Enzymatic Synthesis of ω-Hydroxydodecanoic Acid By Employing a Cytochrome P450 from Limnobacter sp. 105 MED
Abstract
1. Introduction
2. Results and Discussion
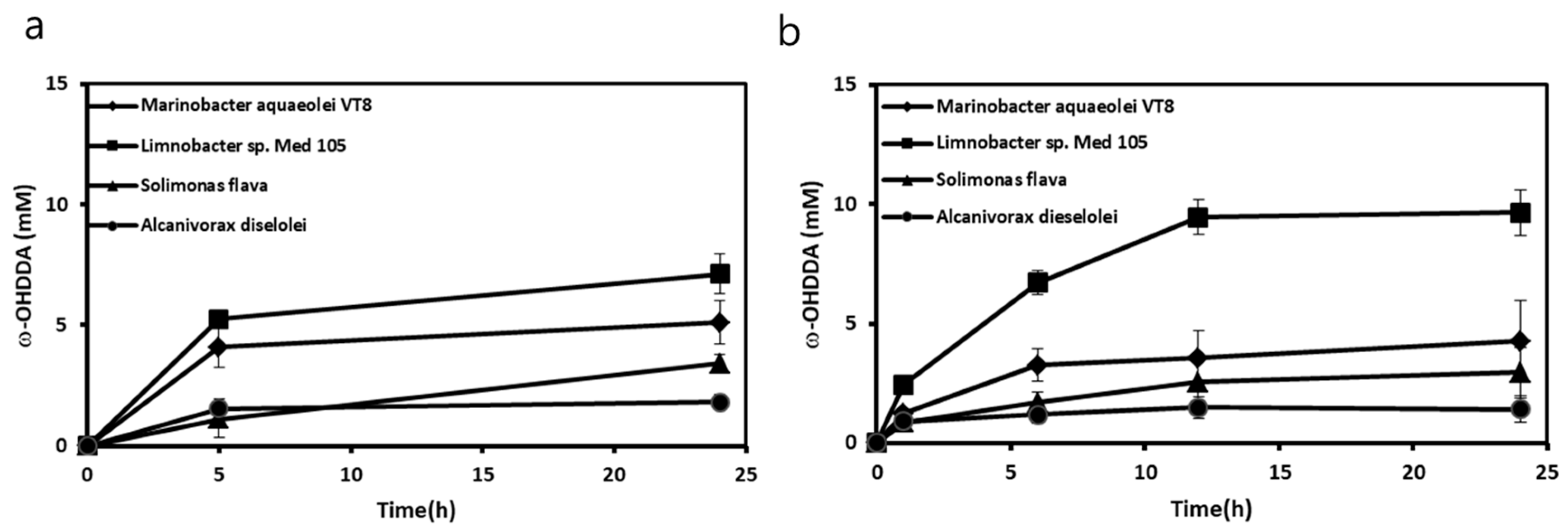
2.1. Construction of a CYP153A Three-Component System
2.2. Use of Solid State Powdered Substrate in Bioconversion
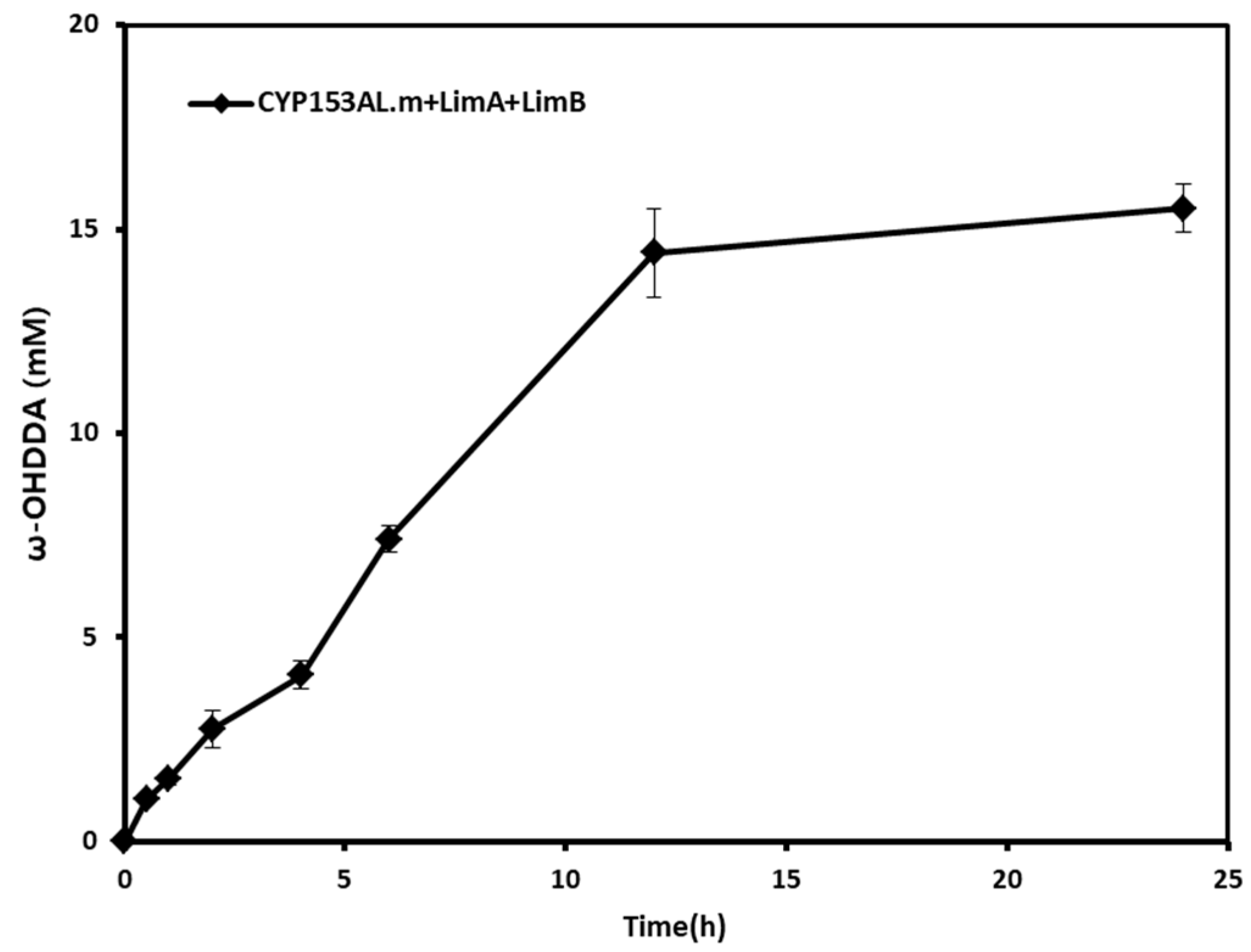
2.3. Effect of Homogeneous Redox Partners
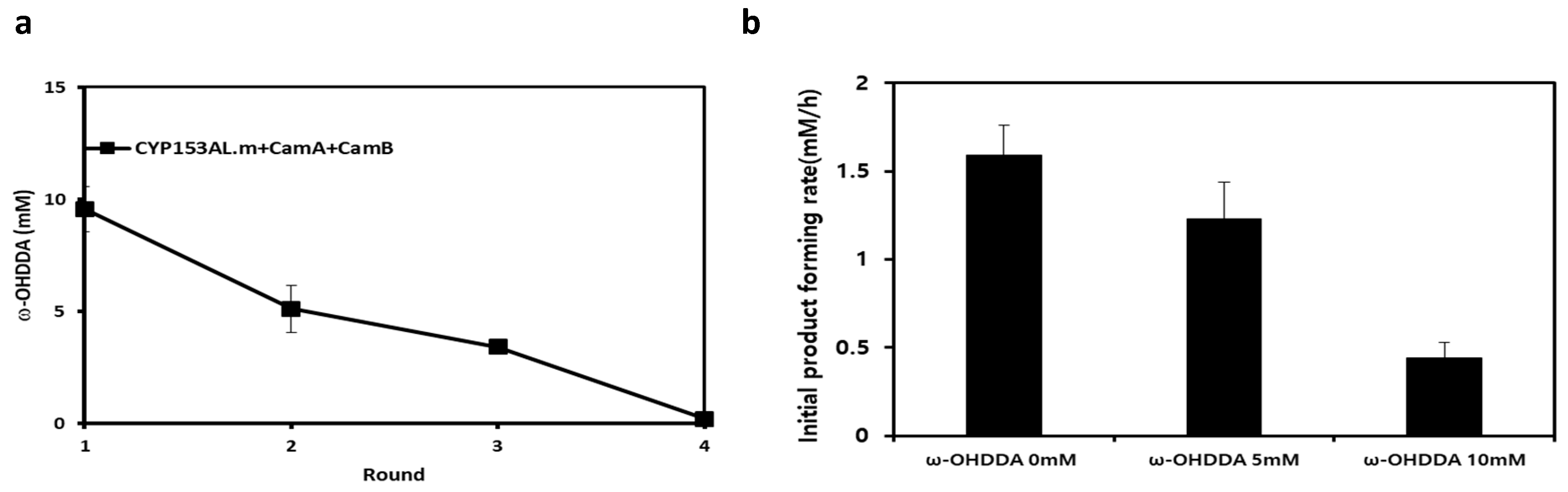
2.4. Limitations of the CYP153AL Three-Component System
3. Materials and Methods
3.1. Chemicals and Media
3.2. Plasmid Construction
3.3. Protein Expression and Purification
3.4. CO-Binding Assay
3.5. In-Vitro Oxidation Assay of CYP153As
3.6. Resting Cell Reaction
3.7. Product Identification and Quantification
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BSTFA | N, O-bis(trimethylsilyl)trifluoroacetamide |
| CDW | cell dry weight |
| CYP | cytochrome P450 enzyme |
| CYP153AL.m | Limnobacter sp. MED 105 CYP153A |
| CYP153AM.aq | Marinobacter aquaeolei CYP153A |
| CYP153AA.d | Alcanivorax dieselolei CYP153A |
| CYP153AS.f | Solimonas flava CYP153A |
| DCA | dicarboxylic acid |
| DDA | dodecanoic acid |
| ω-OHDDA | omega-hydroxydodecanoic acid |
| OHFAs | omega-hydroxy fatty acids |
| TMS | trimethylsilyl |
References
- Soliday, C.L.; Kolattukudy, P.E. Biosynthesis of Cutin ω-Hydroxylation of Fatty Acids by a Microsomal Preparation from Germinating Vicia faba. Plant Physiol. 1977, 59, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, F.; Cai, J.; Xie, W.; Long, T.E.; Turner, S.R.; Lyons, A.; Gross, R.A. Polymers from Fatty Acids: Poly(ω-hydroxyl tetradecanoic acid) Synthesis and Physico-Mechanical Studies. Biomacromolecules 2011, 12, 3291–3298. [Google Scholar] [CrossRef] [PubMed]
