Abstract
Corynebacterium glutamicum is an industrial strain used for the production of valuable chemicals such as L-lysine and L-glutamate. Although C. glutamicum has various industrial applications, a limited number of tunable systems are available to engineer it for efficient production of platform chemicals. Therefore, in this study, we developed a novel tunable promoter system based on repeats of the Vitreoscilla hemoglobin promoter (Pvgb). Tunable expression of green fluorescent protein (GFP) was investigated under one, four, and eight repeats of Pvgb (Pvgb, Pvgb4, and Pvgb8). The intensity of fluorescence in recombinant C. glutamicum strains increased as the number of Pvgb increased from single to eight (Pvgb8) repeats. Furthermore, we demonstrated the application of the new Pvgb promoter-based vector system as a platform for metabolic engineering of C. glutamicum by investigating 5-aminovaleric acid (5-AVA) and gamma-aminobutyric acid (GABA) production in several C. glutamicum strains. The profile of 5-AVA and GABA production by the recombinant strains were evaluated to investigate the tunable expression of key enzymes such as DavBA and GadBmut. We observed that 5-AVA and GABA production by the recombinant strains increased as the number of Pvgb used for the expression of key proteins increased. The recombinant C. glutamicum strain expressing DavBA could produce higher amounts of 5-AVA under the control of Pvgb8 (3.69 ± 0.07 g/L) than the one under the control of Pvgb (3.43 ± 0.10 g/L). The average gamma-aminobutyric acid production also increased in all the tested strains as the number of Pvgb used for GadBmut expression increased from single (4.81–5.31 g/L) to eight repeats (4.94–5.58 g/L).
1. Introduction
Biorefinery processes have been developed to establish a sustainable alternative to produce chemicals, polymers, and fuels, and they currently rely on petroleum-based processes [,,,,,,,,,,,,,,,,]. Development of recombinant microorganisms capable of converting a broader range of renewable biomass feedstock into bio-based chemicals, with properties comparable to those of conventional petrochemical products, have been extensively studied [,,,,,,,,,]. For example, processes for the production of bio-polymers, such as polylactic acid (PLA) and polybutylene succinate (PBS) in a biorefinery have been established [,,]. Polymerization of microbial fermentation derived-monomers, such as diamines, dicarboxylic acids, and amino carboxylic acids, yields these bio-based polymers [,]. Escherichia coli is one of the most commonly used microorganisms for the production of monomers used to prepare bio-based polymers [,,]. However, production of amino acid-derived products such as 5-aminovaleric acid (5-AVA) and gamma-aminobutyric acid (GABA) from glucose using recombinant E. coli is limited, due to its inability to accumulate important precursors such as L-lysine and L-glutamate [,]. In this regard, Corynebacterium glutamicum, which is a generally recognized as safe (GRAS) strain, is an excellent workhorse to develop because of its innate ability to produce large amounts of amino acids such as L-lysine and L-glutamate [,,]. Several studies have demonstrated that C. glutamicum can produce high levels of bio-based nylon monomers such putrescine, succinic acid [], GABA [], cadaverine [,], 5-AVA and glutaric acid, []. Bio-based nylon-4, nylon-5 and nylon-6,5 can be produced from butyrolactam and Δ-valerolactam, which are derived from GABA and 5-AVA, respectively []. Biosynthetic production of both 5-AVA and GABA is more sustainable than chemical production because the fermentative production of 5-AVA and GABA is conducted under mild conditions with high catalytic efficiency [,,,,,,]. In addition, 5-AVA and GABA have been successfully produced from biomass-derived sugars such as Miscanthus hydrolysate and empty fruit bunch biosugar solution [,].
