Enhanced Lycopene Production in Escherichia coli by Expression of Two MEP Pathway Enzymes from Vibrio sp. Dhg
Abstract
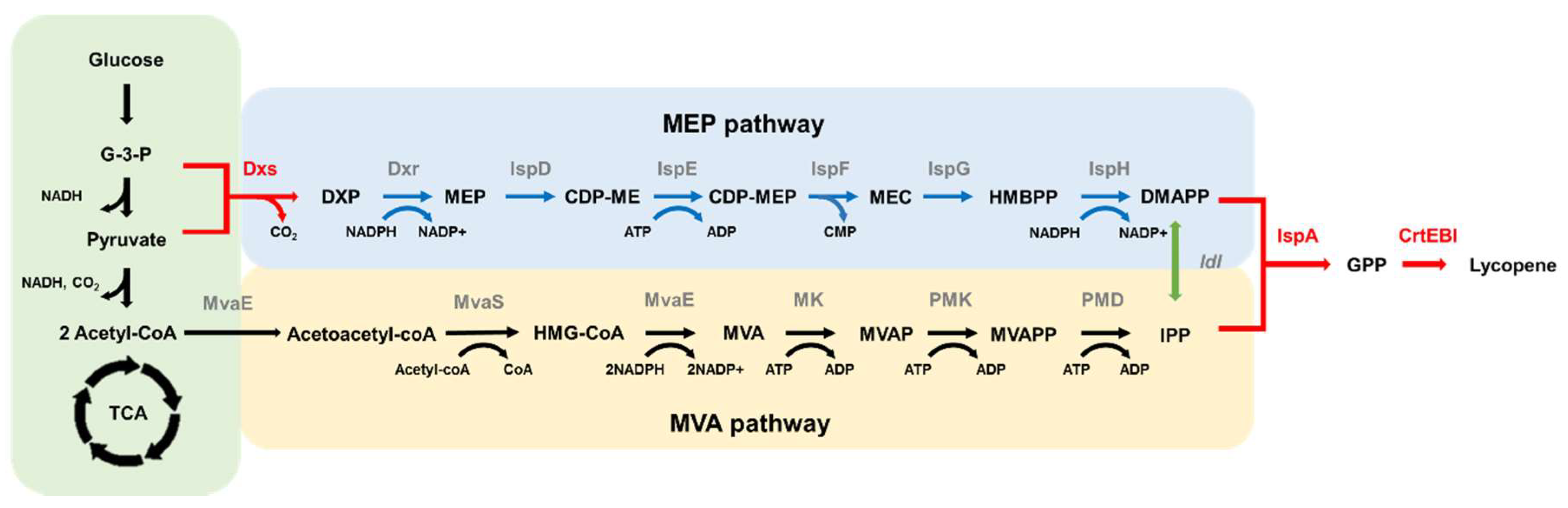
:1. Introduction
2. Results
2.1. Enhanced Lycopene Production with Optimization of crtEBI Expression
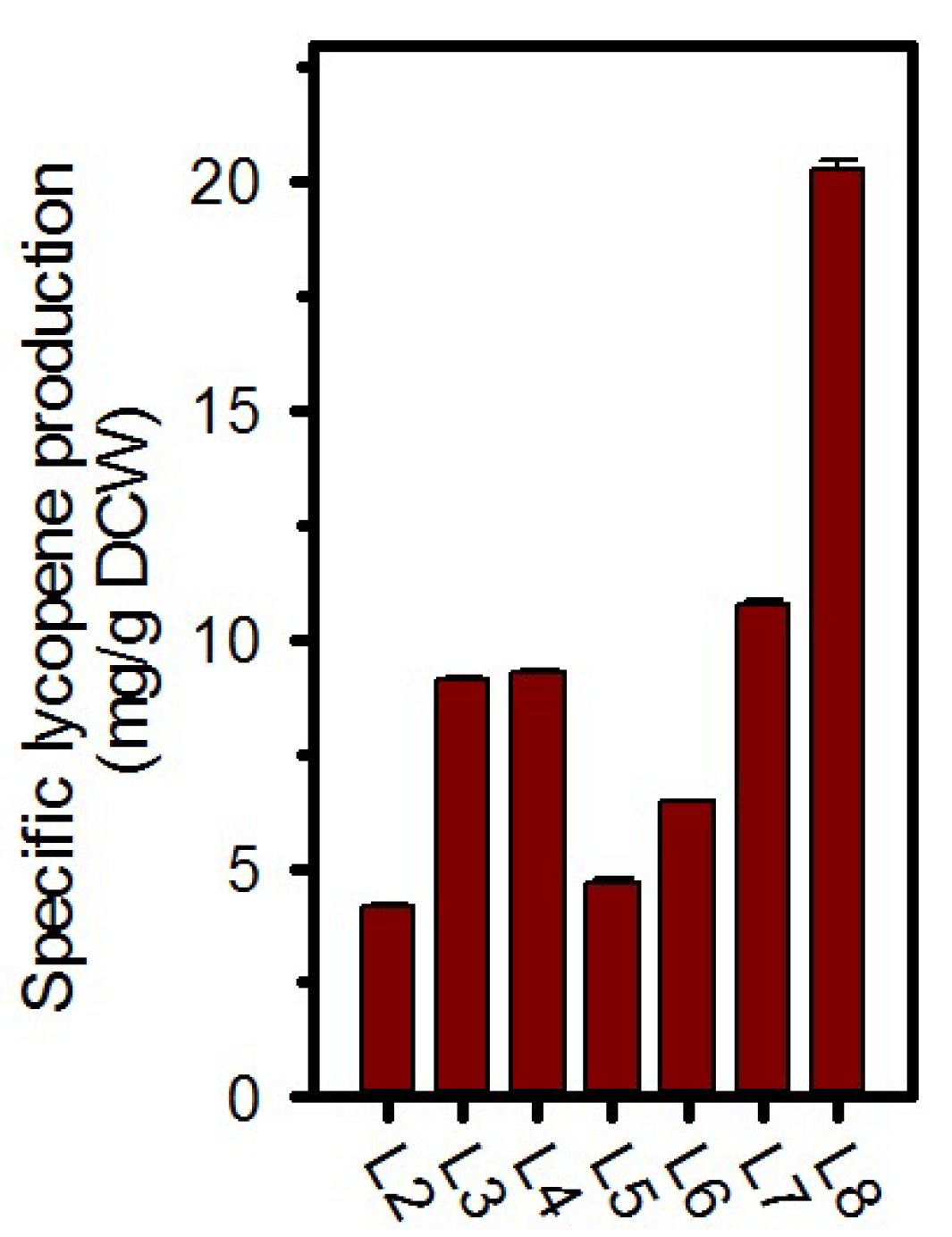
2.2. Validation of Enhanced Lycopene Production with Expression of Dxs and IspA
2.3. Evaluation of Enzyme Activity
3. Discussion
4. Materials and Methods
4.1. Reagents and Oligonucleotides
4.2. Construction of Strains and Plasmids
4.3. Culture Medium and Culture Condition
4.4. Quantification of Lycopene Production
4.5. Enzyme Assay for Dxs
4.6. Enzyme Assay for IspA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hernández-Almanza, A.; Montañez, J.; Martínez, G.; Aguilar-Jiménez, A.; Contreras-Esquivel, J.C.; Aguilar, C.N. Lycopene: Progress in microbial production. Trends Food Sci. Technol. 2016, 56, 142–148. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Jo, H.G.; Kim, M.J.; Kang, M.J.; Shin, H.J. Fruit juice supplementation alters human skin antioxidant levels In vivo: Case study of korean adults by resonance raman spectroscopy. Biotechnol. Bioprocess Eng. 2018, 23, 116–121. [Google Scholar] [CrossRef]
- Jung, J.; Lim, J.H.; Kim, S.Y.; Im, D.K.; Seok, J.Y.; Lee, S.J.V.; Oh, M.K.; Jung, G.Y. Precise precursor rebalancing for isoprenoids production by fine control of gapA expression in Escherichia coli. Metab. Eng. 2016, 38, 401–408. [Google Scholar] [CrossRef]
- Yen, H.W.; Palanisamy, G.; Su, G.C. The influences of supplemental vegetable oils on the growth and β-carotene accumulation of oleaginous yeast-Rhodotorula glutinis. Biotechnol. Bioprocess Eng. 2019, 24, 522–528. [Google Scholar] [CrossRef]
- Chen, H.; Liu, C.; Li, M.; Zhang, H.; Xian, M.; Liu, H. Directed evolution of mevalonate kinase in: Escherichia coli by random mutagenesis for improved lycopene. RSC Adv. 2018, 8, 15021–15028. [Google Scholar] [CrossRef]
- Jin, Y.S.; Stephanopoulos, G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 2007, 9, 337–347. [Google Scholar] [CrossRef]
- Ciriminna, R.; Fidalgo, A.; Meneguzzo, F.; Ilharco, L.M.; Pagliaro, M. Lycopene: Emerging Production Methods and Applications of a Valued Carotenoid. ACS Sustain. Chem. Eng. 2016, 4, 643–650. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Shen, H.J.; Cui, Y.Y.; Chen, S.G.; Weng, Z.M.; Zhao, M.; Liu, J.Z. Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biotechnol. 2013, 13, 6. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Xu, Q.; Zhang, Z.; Jiang, L.; Huang, H. Efficient production of lycopene by engineered E. coli strains harboring different types of plasmids. Bioprocess Biosyst. Eng. 2018, 41, 489–499. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, W.; Wang, Y.; Liu, H.; Li, X.; Yuan, Y. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb. Cell Factories 2016, 15, 113. [Google Scholar] [CrossRef] [PubMed]