- Ebata, H.; Toshima, K.; Matsumura, S. Lipase-Catalyzed Synthesis and Properties of Poly[(12-hydroxydodecanoate)-co-(12-hydroxystearate)] Directed Towards Novel Green and Sustainable Elastomers. Macromol. Biosci. 2008, 8, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Sugiyama, K. Growth inhibition and apoptosis induction of human melanoma cells by omega-hydroxy fatty acids. Anti-Cancer Drugs 2005, 16, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Bordeaux, M.; Galarneau, A.; Fajula, F.; Drone, J. A Regioselective Biocatalyst for Alkane Activation Under Mild Conditions. Angew. Chem. Int. Ed. Engl. 2011, 50, 2075–2079. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Watanabe, A. Process for Producing Omega-Hydroxy Fatty Acids. U.S. Patent 5191096, 2 March 1993. [Google Scholar]
- Labinger, J.A. Selective alkane oxidation: Hot and cold approaches to a hot problem. J. Mol. Catal. A Chem. 2004, 220, 27–35. [Google Scholar] [CrossRef]
- Lu, W.; Ness, J.E.; Xie, W.; Zhang, X.; Minshull, J.; Gross, R.A. Biosynthesis of Monomers for Plastics from Renewable Oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Scheps, D.; Honda Malca, S.; Richter, S.M.; Marisch, K.; Nestl, B.M.; Hauer, B. Synthesis of ω-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb. Boithechnol. 2013, 6, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Funhoff, E.G.; Salzmann, J.; Bauer, U.; Witholt, B.; van Beilen, J.B. Hydroxylation and epoxidation reactions catalyzed by CYP153 enzymes. Enzyme Microb. Technol. 2007, 40, 806–812. [Google Scholar] [CrossRef]
- Scheps, D.; Malca, S.H.; Hoffmann, H.; Nestl, B.M.; Hauer, B. Regioselective ω-hydroxylation of medium-chain n-alkanes and primary alcohols by CYP153 enzymes from Mycobacterium marinum and Polaromonas sp. strain JS666. Org. Biomol. Chem. 2011, 9, 6727–6733. [Google Scholar] [CrossRef] [PubMed]
