patent US10087473B2, 2 October 2018. [Google Scholar]
- Ozaki, A.; Mori, H.; Shibasaki, T.; Ando, K.; Chiba, S. Process for Producing trans-4-hydroxy-L-proline. U.S. Patent US7238501B2, 3 July 2007. [Google Scholar]
- Ozaki, A.; Mori, H.; Shibasaki, T.; Ando, K.; Ochiai, K.; Chiba, S.; Uosaki, Y. Process for Producing cis-3-hydroxy-L-proline. U.S. Patent US6413748B1, 2 July 2002. [Google Scholar]
- Kino, K.; Hara, R. L-proline cis-4-hydroxylase and Use thereof to Produce cis-4-hydroxy-L-proline. U.S. Patent US8541209B2, 24 September 2013. [Google Scholar]
- Chen, H.; Bong, Y.K.; Cabirol, F.; Gohel, A.; Li, T.; Moore, J.C.; Quintanar-Audelo, M.; Yang, H.; Collier, S.J.; Smith, D. Biocatalysts and Methods for Hydroxylation of Chemical Compounds. European Patent EP2847327B1, 7 May 2013. [Google Scholar]
- Kodera, T.; Smirnov, S.V.; Samsonova, N.N.; Kotliarova, V.A.; Rushkevich, N.Y.; Kozlov, Y.I.; Shimizu, S.; Ogawa, J.; Hibi, M. Method for Producing 4-hydroxy-L-isoleucine. U.S. Patent US 8367381B2, 5 February 2013. [Google Scholar]
- Kino, K.; Hara, R.; Miyake, R.; Kawabata, H. Method for Producing hydroxy-L-lysine Employing an L-lysine Hydroxylase and Method for Producing hydroxy-L-pipecolic Acid. European Patent EP2889378B1, 18 February 2014. [Google Scholar]
- Kodera, T.; Smirnov, S.V.; Samsonova, N.N.; Kozlov, Y.I.; Koyama, R.; Hibi, M.; Ogawa, J.; Yokozeki, K.; Shimizu, S. A novel L-isoleucine hydroxylating enzyme, L-isoleucine dioxygenase from Bacillus thuringiensis, produces (2S,3R,4S)-4-hydroxyisoleucine. Biochem. Biophys. Res. Commun. 2009, 390, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Fowden, L.; Pratt, H.M.; Smith, A. 4-hydroxyisoleucine from seed of Trigonella foenum-graecum. Phytochemistry 1973, 12, 1707–1711. [Google Scholar] [CrossRef]
- Cao, H.; Liu, S.; Li, J.; Yue, G.; Zhang, M. Production L-4-hydroxy-isoleucine Conversion Method of a Microbial Enzyme Method. Chinese Patent CN 105779522 B, 17 May 2016. [Google Scholar]
- Broca, C.; Gross, R.; Petit, P.; Sauvaire, Y.; Manteghetti, M.; Tournier, M.; Masiello, P.; Gomis, R.; Ribes, G. 4-Hydroxyisoleucine: Experimental evidence of its insulinotropic and antidiabetic properties. Am. J. Physiol. Endocrinol. Metab. 1999, 277, E617–E623. [Google Scholar] [CrossRef] [PubMed]
- Jette, L.; Harvey, L.; Eugeni, K.; Levens, N. 4-Hydroxyisoleucine: A plant-derived treatment for metabolic syndrome. Curr. Opin. Investig. Drugs 2009, 10, 353–358. [Google Scholar] [PubMed]
- Ogawa, J.; Kodera, T.; Smirnov, S.V.; Hibi, M.; Samsonova, N.N.; Koyama, R.; Yamanaka, H.; Mano, J.; Kawashima, T.; Yokozeki, K.; et al. A novel L-isoleucine metabolism in Bacillus thuringiensis generating (2S,3R,4S)-4-hydroxyisoleucine, a potential insulinotropic and anti-obesity amino acid. Appl. Microbiol. Biotechnol. 2011, 89, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ma, J.; Li, Z.; Liang, Y.; Xu, Q.; Xie, X.; Chen, N. A strategy for L-isoleucine dioxygenase screening and 4-hydroxyisoleucine production by resting cells. Bioengineered 2018, 9, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Kawashima, T.; Kodera, T.; Smirnov, S.V.; Sokolov, P.M.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; Ogawa, J. Characterization of Bacillus thuringiensis L-isoleucine dioxygenase for production of useful amino acids. Appl. Environ. Microbiol. 2011, 77, 6926–6930. [Google Scholar] [CrossRef] [PubMed]
- Enoki, J.; Meisborn, J.; Müller, A.-C.; Kourist, R. A multi-enzymatic cascade reaction for the stereoselective production of γ-oxyfunctionalyzed amino acids. Front. Microbiol. 2016, 7, 425. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, P.M.; Smirnov, S.V.; Kodera, T.; Sugiyama, M.; Yokozeki, K.; Hibi, M.; Ogawa, J.; Shimizu, S. A novel family of bacterial dioxygenases that catalyse the hydroxylation of free L-amino acids. FEMS Microbiol. Lett. 2012, 331, 97–104. [Google Scholar]