- Sevgili, A.; Erkmen, O. Improved lycopene production from different substrates by mated fermentation of Blakeslea Trispora. Foods 2019, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.X.; Lu, Q.; Bu, Y.F.; Liu, J.Z. Metabolic engineering for the microbial production of isoprenoids: Carotenoids and isoprenoid-based biofuels. Synth. Syst. Biotechnol. 2017, 2, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Naviglio, D.; Pizzolongo, F.; Ferrara, L.; Aragòn, A.; Santini, A. Extraction of pure lycopene from industrial tomato by-products in water using a new high-pressure process. J. Sci. Food Agric. 2008, 88, 2414–2420. [Google Scholar] [CrossRef]
- Ilahy, R.; Tlili, I.; Siddiqui, M.W.; Hdider, C.; Lenucci, M.S. Inside and beyond color: Comparative overview of functional quality of tomato and watermelon fruits. Front. Plant Sci. 2019, 10, 769. [Google Scholar] [CrossRef]
- Liu, X.J.; Liu, R.S.; Li, H.M.; Tang, Y.J. Lycopene production from synthetic medium by Blakeslea trispora NRRL 2895 (+) and 2896 (−) in a stirred-tank fermenter. Bioprocess Biosyst. Eng. 2012, 35, 739–749. [Google Scholar] [CrossRef]
- Yamano, S.; Ikenaga, H.; Misawa, N.; Ishii, T.; Nakagawa, M. Metabolic engineering for production of β-carotene and lycopene in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 1994, 58, 1112–1114. [Google Scholar] [CrossRef]
- Farmer, W.R.; Liao, J.C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 2000, 18, 533–537. [Google Scholar] [CrossRef]
- Demissie, Z.A.; Erland, L.A.E.; Rheault, M.R.; Mahmoud, S.S. The biosynthetic origin of irregular monoterpenes in Lavandula. J. Biol. Chem. 2013, 288, 6333–6341. [Google Scholar] [CrossRef]
- Cheng, B.Q.; Wei, L.J.; Lv, Y.B.; Chen, J.; Hua, Q. Elevating limonene production in oleaginous yeast Yarrowia lipolytica via genetic engineering of limonene biosynthesis pathway and optimization of medium composition. Biotechnol. Bioprocess Eng. 2019, 24, 500–506. [Google Scholar] [CrossRef]
