Characterization of β-Glycosidase from Caldicellulosiruptor owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf
Abstract
1. Introduction
2. Results and Discussion
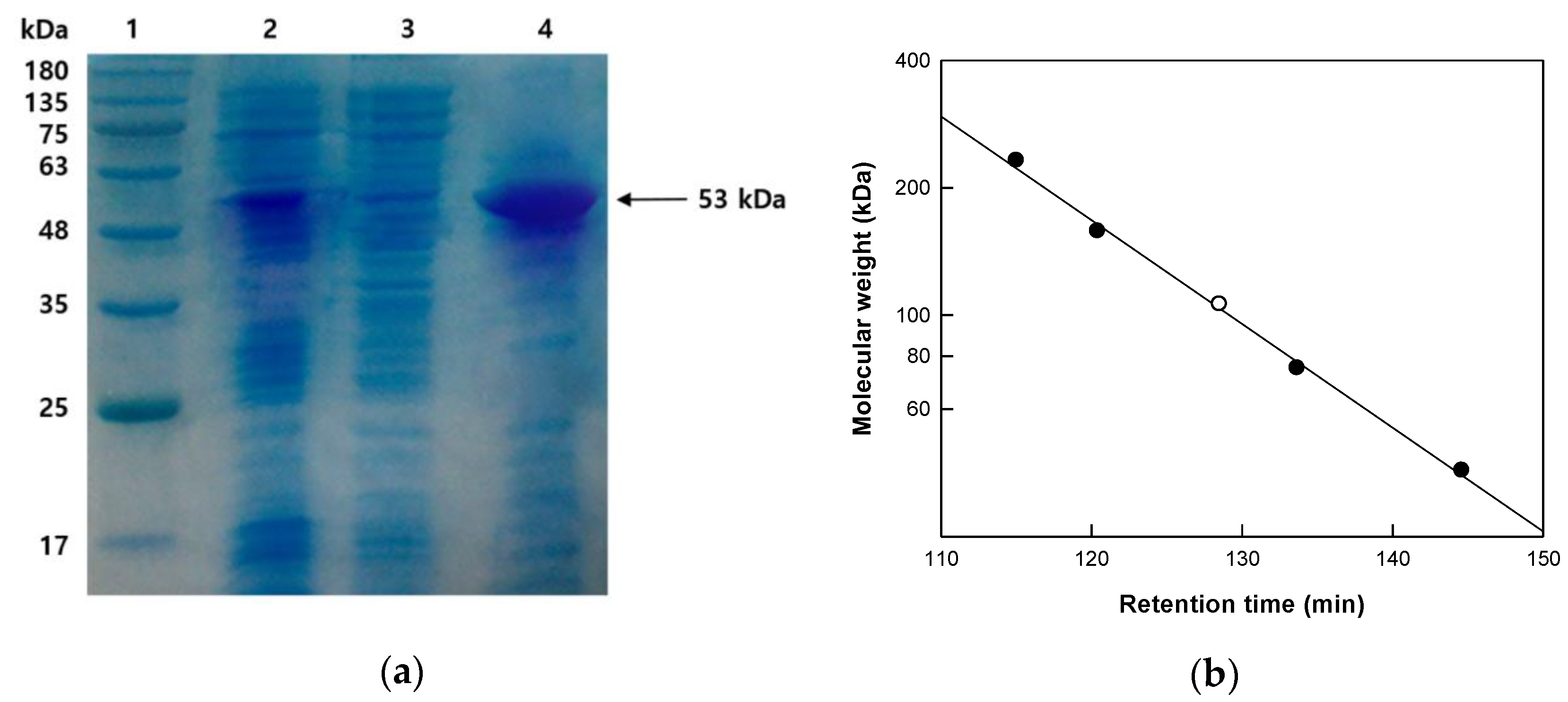
2.1. Cloning of Gene, Purification of Enzyme, and Determination of Molecular Mass
2.2. Hydrolytic Activity According to pH and Temperature Change
2.3. Substrate Specificity
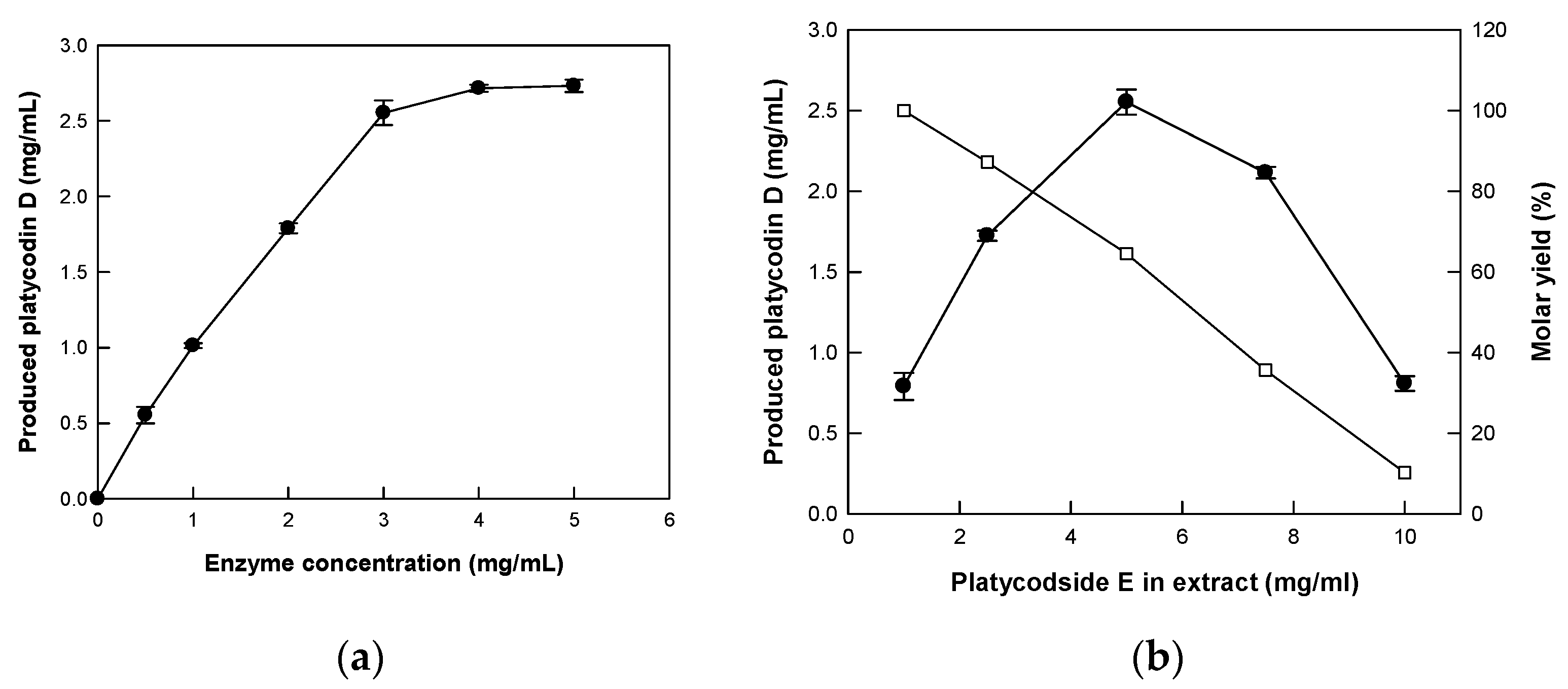
2.4. Optimization of Reaction Conditions
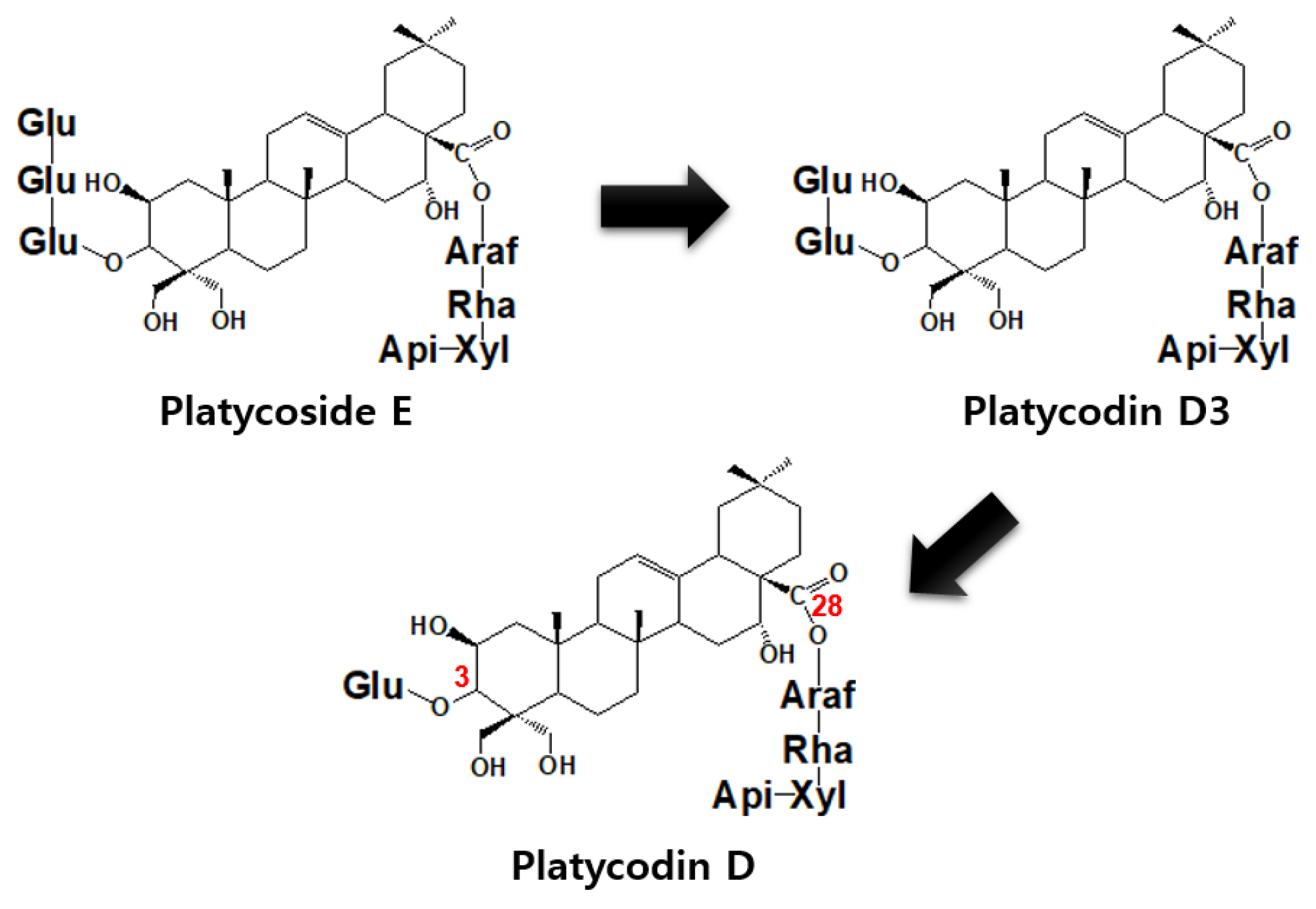
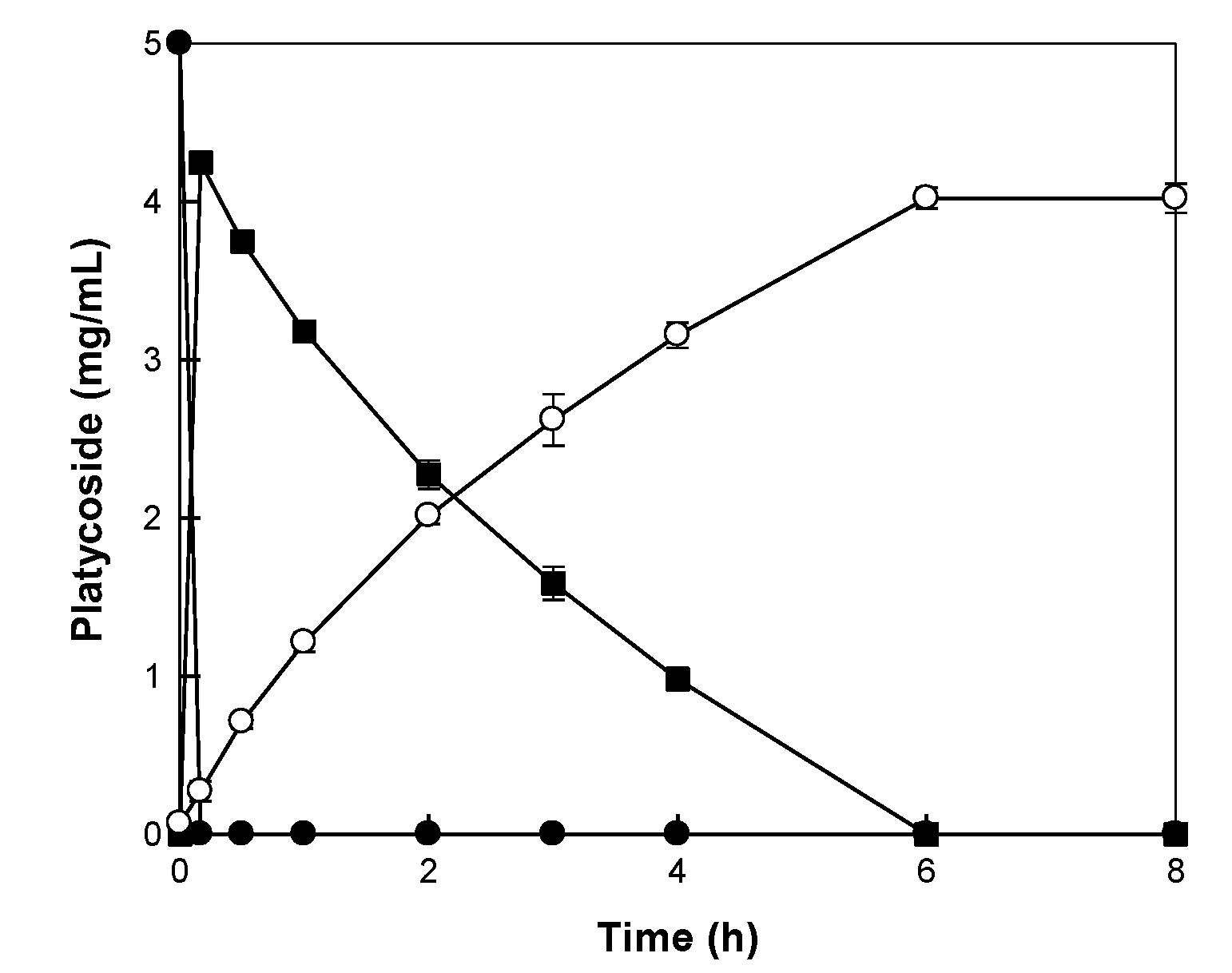
2.5. Production of Platycodin D from BFLE by β-Glycosidase from C. owensensis
3. Materials and Methods
3.1. Bacterial Strains, Plasmid Vector and Gene Cloning
3.2. Bacteria Culture for Enzyme Expression
3.3. Preparation of β-Glycosidase from C. Owensensis
3.4. SDS-PAGE and Gel-Filtration Chromatography
3.5. Preparation of BFLE
3.6. Hydrolytic Activity Assay
3.7. Biotransformation of Platycoside
3.8. HPLC Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Choi, C.Y.; Kim, J.Y.; Kim, Y.S.; Chung, Y.C.; Hahm, K.S.; Jeong, H.G. Augmentation of Macrophage Functions by an Aqueous Extract Isolated from Platycodon grandiflorum. Cancer Lett. 2001, 166, 17–25. [Google Scholar] [CrossRef]