- Narhi, L.O.; Fulco, A.J. Identification and Characterization of Two Functional Domains in Cytochrome P-450BM-3, a Catalytically Self-sufficient Monooxygenase Induced by Barbiturates in Bacillus megaterium. J. Biol. Chem 1987, 262, 6683–6690. [Google Scholar] [PubMed]
- Malca, S.H.; Scheps, D.; Kühnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115–5117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wei, D.-Q. Recent Progress on Structural Bioinformatics Research of Cytochrome P450 and Its Impact on Drug Discovery. Adv. Struct. Eng. 2015, 827, 327–339. [Google Scholar]
- Darabi, M.; Seddigh, S.; Abarshahr, M. Structural, functional, and phylogenetic studies of cytochrome P450 (CYP) enzyme in seed plants by bioinformatics tools. Caryologia 2017, 70, 62–76. [Google Scholar] [CrossRef]
- Fischer, M.; Knoll, M.; Sirim, D.; Wagner, F.; Funke, S.; Pleiss, J. The Cytochrome P450 Engineering Database: A navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics 2007, 23, 2015–2017. [Google Scholar] [CrossRef]
- Ma, Q.H.; Tian, B. Biochemical characterization of a cinnamoyl-CoA reductase from wheat. Biol. Chem. 2005, 386, 553–560. [Google Scholar] [CrossRef]
- von Bühler, C.; Le-Huu, P.; Urlacher, V.B. Cluster Screening: An Effective Approach for Probing the Substrate Space of Uncharacterized Cytochrome P450s. ChemBioChem 2013, 14, 2189–2198. [Google Scholar] [CrossRef]
- Donova, M.V.; Nikolayeva, V.M.; Dovbnya, D.V.; Gulevskaya, S.A.; Suzina, N.E. Methyl-beta-cyclodextrin alters growth, activity and cell envelope features of sterol-transforming mycobacteria. Microbiology 2007, 153, 1981–1992. [Google Scholar] [CrossRef]
- Kühnel, K.; Maurer, S.C.; Galeyeva, Y.; Frey, W.; Laschat, S.; Urlacher, V.B. Hydroxylation of Dodecanoic Acid and (2R,4R,6R,8R)-Tetramethyldecanol on a Preparative Scale using an NADH-Dependent CYP102A1 Mutant. Adv. Synth. Catal. 2007, 349, 1451–1461. [Google Scholar] [CrossRef]