To engineer C. glutamicum for biochemical production, its metabolic pathway has been manipulated using a plasmid-based expression system, which establishes the synthetic pathway for the production of biochemicals []. This is an important step for evaluating the success of the constructed pathway and for establishing the target metabolite production in recombinant strains. This also helps identify which key reactions can be improved in order to drive metabolic flux toward optimized chemical production [,,]. For example, the production of GABA using C. glutamicum strains was established by using a synthetic promoter-based expression vector system (PL26 < PI16 < PH36) capable of low, intermediate, and high-strength glutamate decarboxylase (GadBmut) expression. It was demonstrated that the use of the high-strength promoter PH36 in the pHGmut strain (5.89 ± 0.35 g/L) enabled higher production of GABA than in pIGmut (5.32 ± 0.04 g/L) and pLGmut (4.87 ± 0.15 g/L) strains with expression of GadBmut under intermediate (PI16) and low (PL26) strength promoters, respectively []. Cadaverine production using a synthetic promoter-based expression system (PL10 < PL26 < PI1 6 < PI64 < PH30 < PH36) was also evaluated, and it was observed that a high level of protein expression using the strongest promoter (PH36) does not necessarily result in the highest level of metabolite production [,]. In case of batch fermentation for cadaverine production using recombinant C. glutamicum strains, the use of PH30 (23.8 g/L) for the expression of lysine decarboxylase produced a higher titer than that when the PH36 promoter was used (21.3 g/L) []. However, the repertoire of vector systems capable of different levels of protein expression in C. glutamicum is still limited []. Most promoters in plasmids derived from E. coli plasmids such as Ptac, Ptrc, PlacUV5, PR, and PL have been evaluated [,]. However, despite the adoption of the promoters from E. coli plasmids, the variety of genetic engineering tools applicable to C. glutamicum is still less compared to the tools available for E. coli [,,]. Therefore, the discovery or creation of a new vector system and evaluation of its capability for tunable protein expression in a target host strain are important factors in developing recombinant strains for the production of biochemicals in biorefineries [,].
Recently, the use of five repeats of the Ptac promoter successfully enabled stable overexpression of phaCAB genes in recombinant E. coli and enhanced poly(R-3-hydroxybutyrate) (PHB) accumulation 5.6 times that of the control strain, which had a single copy of the Ptac promoter []. In another report, the use of the promoter from Vitreoscilla hemoglobin protein (Pvgb) was successfully demonstrated in E. coli. It was found that the expression vectors with increasing repeats of the Pvgb promoter enabled tunable expression of the PHB synthesis operon (phaCAB) and allowed enhanced accumulation of poly(hydroxybutyrate) (PHB) []. The highest accumulation of 90% PHB in 5.37 g/L CDW was achieved by the recombinant strain with eight repeats of the Pvgb promoter. The use of the Pvgb promoter system was also successfully demonstrated in recombinant C. glutamicum. It was used to investigate the effect of VHb protein expression on L-glutamate and L-glutamine production in the recombinant strains. However, it was observed that the expression of the VHb protein under the Ptac promoter was better than that with the use of a single repeat of Pvgb promoter (1.71 ± 0.08 nmol/g > 0.69 ± 0.10 nmol/g) []. Therefore, in this study, new tunable gene expression systems were developed based on repeats of Pvgb. The strength of the Pvgb promoter-based expression systems with increasing repeats of Pvgb were investigated by evaluating the expression level of GFP in recombinant strains. To demonstrate the use of the new tunable Pvgb promoter-based expression system as a novel platform for metabolic engineering of C. glutamicum, 5-AVA and GABA production were established in different strains of C. glutamicum. 5-AVA and GABA production were selected as model compounds, because they are derived from L-lysine and L-glutamate, respectively.