- Li, W.; Tian, Y.H.; Liu, Y.; Wang, Z.; Tang, S.; Zhang, J.; Wang, Y.P. Platycodin D Exerts Anti-Tumor Efficacy in H22 Tumor-Bearing Mice Via Improving Immune Function and Inducing Apoptosis. J. Toxicol. Sci. 2016, 41, 417–428. [Google Scholar] [CrossRef]
- Kim, J.Y.; Hwang, Y.P.; Kim, D.H.; Han, E.H.; Chung, Y.C.; Roh, S.H.; Jeong, H.G. Inhibitory Effect of the Saponins Derived from Roots of Platycodon grandiflorum on Carrageenan-Induced Inflammation. Biosci. Biotechnol. Biochem. 2006, 70, 858–864. [Google Scholar] [CrossRef]
- Ye, Y.; Pei, L.; Ding, J.; Wu, C.; Sun, C.; Liu, S. Effects of Platycodin D on S100a8/A9-Induced Inflammatory Response in Murine Mammary Carcinoma 4t1 Cells. Int. Immunopharmacol. 2019, 67, 239–247. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative Stress and Neurodegeneration: Where Are We Now? J. Neurochem. 2006, 97, 1634–1658. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, S.; Du, J.; Jinfu, Y.; Shumin, W. Platycodin D Attenuates Airway Inflammation in a Mouse Model of Allergic Asthma by Regulation Nf-Kappab Pathway. Inflammation 2015, 38, 1221–1228. [Google Scholar] [CrossRef]