- Smirnov, S.V.; Sokolov, P.M.; Kotlyarova, V.A.; Samsonova, N.N.; Kodera, T.; Sugiyama, M.; Torii, T.; Hibi, M.; Shimizu, S.; Yokozeki, K.; et al. A novel L-isoleucine-4′-dioxygenase and L-isoleucine dihydroxylation cascade in Pantoea ananatis. Microbiologyopen 2013, 2, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Kawashima, T.; Sokolov, P.M.; Smirnov, S.V.; Kodera, T.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; Ogawa, J. L-Leucine 5-hydroxylase of Nostoc punctiforme is a novel type of Fe(II)/α-ketoglutarate-dependent dioxygenase that is useful as a biocatalyst. Appl. Microbiol. Biotechnol. 2013, 97, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Gao, D.; Xu, P.; Guo, Q.; Zhu, Z.; Cheng, X.; Bai, S.; Qin, H.-M.; Lu, F. A novel L-leucine 5-hydroxylase from Nostoc piscinale unravels unexpected sulfoxidation activity toward L-methionine. Protein Expr. Purif. 2018, 149, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Correia Cordeiro, R.S.; Enoki, J.; Busch, F.; Mügge, C.; Kourist, R. Cloning and characterization of a new delta-specific L-leucine dioxygenase from Anabaena variabilis. J. Biotechnol. 2018, 284, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Kanda, F.; Ishibashi, M.; Shigemori, H. Manzacidins A-C, novel tetrahydropyrimidine alkaloids from the Okinawan marine sponge Hymeniacidon sp. J. Org. Chem. 1991, 56, 4574–4576. [Google Scholar] [CrossRef]
- Hashimoto, T.; Maruoka, K. Syntheses of manzacidins: A stage for the demonstration of synthetic methodologies. Org. Biomol. Chem. 2008, 6, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Zwick, C.R.; Renata, H. Remote C–H hydroxylation by an α-ketoglutarate-dependent dioxygenase enables efficient chemoenzymatic synthesis of manzacidin C and proline analogs. J. Am. Chem. Soc. 2018, 140, 1165–1169. [Google Scholar] [CrossRef] [PubMed]
- Kling, A.; Lukat, P.; Almeida, D.V.; Bauer, A.; Fontaine, E.; Sordello, S.; Zaburannyi, N.; Herrmann, J.; Wenzel, S.C.; König, C.; et al. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Lukat, P.; Katsuyama, Y.; Wenzel, S.; Binz, T.; König, C.; Blankenfeldt, W.; Brönstrup, M.; Müller, R. Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity. Chem. Sci. 2017, 8, 7521–7527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwick, C.R.; Renata, H. Evolution of biocatalytic and chemocatalytic C–H functionalization strategy in the synthesis of manzacidin C. J. Org. Chem. 2018, 83, 7407–7415. [Google Scholar] [CrossRef] [PubMed]
- Zwick, C.R.; Renata, H. A one-pot chemoenzymatic synthesis of (2S, 4R)-4-methylproline enables the first total synthesis of antiviral lipopeptide cavinafungin B. Tetrahedron 2018, 74, 6469–6473. [Google Scholar] [CrossRef]
- Ortíz-López, F.J.; Monteiro, M.C.; González-Menéndez, V.; Tormo, J.R.; Genilloud, O.; Bills, G.F.; Vicente, F.; Zhang, C.; Roemer, T.; Singh, S.B.; et al. Cyclic colisporifungin and linear cavinafungins, antifungal lipopeptides isolated from Colispora cavincola. J. Nat. Prod. 2015, 78, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Estoppey, D.; Lee, C.M.; Janoschke, M.; Lee, B.H.; Wan, K.F.; Dong, H.; Mathys, P.; Filipuzzi, I.; Schuhmann, T.; Riedl, R.; et al. The natural product cavinafungin selectively interferes with Zika and Dengue virus replication by inhibition of the host signal peptidase. Cell Rep. 2017, 19, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-M.; Miyakawa, T.; Nakamura, A.; Xue, Y.