2. Results and Discussion
2.1. Construction of Pvgb Promoter-Based Tunable Expression Systems for C. glutamicum and Evaluation of Green Fluorescent Protein in the Recombinant Strains
To investigate the tunability of protein expression using Pvgb in recombinant C. glutamicum, several expression vector systems based on pCES208 plasmids were constructed. GFP was expressed in C. glutamicum under the control of increasing repeats of Pvgb (Pvgb, Pvgb4, Pvgb8). The strength of protein expression in the three constructs was investigated by measuring the intensity of fluorescence in the recombinant strains, by using the fluorescent activated cell sorting analysis (FACS) (Figure 1) []. The resulting recombinant C. glutamicum EGFPV1, EGFPV4, and EGPV8 strains harbored one (Pvgb), four (Pvgb4), and eight (Pvgb8) repeats of Pvgb, respectively. C. glutamicum KCTC 1857 was cultured and used as the negative control. Colonies were randomly picked and GFP expression in each cell was evaluated by using FACS. The measured fluorescence intensity obtained from colonies with Pvgb-promoter-based vectors ranged from 102–103, whereas the negative control cells did not show significant fluorescence. The fluorescence by clones with Pvgb8 were more intense than the clones with Pvgb4 and Pvgb, after 24 h of cultivation (Figure 2). However, after 48 h, no significant difference was observed in GFP expression between clones with Pvgb4 and those with Pvgb8 (data not shown). The increasing number of Pvgb in the constructed expression system was directly proportional to the intensity of the GFP fluorescence in the resulting recombinant strains (Figure 2). These observations were similar to the PHB production enhancement in recombinant E. coli strains harboring the PHB operon (phaCAB) under the control of Pvgb8 compared to the use of Pvgb. Based on these results, we have demonstrated that the strength of protein expression using Pvgb was tunable by modulating the number of its copies in the constructed expression system. To further demonstrate tunability of the Pvgb promoter-based expression system in recombinant C. glutamicum, the developed constructs were used to produce 5-AVA and GABA from L-lysine and L-glutamate, respectively. 5-AVA and GABA were selected as model compounds to test the application of the Pvgb promoter-based expression system because their production in C. glutamicum has been extensively studied in recent times [,,,,,].
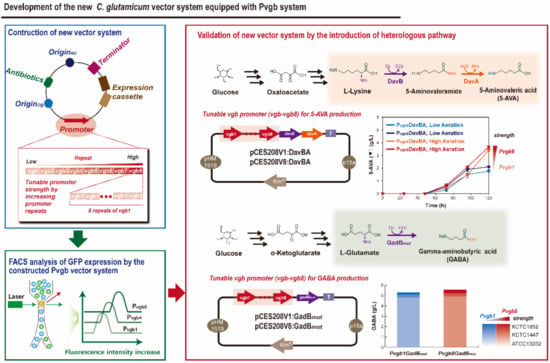
Figure 1.
Schematic representation of the development of the C. glutamicum Pvgb promoter-based vector system equipped with Pvgb promoter.
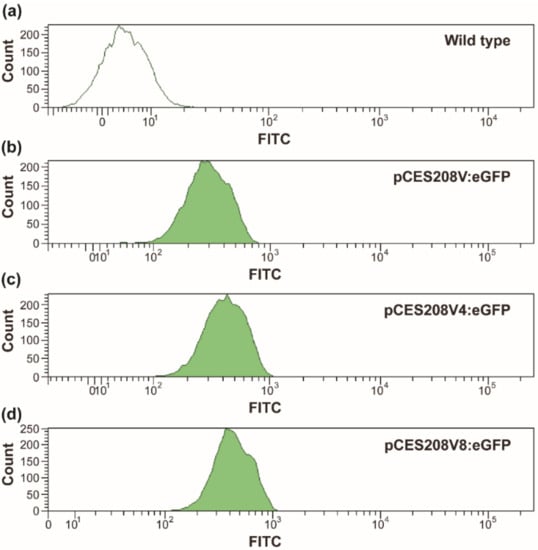
Figure 2.
The fluorescence intensity of the recombinant EGFPV1 (b), EGFPV4 (c), and EGPV8 (d) strains after 24 h of cultivation. The wild type strain, C. glutamicum KCTC 1857 (a) was used as negative control.