- Zhao, H.L.; Harding, S.V.; Marinangeli, C.P.; Kim, Y.S.; Jones, P.J. Hypocholesterolemic and Anti-Obesity Effects of Saponins from Platycodon grandiflorum in Hamsters Fed Atherogenic Diets. J. Food Sci. 2008, 73, H195–H200. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lin, Y.C.; Kuo, W.W.; Shen, C.Y.; Cheng, Y.C.; Lin, Y.M.; Chang, R.L.; Padma, V.V.; Huang, C.Y.; Huang, C.Y. Platycodin D Reverses Pathological Cardiac Hypertrophy and Fibrosis in Spontaneously Hypertensive Rats. Am. J. Chin. Med. 2018, 46, 537–549. [Google Scholar] [CrossRef]
- Yoo, D.S.; Choi, Y.H.; Cha, M.R.; Lee, B.H.; Kim, S.J.; Yon, G.H.; Hong, K.S.; Jang, Y.S.; Lee, H.S.; Kim, Y.S.; et al. Hplc-Elsd Analysis of 18 Platycosides from Balloon Flower Roots (Platycodi Radix) Sourced from Various Regions in Korea and Geographical Clustering of the Cultivation Areas. Food Chem. 2011, 129, 645–651. [Google Scholar] [CrossRef]
- Park, C.S.; Yoo, M.H.; Noh, K.H.; Oh, D.K. Biotransformation of Ginsenosides by Hydrolyzing the Sugar Moieties of Ginsenosides Using Microbial Glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef]