- Kuzuyama, T.; Seto, H. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012, 88, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Nian, R.; Xian, M.; Zhang, H. Metabolic engineering for the production of isoprene and isopentenol by Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 7725–7738. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Kwak, D.H.; Park, S.; Woo, S.; Yang, J.S.; Kang, C.W.; Kim, B.; Noh, M.H.; Seo, S.W.; Jung, G.Y. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 2019, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Kondo, K.; Saito, T.; Shimada, H.; Fraser, P.D.; Misawa, N. Production of the carotenoids lycopene, β-carotene, and astaxanthin in the food yeast Candida utilis. Appl. Environ. Microbiol. 1998, 64, 1226–1229. [Google Scholar] [PubMed]
- Kim, S.W.; Keasling, J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001, 72, 408–415. [Google Scholar] [CrossRef]
- Banerjee, A.; Wu, Y.; Banerjee, R.; Li, Y.; Yan, H.; Sharkey, T.D. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 2013, 288, 16926–16936. [Google Scholar] [CrossRef]
- Kang, M.J.; Yoon, S.H.; Lee, Y.M.; Lee, S.H.; Kim, J.E.; Jung, K.H.; Shin, Y.C.; Kim, S.W. Enhancement of lycopene production in Escherichia coli by optimization of the lycopene synthetic pathway. J. Microbiol. Biotechnol. 2005, 15, 880–886. [Google Scholar]
- Li, M.; Hou, F.; Wu, T.; Jiang, X.; Li, F.; Liu, H.; Xian, M.; Zhang, H. Recent advances of metabolic engineering strategies in natural isoprenoid production using cell factories. Nat. Prod. Rep. 2019. [Google Scholar] [CrossRef]
- Glazunova, O.A.; Trushkin, N.A.; Moiseenko, K.V.; Filimonov, I.S.; Fedorova, T.V. Catalytic efficiency of basidiomycete laccases: Redox potential versus substrate-binding pocket structure. Catalysts 2018, 8, 152. [Google Scholar] [CrossRef]
- Chang, W.; Song, H.; Liu, H.; Liu, P. Current development in isoprenoid precursor biosynthesis and regulation. Curr. Opin. Chem. Biol. 2013, 17, 571–579. [Google Scholar] [CrossRef]
- Martínez-De Drets, G.; Arias, A. Enzymatic basis for differentiation of Rhizobium into fast- and slow-growing groups. J. Bacteriol. 1972, 109, 467–470. [Google Scholar] [PubMed]
- Scholtz, R.; Wackett, L.P.; Egli, C.; Cook, A.M.; Leisinger, T. Dichloromethane dehalogenase with improved catalytic activity isolated from a fast-growing dichloromethane-utilizing bacterium. J. Bacteriol. 1988, 170, 5698–5704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Des Soye, B.J.; Davidson, S.R.; Weinstock, M.T.; Gibson, D.G.; Jewett, M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 2018, 7, 2245–2255. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.W.; Lim, H.G.; Yang, J.; Noh, M.H.; Seo, S.W.; Jung, G.Y. Synthetic auxotrophs for stable and tunable maintenance of plasmid copy number. Metab. Eng. 2018, 48, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Matthäus, F.; Ketelhot, M.; Gatter, M.; Barth, G. Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2014, 80, 1660–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, K.L.; Kim, S.W.; Keasling, J.D. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2000, 2, 328–338. [Google Scholar] [CrossRef]