-L.; Kawashima, T.; Kasahara, T.; Hibi, M.; Ogawa, J.; Tanokura, M. Expression, purification, crystallization and preliminary X-ray analysis of a novel N-substituted branched-chain L-amino-acid dioxygenase from Burkholderia ambifaria AMMD. Acta Crystallogr. F 2012, 68, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Kawashima, T.; Kasahara, T.; Sokolov, P.M.; Smirnov, S.V.; Kodera, T.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; Ogawa, J. A novel Fe(II)/α-ketoglutarate-dependent dioxygenase from Burkholderia ambifaria has β-hydroxylating activity of N-succinyl L-leucine. Lett. Appl. Microbiol. 2012, 55, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.M.; Miyakawa, T.; Jia, M.Z.; Nakamura, A.; Ohtsuka, J.; Xue, Y.L.; Kawashima, T.; Kasahara, T.; Hibi, M.; Ogawa, J.; et al. Crystal structure of a novel N-substituted L-amino acid dioxygenase from Burkholderia ambifaria AMMD. PLoS ONE 2013, 8, e63996. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-M.; Miyakawa, T.; Nakamura, A.; Hibi, M.; Ogawa, J.; Tanokura, M. Structural optimization of SadA, an Fe(II)- and α-ketoglutarate-dependent dioxygenase targeting biocatalytic synthesis of N-succinyl-L-threo-3,4-dimethoxyphenylserine. Biochem. Biophys. Res. Commun. 2014, 450, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Kasahara, T.; Kawashima, T.; Yajima, H.; Kozono, S.; Smirnov, S.V.; Kodera, T.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; et al. Multi-enzymatic synthesis of optically pure β-hydroxy α-amino acids. Adv. Synth. Catal. 2015, 357, 767–774. [Google Scholar] [CrossRef]
- Yan, W.; Song, H.; Song, F.; Guo, Y.; Wu, C.-H.; Sae Her, A.; Pu, Y.; Wang, S.; Naowarojna, N.; Weitz, A.; et al. Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme. Nature 2015, 527, 539. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.-N.; Chen, S.-L. Asymmetric abstraction of two chemically-equivalent methylene hydrogens: Significant enantioselectivity of endoperoxide presented by fumitremorgin B endoperoxidase. Phys. Chem. Chem. Phys. 2018, 20, 26500–26505. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, J.E.; Field, R.A.; Lawrence, C.C.; Lee, V.; Robinson, J.K.; Schofield, C.J. Substrate specificity of proline-4-hydroxylase: Chemical and enzymatic synthesis of 2S,3R,4S-epoxyproline. Tetrahedron Lett. 1994, 35, 4649–4652. [Google Scholar] [CrossRef]
- Mattay, J.; Houwaart, S.; Huttel, W. Cryptic production of trans-3-hydroxyproline in echinocandin B biosynthesis. Appl. Environ. Microbiol. 2018, 84, e02370-17. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Shibasaki, T.; Uozaki, Y.; Ochiai, K.; Ozaki, A. Detection of novel proline 3-hydroxylase activities in Streptomyces and Bacillus spp. by regio- and stereospecific hydroxylation of L-proline. Appl. Environ. Microbiol. 1996, 62, 1903–1907. [Google Scholar] [PubMed]
- Mori, H.; Shibasaki, T.; Yano, K.; Ozaki, A. Purification and cloning of a proline 3-hydroxylase, a novel enzyme which hydroxylates free L-proline to cis-3-hydroxy-L-proline. J. Bacteriol. 1997, 179, 5677–5683. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, T.; Mori, H.; Ozaki, A. Cloning of an isozyme of proline 3-hydroxylase and its purification from recombinant Escherichia coli. Biotechnol. Lett. 2000, 22, 1967–1973. [Google Scholar] [CrossRef]