- Shin, K.C.; Oh, D.K. Classification of Glycosidases That Hydrolyze the Specific Positions and Types of Sugar Moieties in Ginsenosides. Crit. Rev. Biotechnol. 2016, 36, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.J.; You, H.J.; Park, M.S.; Johnston, T.V.; Ku, S.; Ji, G.E. Biocatalysis of Platycoside E and Platycodin D3 Using Fungal Extracellular Beta-Glucosidase Responsible for Rapid Platycodin D Production. Int. J. Mol. Sci. 2018, 19, 2671. [Google Scholar] [CrossRef]
- Wie, H.J.; Zhao, H.L.; Chang, J.H.; Kim, Y.S.; Hwang, I.K.; Ji, G.E. Enzymatic Modification of Saponins from Platycodon grandiflorum with Aspergillus niger. J. Agric. Food Chem. 2007, 55, 8908–8913. [Google Scholar] [CrossRef]
- Li, W.; Zhao, L.C.; Wang, Z.; Zheng, Y.N.; Liang, J.; Wang, H. Response Surface Methodology to Optimize Enzymatic Preparation of Deapio-Platycodin D and Platycodin D from Radix Platycodi. Int. J. Mol. Sci. 2012, 13, 4089–4100. [Google Scholar] [CrossRef]
- Jeong, E.K.; Ha, I.J.; Kim, Y.S.; Na, Y.C. Glycosylated Platycosides: Identification by Enzymatic Hydrolysis and Structural Determination by Lc-Ms/Ms. J. Sep. Sci. 2014, 37, 61–68. [Google Scholar] [CrossRef]