2.2. 5-AVA Production Using Recombinant C. glutamicum with Tunable Pvgb Promoter-Based Expression Systems
To demonstrate that the Pvgb promoter-based expression system is a new platform for engineering better C. glutamicum strains, its feasibility to produce 5-AVA in C. glutamicum KCTC 1857 was evaluated [,,]. Tunability of protein expression by Pvgb was verified by developing C. glutamicium 5AVA1 and 5AVA8 strains for the expression of lysine 2-monooxygenase (DavB) and delta-aminovaleramidase (DavA) under the control of single (Pvgb) and eight repeats (Pvgb8) of Pvgb [,,]. 5-AVA production using the recombinant strains was evaluated in flask cultivation under low and high aeration conditions, as it was previously demonstrated that protein expression using Pvgb was enhanced under microaerobic conditions in recombinant E. coli strains []. The production of 5-AVA by the recombinant C. glutamicum 5AVA1 (3.43 ± 0.10 g/L) and 5AVA8 (3.68 ± 0.07 g/L) strains was better under high aeration conditions than at low aeration conditions (2.10–1.76 g/L) (Figure 3). Higher accumulation of L-lysine and glutarate was also detected after flask cultivation of 5AVA1 and 5AVA8 strains under high aeration conditions (4.41–4.46 g/L of L-lysine and 0.71–0.74 g/L of glutarate) than that at low aeration (3.99–4.58 g/L of L-lysine and 0.49–0.57 g/L of glutarate) (Figure 3c,d). 5-AVA production by all the recombinant strains were higher under high aeration conditions than that under flask cultivation with low aeration conditions. These results were similar to the trend of L-glutamate and L-glutamine production by recombinant C. glutamicum strains, which expressed the Vitreoscilla hemoglobin gene under Pvgb and Ptac, wherein chemical production was higher under high aeration than at low aeration []. This is because C. glutamicum is an obligate aerobic microorganism, and it requires high aeration for efficient amino acid production [,,]. Additionally, because DavB requires oxygen as a co-substrate, higher aeration conditions are preferred for 5-AVA production [].
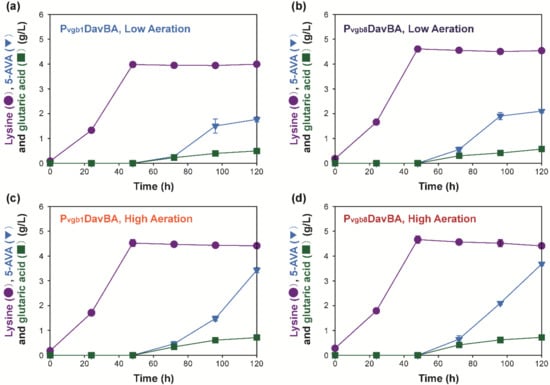
Figure 3.
Time profile of 5-aminovaleric acid, L-lysine and glutaric acid production by recombinant C. glutamicum 5AVA1 (a, c) and 5AVA8 (b,d) strains under low (a,b) and high (c,d) aeration.
Furthermore, the effect of eight repeats of Pvgb on 5-AVA production was evaluated to demonstrate the tunability of protein expression using the constructed Pvgb promoter-based vector system (Figure 3). Different levels of 5-AVA production were achieved when the number of Pvgb increased from single (Pvgb) to eight repeats (Pvgb8) (Figure 3). It was observed that 5-AVA production increased based on the number of repeats of Pvgb. 5-AVA production by the 5AVA8 strain (Figure 3d, 3.69 ± 0.07 g/L), which expressed DavBA under Pvgb8, was higher than that of the 5AVA1 strain (Figure 3c, 3.43 ± 0.10 g/L)), which harbored Pvgb. 5-AVA production by 5AVA1 and 5AVA8 strains (3.43–3.69 g/L) were comparably higher than that of the H30_AVA strain, which expressed DavBA under the strong synthetic promoter PH30 (1.4 ± 0.3 g/L g/L) (Table S1) []. This effect was similar to the observed trend of enhanced PHB accumulation in recombinant E. coli strains when the number of Ptac and Pvgb repeats increased to five and eight, respectively [,].