- Kim, S.G.; Noh, M.H.; Lim, H.G.; Jang, S.; Jang, S.; Koffas, M.A.G.; Jung, G.Y. Molecular parts and genetic circuits for metabolic engineering of microorganisms. FEMS Microbiol. Lett. 2018, 365, fny187. [Google Scholar] [CrossRef]
- Seo, S.W.; Yang, J.S.; Kim, I.; Yang, J.; Min, B.E.; Kim, S.; Jung, G.Y. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013, 15, 67–74. [Google Scholar] [CrossRef]
- Gallego-Jara, J.; de Diego, T.; del Real, Á.; Écija-Conesa, A.; Manjón, A.; Cánovas, M. Lycopene overproduction and in situ extraction in organic-aqueous culture systems using a metabolically engineered Escherichia coli. AMB Express 2015, 5, 65. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.Z.; Rouvière, P.E.; LaRossa, R.A.; Suh, W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 2006, 8, 79–90. [Google Scholar] [CrossRef]
- Kuzuyama, T.; Takagi, M.; Takahashi, S.; Seto, H. Cloning and characterization of 1-deoxy-D-xylulose 5-phosphate synthase from Streptomyces sp. Strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J. Bacteriol. 2000, 182, 891–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albe, K.R.; Butler, M.H.; Wright, B.E. Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 1990, 143, 163. [Google Scholar] [CrossRef]
- Hahn, F.M.; Eubanks, L.M.; Testa, C.A.; Blagg, B.S.J.; Baker, J.A.; Poulter, C.D. 1-deoxy-D-xylulose 5-phosphate synthase, the gene product of open reading frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 2001, 183, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Zheng, Y.; Tang, H.; Long, Z.; Li, J.; Zhang, Z.; Liu, S.; Mao, D.; Wei, T. Highly efficient synthesis of 2,5-dihydroxypyridine using Pseudomonas sp. ZZ-5 nicotine hydroxylase immobilized on immobead 150. Catalysts 2018, 8, 548. [Google Scholar] [CrossRef] [Green Version]
- Seok, J.Y.; Yang, J.; Choi, S.J.; Lim, H.G.; Choi, U.J.; Kim, K.J.; Park, S.; Yoo, T.H.; Jung, G.Y. Directed evolution of the 3-hydroxypropionic acid production pathway by engineering aldehyde dehydrogenase using a synthetic selection device. Metab. Eng. 2018, 47, 113–120. [Google Scholar] [CrossRef]
- Currin, A.; Swainston, N.; Day, P.J.; Kell, D.B. Synthetic biology for the directed evolution of protein biocatalysts: Navigating sequence space intelligently. Chem. Soc. Rev. 2015, 44, 1172–1239. [Google Scholar] [CrossRef] [Green Version]