- Ha, I.J.; Ha, Y.W.; Kang, M.; Lee, J.; Park, D.; Kim, Y.S. Enzymatic Transformation of Platycosides and One-Step Separation of Platycodin D by High-Speed Countercurrent Chromatography. J. Sep. Sci. 2010, 33, 1916–1922. [Google Scholar] [CrossRef]
- Kil, T.G.; Kang, S.H.; Kim, T.H.; Shin, K.C.; Oh, D.K. Enzymatic Biotransformation of Balloon Flower Root Saponins into Bioactive Platycodin D by Deglucosylation with Caldicellulosiruptor bescii Beta-Glucosidase. Int. J. Mol. Sci. 2019, 20, 3854. [Google Scholar] [CrossRef]
- Kang, S.H.; Kim, T.H.; Shin, K.C.; Ko, Y.J.; Oh, D.K. Biotransformation of Food-Derived Saponins, Platycosides, into Deglucosylated Saponins Including Deglucosylated Platycodin D and Their Anti-Inflammatory Activities. J. Agric. Food Chem. 2019, 67, 1470–1477. [Google Scholar] [CrossRef]
- Ha, Y.W.; Na, Y.C.; Seo, J.J.; Kim, S.N.; Linhardt, R.J.; Kim, Y.S. Qualitative and Quantitative Determination of Ten Major Saponins in Platycodi Radix by High Performance Liquid Chromatography with Evaporative Light Scattering Detection and Mass Spectrometry. J. Chromatogr. A 2006, 1135, 27–35. [Google Scholar] [CrossRef]