- Rammler, D.H.; Zaffaroni, A. Biological implications of DMSO based on a review of its chemical properties. Ann. N. Y. Acad. Sci. 1967, 141, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Lundemo, M.T.; Notonier, S.; Striedner, G.; Hauer, B.; Woodley, J.M. Process limitations of a whole-cell P450 catalyzed reaction using a CYP153A-CPR fusion construct expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 2016, 100, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Rimal, H.; Lee, S.W.; Lee, J.H.; Oh, T.J. Understanding of real alternative redox partner of Streptomyces peucetius DoxA: Prediction and validation using in silico and in vitro analyses. Arch. Biochem. Biophys. 2015, 585, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Fact. 2017, 16, 106. [Google Scholar] [CrossRef] [PubMed]
- Wachtmeister, J.; Rother, D. Recent advances in whole cell biocatalysis techniques bridging from investigative to industrial scale. Curr. Opin. Biotechnol. 2016, 42, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, L.; Li, F.; Zhang, X.; Qu, Z.; Han, L.; Li, Z.; Sun, J.; Qi, F.; Yao, Q.; et al. Mechanistic Insights into Interactions between Bacterial Class I P450 Enzymes and Redox Partners. ACS Catal. 2018, 8, 9992–10003. [Google Scholar] [CrossRef]
- Pandey, B.P.; Lee, N.; Choi, K.Y.; Kim, J.N.; Kim, E.J.; Kim, B.G. Identification of the specific electron transfer proteins, ferredoxin, and ferredoxin reductase, for CYP105D7 in Streptomyces avermitilis MA4680. Appl. Microbiol. Biotechnol. 2014, 98, 5009–5017. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic. Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Park, B.G.; Ahsan, M.M.; Kim, J.; Yun, H.; Choi, K.Y.; Kim, B.G. Production of ω-hydroxy palmitic acid using CYP153A35 and comparison of cytochrome P450 electron transfer system in vivo. Appl. Microbiol. Biotechnol. 2016, 100, 10375–10384. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, R.; Urlacher, V.B. Cytochromes P450 as promising catalysts for biotechnological application: Chances and limitations. Appl. Microbiol. Biotechnol. 2014, 98, 6185–6203. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.H.; Park, B.G.; Jung, E.; Lee, P.-G.; Kim, B.-G. fadD deletion and fadL overexpression in Escherichia coli increase hydroxy long-chain fatty acid productivity. Appl. Microbiol. Biotechnol. 2014, 98, 8917–8925. [Google Scholar] [CrossRef] [PubMed]

| Entry | CYP153AM.aq CamA + CamB | CYP153AL.m CamA + CamB | CYP153AL.m LimA + LimB |
|---|---|---|---|
| Coupling efficiency (%) | 53.6 ± 5.5 | 52.3 ± 3.6 | 48.4 ± 5.4 |
| Initial product forming rate (µM/min) | 7.41 ± 0.70 | 8.91 ± 0.89 | 9.81 ± 1.60 |
| NADH consumption rate (µM/min) | 13.83 ± 1.16 | 17.04 ± 0.32 | 20.26 ± 0.57 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, S.-Y.; Yoo, H.-W.; Sarak, S.; Kim, B.-G.; Yun, H. Enzymatic Synthesis of ω-Hydroxydodecanoic Acid By Employing a Cytochrome P450 from Limnobacter sp. 105 MED. Catalysts 2019, 9, 54. https://doi.org/10.3390/catal9010054
Joo S-Y, Yoo H-W, Sarak S, Kim B-G, Yun H. Enzymatic Synthesis of ω-Hydroxydodecanoic Acid By Employing a Cytochrome P450 from Limnobacter sp. 105 MED. Catalysts. 2019; 9(1):54. https://doi.org/10.3390/catal9010054
Chicago/Turabian StyleJoo, Sung-Yeon, Hee-Wang Yoo, Sharad Sarak, Byung-Gee Kim, and Hyungdon Yun. 2019. "Enzymatic Synthesis of ω-Hydroxydodecanoic Acid By Employing a Cytochrome P450 from Limnobacter sp. 105 MED" Catalysts 9, no. 1: 54. https://doi.org/10.3390/catal9010054
APA StyleJoo, S.-Y., Yoo, H.-W., Sarak, S., Kim, B.-G., & Yun, H. (2019). Enzymatic Synthesis of ω-Hydroxydodecanoic Acid By Employing a Cytochrome P450 from Limnobacter sp. 105 MED. Catalysts, 9(1), 54. https://doi.org/10.3390/catal9010054