- Weinstock, M.T.; Hesek, E.D.; Wilson, C.M.; Gibson, D.G. Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 2016, 13, 849–851. [Google Scholar] [CrossRef]
- Querol, J.; Besumbes, O.; Maria Lois, L.; Boronat, A.; Imperial, S. A fluorometric assay for the determination of 1-deoxy-D-xylulose 5-phosphate synthase activity. Anal. Biochem. 2001, 296, 101–105. [Google Scholar] [CrossRef]
- Noh, M.H.; Lim, H.G.; Park, S.; Seo, S.W.; Jung, G.Y. Precise flux redistribution to glyoxylate cycle for 5-aminolevulinic acid production in Escherichia coli. Metab. Eng. 2017, 43, 1–8. [Google Scholar] [CrossRef]
- Dozier, J.K.; Distefano, M.D. An enzyme-coupled continuous fluorescence assay for farnesyl diphosphate synthases. Anal. Biochem. 2012, 421, 158–163. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Hamano, Y.; Kuzuyama, T.; Itoh, N.; Seto, H.; Dairi, T. Interconversion of the product specificity of type I eubacterial farnesyl diphosphate synthase and geranylgeranyl diphosphate synthase through one amino acid substitution. J. Biochem. 2003, 133, 83–91. [Google Scholar] [CrossRef] [PubMed]

| Name | Description | Source |
|---|---|---|
| Strains | ||
| E. coli Mach-T1R | Cloning host | Invitrogen |
| E. coli W3110 | Production host, source for dxsEC and ispAEC | ATCC 9637 |
| Vibrio sp. dhg | Recently isolated fast-growing strain, source for dxsVDHG and ispAVDHG | [23] |
| JYJ0 | E. coli W3110/pCDF_crtEBI | [4] |
| L1 | E. coli W3110/p1EBI | This study |
| L2 | E. coli W3110/p2EBI | This study |
| L3 | E. coli W3110/pdE | This study |
| L4 | E. coli W3110/pdV | This study |
| L5 | E. coli W3110/piE | This study |
| L6 | E. coli W3110/piV | This study |
| L7 | E. coli W3110/pdEiE | This study |
| L8 | E. coli W3110/pdViV | This study |
| D1 | E. coli W3110/pCdEH | This study |
| D2 | E. coli W3110/pCdVH | This study |
| V1 | E. coli W3110/pCiEH | This study |
| V2 | E. coli W3110/pCiVH | This study |
| P | E. coli W3110/pCPFT | This study |
| Plasmids | ||
| pACYCduet_1 | p15A, LacI, CmR, E. coli expression vector | Novagen |
| p1EBI | p15A, LacI, CmR, Ptac_synUTRcrtE_crtE_ crtB_crtI_TBBa_B1005 | This study |