- Lee, N.K.; Nyakudya, E.; Jeong, Y.-S. Bioconversion of Platycodon grandiflorum Saponins by the Platycodin D-Converting Microorganism, Yeast Cyberlindnera fabianii. J. Food Biochem. 2016, 40, 358–365. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
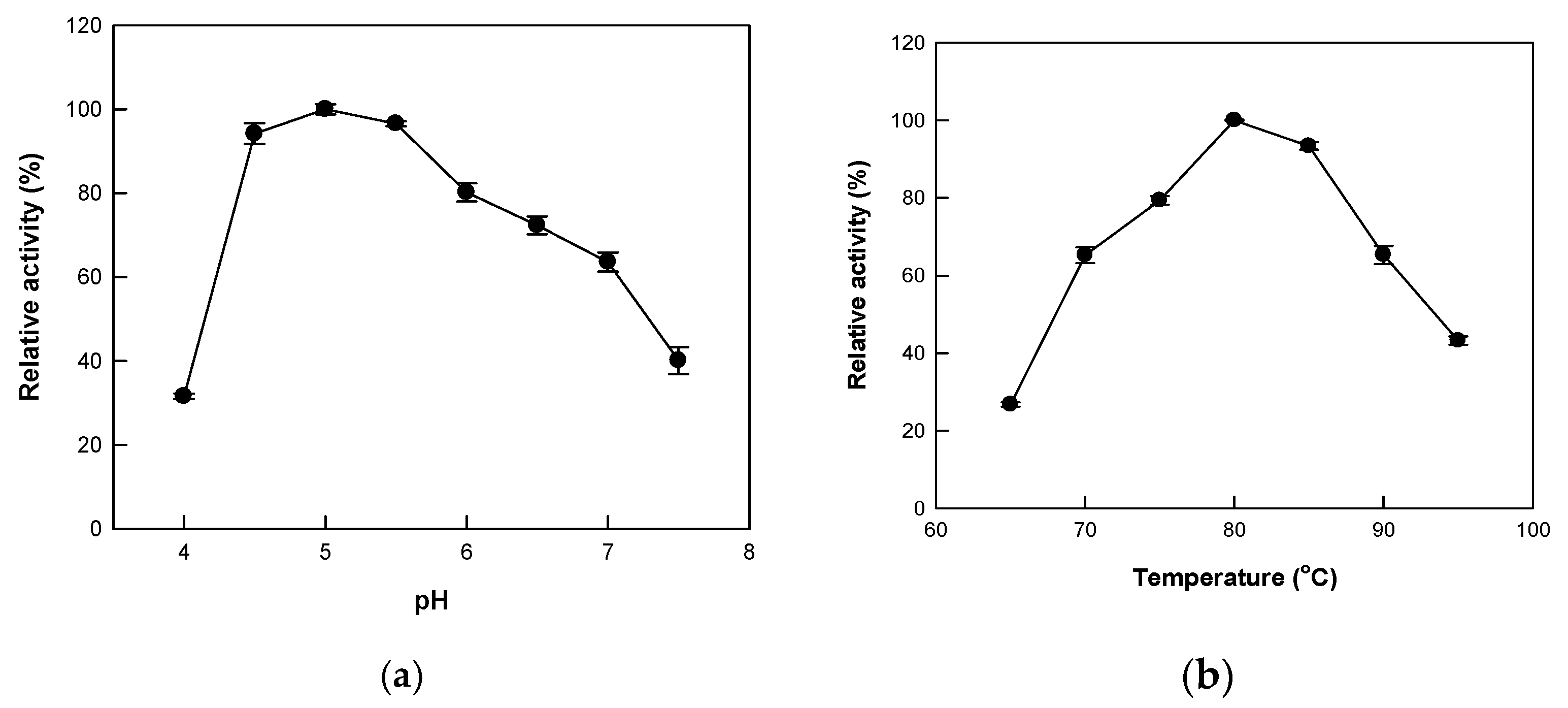

| Substrates | Specific Activity (μmol/min/mg) | |
|---|---|---|
| Aryl-glycoside | oNP-β-d-glucopyranoside | 46.0 |
| oNP-β-d-galactopyranoside | 21.0 | |
| oNP-β-d-xylopyranoside | 6.6 | |
| pNP-β-d-glucopyranoside | 51.6 | |
| pNP-β-d-galactopyranoside | 35.7 | |
| pNP-β-d-xylopyranoside | 5.5 | |
| pNP-α-l-arabinopyranoside | 11.5 | |
| pNP-α-l-rhamnopyranoside | 8.6 | |
| Platycoside | Platycoside E | 72.5 |
| Deapi-platycoside E | 60.6 | |
| Platycodin D3 | 2.5 | |
| Deapi-platycodin D3 | 1.1 | |
| Platycodin D | ND | |
| Deapi-platycodin D | ND | |
| Platycoside | Content (%, w/w) | Concentration (mg/mL) |
|---|---|---|
| Deapi-platycoside E | 2.89 | 0.16 |
| Platycoside E | 92.31 | 5.19 |
| Deapi-platycodin D | 0.53 | 0.03 |
| Platycodin D | 1.34 | 0.07 |
| Polygalacin D | 2.12 | 0.12 |
| 3″-O-Acetyl polygalacin D | 0.45 | 0.03 |