- Mattay, J.; Hüttel, W. Pipecolic acid hydroxylases: A monophyletic clade among cis-selective bacterial proline hydroxylases that discriminates L-proline. ChemBioChem 2017, 18, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- Hara, R.; Kino, K. Characterization of novel 2-oxoglutarate dependent dioxygenases converting L-proline to cis-4-hydroxy-L-proline. Biochem. Biophys. Res. Commun. 2009, 379, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Koketsu, K.; Shomura, Y.; Moriwaki, K.; Hayashi, M.; Mitsuhashi, S.; Hara, R.; Kino, K.; Higuchi, Y. Refined regio- and stereoselective hydroxylation of L-pipecolic acid by protein engineering of L-proline cis-4-hydroxylase based on the x-ray crystal structure. ACS Synth. Biol. 2015, 4, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Hara, R.; Uchiumi, N.; Kino, K. Identification and characterization of 2-oxoglutarate-dependent dioxygenases catalyzing selective cis-hydroxylation of proline and pipecolinic acid from actinomycetes. J. Biotechnol. 2014, 172, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, J.; Liu, J.; Guo, X.; Rao, D.; Liu, H.; Zheng, P.; Sun, J.; Ma, Y. Simultaneously improving the activity and thermostability of a new proline 4-hydroxylase by loop grafting and site-directed mutagenesis. Appl. Microbiol. Biotechnol. 2019, 103, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, T.; Mori, H.; Chiba, S.; Ozaki, A. Microbial proline 4-hydroxylase screening and gene cloning. Appl. Environ. Microbiol. 1999, 65, 4028–4031. [Google Scholar] [PubMed]
- Shibasaki, T.; Mori, H.; Ozaki, A. Enzymatic production of trans-4-hydroxy-L-proline by regio- and stereospecific hydroxylation of L-proline. Biosci. Biotechnol. Biochem. 2000, 64, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.; Olewinski, R.; Salmon, P.; Connors, N. Novel proline hydroxylase activities in the pneumocandin-producing fungus Glarea lozoyensis responsible for the formation of trans 3- and trans 4-hydroxyproline. Appl. Microbiol. Biotechnol. 2003, 62, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Houwaart, S.; Youssar, L.; Hüttel, W. Pneumocandin biosynthesis: Involvement of a trans-selective proline hydroxylase. ChemBioChem 2014, 15, 2365–2369. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Sheng, H.; Li, Z.; Ye, Q. Biosynthesis of trans-4-hydroxyproline by recombinant strains of Corynebacterium glutamicum and Escherichia coli. BMC Biotechnol. 2014, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Hüttel, W. A simple procedure for selective hydroxylation of L-proline and L-pipecolic acid with recombinantly expressed proline hydroxylases. Adv. Synth. Catal. 2011, 353, 1375–1383. [Google Scholar] [CrossRef]
- Hojati, Z.; Milne, C.; Harvey, B.; Gordon, L.; Borg, M.; Flett, F.; Wilkinson, B.; Sidebottom, P.J.; Rudd, B.A.M.; Hayes, M.A.; et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 2002, 9, 1175–1187. [Google Scholar] [CrossRef]
- Baltz, R.H.; Miao, V.; Wrigley, S.K. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005, 22, 717–741. [Google Scholar] [CrossRef] [PubMed]
- Neary, J.M.; Powell, A.; Gordon, L.; Milne, C.; Flett, F.; Wilkinson, B.; Smith, C.P.; Micklefield, J. An asparagine oxygenase (AsnO) and a 3-hydroxyasparaginyl phosphotransferase (HasP) are involved in the biosynthesis of calcium-dependent lipopeptide antibiotics. Microbiology 2007, 153, 768–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strieker, M.; Kopp, F.; Mahlert, C.; Essen, L.-O.; Marahiel, M.A. Mechanistic and structural basis of stereospecific Cβ-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide. ACS Chem. Biol. 2007, 2, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Essen, L.-O.; Walsh, C.T.; Marahiel, M.A. Non-heme hydroxylase engineering for simple enzymatic synthesis of L-threo-hydroxyaspartic acid. ChemBioChem 2008, 9, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Marrero, P.; Cabañas, M.J.; Modolell, J. Induction of translational errors (misreading) by tuberactinomycins and capreomycins. Biochem. Biophys. Res. Commun. 1980, 97, 1047–1052. [Google Scholar] [CrossRef]