| p2EBI | p15A, LacI, CmR, Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| pdE | p15A, LacI, CmR, Ptac_synUTRdxsEC_dxsEC_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| pdV | p15A, LacI, CmR, Ptac_synUTRdxsVDHG_dxsVDHG_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| piE | p15A, LacI, CmR, Ptac_synUTRispAEC_ispA EC_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| piV | p15A, LacI, CmR, Ptac_synUTRispAVDHG_ispAVDHG_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| pdEiE | p15A, LacI, CmR, Ptac_synUTRispAEC_ ispA EC_ Ptac_synUTRdxsEC_dxsEC_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| pdViV | p15A, LacI, CmR, Ptac_synUTRispAVDHG_ ispA VDHG_ Ptac_synUTRdxsVDHG_dxsVDHG_ Ptac_synUTRcrtE_crtE_Ptac_synUTRcrtB_crtB_Ptac_synUTRcrtI_crtI_TBBa_B1005 | This study |
| pCDFduet_1 | CloDF13, LacI, SmR, E. coli expression vector | Novagen |
| pCDF_crtEBI | CloDF13, LacI, SmR, PBBa_J23100_ synUTRcrtE_crtE_crtB_crtI_TBBa_B1005 | [4] |
| pCdEH | CloDF13, LacI, SmR, Ptac_synUTRdxsEC_dxsEC_6X His_TBBa_B1001 | This study |
| pCdVH | CloDF13, LacI, SmR, Ptac_synUTRdxsVDHG_dxsVDHG_6X His_TBBa_B1001 | This study |
| pCiEH | CloDF13, LacI, SmR, Ptac_synUTRispAEC_ispAEC_6X His_TBBa_B1001 | This study |
| pCiVH | CloDF13, LacI, SmR, Ptac_synUTRispAVDHG_ispAVDHG_6X His_TBBa_B1001 | This study |
| pCPFT | CloDF13, LacI, SmR, Ptac_synUTRPFTase_RAM1_ TBBa_B1001 | This study |
| Enzyme | Substrate | kcat | Km | kcat/Km |
|---|---|---|---|---|
| DxsEC | DL-glyceraldehyde * | 3.78 (±0.53) s−1 | 54.48 (±9.42) mM | 69.38 (±5.33) μM·s |
| DxsVDHG | DL-glyceraldehyde * | 5.74 (±0.23) s−1 | 76.50 (±4.87) mM | 75.05 (±3.82) μM·s |
| IspAEC | DMAPP | 3.02 (±0.16) s−1 | 8.79 (±0.42) μM | 0.34 (±0.04) μM·s |
| IspAVDHG | DMAPP | 4.14 (±0.18) s−1 | 8.73 (±0.44) μM | 0.47 (±0.02) μM·s |
| IspAEC | IPP | 3.11 (±0.11) s−1 | 9.12 (±0.39) μM | 0.34 (±0.01) μM·s |
| IspAVDHG | IPP | 4.11 (±0.11) s−1 | 8.45 (±0.43) μM | 0.49 (±0.02) μM·s |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.J.; Noh, M.H.; Woo, S.; Lim, H.G.; Jung, G.Y. Enhanced Lycopene Production in Escherichia coli by Expression of Two MEP Pathway Enzymes from Vibrio sp. Dhg. Catalysts 2019, 9, 1003. https://doi.org/10.3390/catal9121003
Kim MJ, Noh MH, Woo S, Lim HG, Jung GY. Enhanced Lycopene Production in Escherichia coli by Expression of Two MEP Pathway Enzymes from Vibrio sp. Dhg. Catalysts. 2019; 9(12):1003. https://doi.org/10.3390/catal9121003
Chicago/Turabian StyleKim, Min Jae, Myung Hyun Noh, Sunghwa Woo, Hyun Gyu Lim, and Gyoo Yeol Jung. 2019. "Enhanced Lycopene Production in Escherichia coli by Expression of Two MEP Pathway Enzymes from Vibrio sp. Dhg" Catalysts 9, no. 12: 1003. https://doi.org/10.3390/catal9121003
APA StyleKim, M. J., Noh, M. H., Woo, S., Lim, H. G., & Jung, G. Y. (2019). Enhanced Lycopene Production in Escherichia coli by Expression of Two MEP Pathway Enzymes from Vibrio sp. Dhg. Catalysts, 9(12), 1003. https://doi.org/10.3390/catal9121003