| Platycodin A | 0.36 | 0.02 |
| Total | 100 | 5.62 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, K.-C.; Seo, M.-J.; Kim, D.-W.; Yeom, S.-J.; Kim, Y.-S. Characterization of β-Glycosidase from Caldicellulosiruptor owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf. Catalysts 2019, 9, 1025. https://doi.org/10.3390/catal9121025
Shin K-C, Seo M-J, Kim D-W, Yeom S-J, Kim Y-S. Characterization of β-Glycosidase from Caldicellulosiruptor owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf. Catalysts. 2019; 9(12):1025. https://doi.org/10.3390/catal9121025
Chicago/Turabian StyleShin, Kyung-Chul, Min-Ju Seo, Dae-Wook Kim, Soo-Jin Yeom, and Yeong-Su Kim. 2019. "Characterization of β-Glycosidase from Caldicellulosiruptor owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf" Catalysts 9, no. 12: 1025. https://doi.org/10.3390/catal9121025
APA StyleShin, K.-C., Seo, M.-J., Kim, D.-W., Yeom, S.-J., & Kim, Y.-S. (2019). Characterization of β-Glycosidase from Caldicellulosiruptor owensensis and Its Application in the Production of Platycodin D from Balloon Flower Leaf. Catalysts, 9(12), 1025. https://doi.org/10.3390/catal9121025