- Yin, X.; O’Hare, T.; Gould, S.J.; Zabriskie, T.M. Identification and cloning of genes encoding viomycin biosynthesis from Streptomyces vinaceus and evidence for involvement of a rare oxygenase. Gene 2003, 312, 215–224. [Google Scholar] [CrossRef]
- Yin, X.; Zabriskie, T.M. VioC is a non-heme iron, α-ketoglutarate-dependent oxygenase that catalyzes the formation of 3S-hydroxy-L-arginine during viomycin biosynthesis. ChemBioChem 2004, 5, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; McPhail, K.L.; Kim, K.-J.; Zabriskie, T.M. Formation of the nonproteinogenic amino acid 2S,3R-capreomycidine by VioD from the viomycin biosynthesis pathway. ChemBioChem 2004, 5, 1278–1281. [Google Scholar] [CrossRef] [PubMed]
- Helmetag, V.; Samel, S.A.; Thomas, M.G.; Marahiel, M.A.; Essen, L.-O. Structural basis for the erythro-stereospecificity of the L-arginine oxygenase VioC in viomycin biosynthesis. FEBS J. 2009, 276, 3669–3682. [Google Scholar] [CrossRef] [PubMed]
- Dunham, N.P.; Mitchell, A.J.; Del Rio Pantoja, J.M.; Krebs, C.; Bollinger, J.M., Jr.; Boal, A.K. α-Amine desaturation of D-arginine by the iron(II)- and 2-(Oxo)glutarate-dependent L-arginine 3-hydroxylase, VioC. Biochemistry 2018, 57, 6479–6488. [Google Scholar] [CrossRef] [PubMed]
- Hara, R.; Kitatsuji, S.; Yamagata, K.; Kino, K. Development of a multi-enzymatic cascade reaction for the synthesis of trans-3-hydroxy-L-proline from L-arginine. Appl. Microbiol. Biotechnol. 2016, 100, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Izumikawa, M.; Kawahara, T.; Kagaya, N.; Yamamura, H.; Hayakawa, M.; Takagi, M.; Yoshida, M.; Doi, T.; Shin-ya, K. Pyrrolidine-containing peptides, JBIR-126, -148, and -149, from Streptomyces sp. NBRC 111228. Tetrahedron Lett. 2015, 56, 5333–5336. [Google Scholar] [CrossRef]
- Goering, A.W.; McClure, R.A.; Doroghazi, J.R.; Albright, J.C.; Haverland, N.A.; Zhang, Y.; Ju, K.-S.; Thomson, R.J.; Metcalf, W.W.; Kelleher, N.L. Metabologenomics: Correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2016, 2, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Saaidi, P.-L.; Monfleur, A.; Harari, M.; Cuccaro, J.; Fossey, A.; Besnard, M.; Debard, A.; Mariage, A.; Pellouin, V.; et al. Synthesis of mono- and dihydroxylated amino acids with new α-ketoglutarate-dependent dioxygenases: biocatalytic oxidation of C-H bonds. ChemCatChem 2014, 6, 3012–3017. [Google Scholar] [CrossRef]
- Bastard, K.; Isabet, T.; Stura, E.A.; Legrand, P.; Zaparucha, A. Structural studies based on two lysine dioxygenases with distinct regioselectivity brings insights into enzyme specificity within the clavaminate synthase-like family. Sci. Rep. 2018, 8, 16587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; King-Smith, E.; Renata, H. Total synthesis of tambromycin by combining chemocatalytic and biocatalytic C−H functionalization. Angew. Chem. Int. Ed. Engl. 2018, 57, 5037–5041. [Google Scholar] [CrossRef] [PubMed]
- Miley, G.P.; Rote, J.C.; Silverman, R.B.; Kelleher, N.L.; Thomson, R.J. Total synthesis of tambromycin enabled by indole C–H functionalization. Org. Lett. 2018, 20, 2369–2373. [Google Scholar] [CrossRef] [PubMed]
- Hara, R.; Yamagata, K.; Miyake, R.; Kawabata, H.; Uehara, H.; Kino, K. Discovery of lysine hydroxylases in the clavaminic acid synthase-like superfamily for efficient hydroxylysine bioproduction. Appl. Environ. Microbiol. 2017, 83, e00693-17. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Nolan, E.M.; Walsh, C.T.; Marahiel, M.A. Stereospecific synthesis of threo- and erythro-beta-hydroxyglutamic acid during kutzneride biosynthesis. J. Am. Chem. Soc. 2009, 131, 13523–13530. [Google Scholar] [CrossRef] [PubMed]
- Saudagar, P.S.; Survase, S.A.; Singhal, R.S. Clavulanic acid: A review. Biotechnol. Adv. 2008, 26, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Pavel, E.G.; Zhou, J.; Busby, R.W.; Gunsior, M.; Townsend, C.A.; Solomon, E.I. Circular dichroism and magnetic circular dichroism spectroscopic studies of the non-heme ferrous active site in clavaminate synthase and its interaction with α-ketoglutarate cosubstrate. J. Am. Chem. Soc. 1998, 120, 743–753. [Google Scholar] [CrossRef]
- Zhou, J.; Kelly, W.L.; Bachmann, B.O.; Gunsior, M.; Townsend, C.A.; Solomon, E.I. Spectroscopic studies of substrate interactions with clavaminate synthase 2, a multifunctional α-KG-dependent non-heme iron enzyme: Correlation with mechanisms and reactivities. J. Am. Chem. Soc. 2001, 123, 7388–7398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ren, J.; Stammers, D.K.; Baldwin, J.E.; Harlos, K.; Schofield, C.J. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 2000, 7, 127. [Google Scholar] [PubMed]
- Zhang, Z.; Ren, J.-S.; Harlos, K.; McKinnon, C.H.; Clifton, I.J.; Schofield, C.J. Crystal structure of a clavaminate synthase–Fe(II)–2-oxoglutarate–substrate–NO complex: Evidence for metal centred rearrangements. FEBS Lett. 2002, 517, 7–12. [Google Scholar] [CrossRef]
- Ser, H.-L.; Law, J.W.-F.; Chaiyakunapruk, N.; Jacob, S.A.; Palanisamy, U.D.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: A systematic review. Front. Microbiol. 2016, 7, 522. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Malule, H.; Junne, S.; Nicolas Cruz-Bournazou, M.; Neubauer, P.; Rios-Estepa, R. Streptomyces clavuligerus shows a strong association between TCA cycle intermediate accumulation and clavulanic acid biosynthesis. Appl. Environ. Microbiol. 2018, 102, 4009–4023. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, K.C.d.S.; Souza, A.T.d.; Badino, A.C.; Pedrolli, D.B.; Cerri, M.O. Screening of medium constituents for clavulanic acid production by Streptomyces clavuligerus. Braz. J. Microbiol. 2018, 49, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P. Cephalosporins 1945–1986. Drugs 1987, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Samson, S.M.; Dotzlaf, J.E.; Slisz, M.L.; Becker, G.W.; Van Frank, R.M.; Veal, L.E.; Yeh, W.-K.; Miller, J.R.; Queener, S.W.; Ingolia, T.D. Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Biotechnology 1987, 5, 1207–1214. [Google Scholar] [CrossRef]
- Kovacevic, S.; Weigel, B.J.; Tobin, M.B.; Ingolia, T.D.; Miller, J.R. Cloning, characterization, and expression in Escherichia coli of the Streptomyces clavuligerus gene encoding deacetoxycephalosporin C synthetase. J. Bacteriol. 1989, 171, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, S.; Miller, J.R. Cloning and sequencing of the beta-lactam hydroxylase gene (cefF) from Streptomyces clavuligerus: Gene duplication may have led to separate hydroxylase and expandase activities in the Actinomycetes. J. Bacteriol. 1991, 173, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Lin, B.; Tao, Y.; Yang, K. Engineering deacetoxycephalosporin C synthase as a catalyst for the bioconversion of penicillins. J. Ind. Microbiol. Biotechnol. 2017, 44, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Goo, K.-S.; Chua, C.-S.; Sim, T.-S. Directed evolution and rational approaches to improving Streptomyces clavuligerus deacetoxycephalosporin C synthase for cephalosporin production. J. Ind. Microbiol. Biotechnol. 2009, 36, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Fan, K.; Tian, X.; Zhang, X.; Zhang, Y.; Yang, K. Iterative combinatorial mutagenesis as an effective strategy for generation of deacetoxycephalosporin c synthase with improved activity toward penicillin G. Appl. Environ. Microbiol. 2012, 78, 7809–7812. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-L.; Yang, Y.-B.; Wang, W.-C.; Liu, W.-C.; Hsu, J.-S.; Tsai, Y.-C. Engineering Streptomyces clavuligerus deacetoxycephalosporin C synthase for optimal ring expansion activity toward penicillin G. Appl. Environ. Microbiol. 2003, 69, 2306–2312. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-L.; Yang, Y.-B.; Deng, C.-H.; Liu, W.-C.; Hsu, J.-S.; Lin, Y.-C.; Liaw, S.-H.; Tsai, Y.-C. Directed evolution of Streptomyces clavuligerus deacetoxycephalosporin C synthase for enhancement of penicillin G expansion. Appl. Environ. Microbiol. 2005, 71, 8873–8880. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.-S.; Yang, Y.-B.; Deng, C.-H.; Wei, C.-L.; Liaw, S.-H.; Tsai, Y.-C. Family shuffling of expandase genes to enhance substrate specificity for penicillin G. Appl. Environ. Microbiol. 2004, 70, 6257–6263. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, B.; Jia, X.; Kim, K.H.; Jeon, C.O. Integrative view of 2-oxoglutarate/Fe(II)-dependent oxygenase diversity and functions in bacteria. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.L.; Tang, K.; Chun, B.H.; Jeon, C.O. Large-scale examination of functional and sequence diversity of 2-oxoglutarate/Fe(II)-dependent oxygenases in Metazoa. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2922–2933. [Google Scholar] [CrossRef] [PubMed]
- Zeymer, C.; Hilvert, D. directed evolution of protein catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Directed evolution: Bringing new chemistry to life. Angew. Chem. Int Ed. Engl. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [PubMed]
- Frey, R.; Hayashi, T.; Buller, R.M. Directed evolution of carbon–hydrogen bond activating enzymes. Curr. Opin. Biotechnol. 2019, 60, 29–38. [Google Scholar] [CrossRef] [PubMed]


| Product | Name | Source Organism |
|---|---|---|
| cis-3-hydroxy-L-proline | Cis-P3H1 (type I) | Streptomyces sp. Th1 [92,93] |
| Cis-P3H2 (type II) | Streptomyces sp. Th1 [94] | |
| GetF | Streptomyces sp. [95] | |
| PiFa | Frankia alni [95] | |
| cis-4-hydroxy-L-proline | SmP4H | Sinorhizobium meliloti [96,97] |
| MlP4H | Mesorhizobium loti [96] | |
| SrPH | Streptosporangium roseum [98] 1 | |
| CaPH | Catenulispora acidiphila [98] 1 | |
| trans-4-hydroxy-L-proline | AlP4H | Alteromonas mediterranea [29] |
| MiP4H | Micromonospora sp. CNB394 [29] | |
| ScP4H | Sorangium cellulosum [29] | |
| UbP4H | Uncultured bacterium esnapd13 [99] | |
| DaP4H | Dactylosporangium sp. strain RH1 [100,101] | |
| HtyE | Aspergillus pachycristatus [91] 2 | |
| GloF | Glarea lozoyensis [91,102,103] 2 | |
| P4HP | Pseudomonas stutzeri [104] | |
| P4HB | Bordetella bronchiseptica [104] | |
| trans-3-hydroxy-L-proline | HtyE | Aspergillus pachycristatus [91] 2 |
| GloF | Glarea lozoyensis [91,102,103] 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peters, C.; Buller, R.M. Industrial Application of 2-Oxoglutarate-Dependent Oxygenases. Catalysts 2019, 9, 221. https://doi.org/10.3390/catal9030221
Peters C, Buller RM. Industrial Application of 2-Oxoglutarate-Dependent Oxygenases. Catalysts. 2019; 9(3):221. https://doi.org/10.3390/catal9030221
Chicago/Turabian StylePeters, Christin, and Rebecca M. Buller. 2019. "Industrial Application of 2-Oxoglutarate-Dependent Oxygenases" Catalysts 9, no. 3: 221. https://doi.org/10.3390/catal9030221
APA StylePeters, C., & Buller, R. M. (2019). Industrial Application of 2-Oxoglutarate-Dependent Oxygenases. Catalysts, 9(3), 221. https://doi.org/10.3390/catal9030221