Finally, the effect of his-tagged DavB on 5-AVA production was also investigated, because we have previously reported that it enhanced 5-AVA production in C. glutamcium H30_AVAHis. In our previous study, we demonstrated enhanced 5AVA production by expressing his-tagged DavB along with DavA under the strong synthetic promoter PH30 in H30_AVAHis strain, which produced 4.2 ± 0.9 g/L of 5-AVA []. This significantly increased 5-AVA production compared to that by the H30_AVA strain, which expressed DavB without his-tag (4.2 ± 0.9 g/L >1. 4 ± 0.3 g/L) (Figure 4) []. Therefore, to investigate the effect of this additional his-tag on 5-AVA production under the control of the Pvgb promoter system, recombinant strains 5AVA1His and 5AVA8His were constructed by inserting his-tagged DavB into the Pvgb promoter system with single (Pvgb) and eight (Pvgb8) repeats, respectively. 5-AVA production by the resulting recombinant strains were only evaluated under high aeration conditions, as the low aeration condition did not increase 5-AVA production in 5AVA1 and 5AVA8 strains (Figure 3). Under high aeration conditions, the 5AVA8His (3.31 ± 0.08 g/L) strain produced higher concentrations of 5-AVA than the 5AVA1His (2.82 ± 0.03 g/L) strain (Figure 4, Table S1). As shown in Figure 4, 5-AVA production increased as the number of Pvgb repeats increased from single to eight in 5AVA1His and 5AVA8His (3.31 ± 0.08 g/L > 2.82 ± 0.03 g/L). Glutaric acid production also increased in both 5AVA1His (0.64 ± 0.01 g/L) and 5AVA8His (0.67 ± 0.05g/L). Interestingly, both L-lysine accumulation and 5-AVA production increased as the number of repeats of Pvgb increased from a single repeat in 5AVA1His (4.00 ± 0.06 g/L) to eight repeats in the 5AVA8His (4.38 ± 0.03 g/L) strain. Based on these observations, the Pvgb promoter system was successfully used as a new platform for 5-AVA production in C. glutamicum.
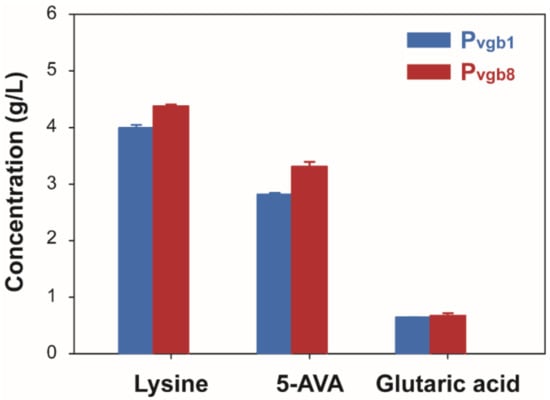
Figure 4.
5-Aminovaleric acid, L-lysine, and glutaric acid production by recombinant C. glutamicum 5AVA1His and 5AVA8His strains under high aeration conditions after 120 h flask cultivation.
2.3. Gamma-Aminobutyric Acid Production Using Recombinant C. glutamicum with Tunable Pvgb Promoter-Based Expression Systems
The constructed Pvgb promoter-based vector system was also used for the expression of mutated glutamate decarboxylase (GadBmut) in order to investigate GABA production in the C. glutamicum strain ATCC 13032 and in high-L-glutamate producing strains C. glutamicum KCTC 1447 and C. glutamicum KCTC 1852 as host strains (Figure 5, Table S2) [,]. C. glutamicum KCTC 1447 and C. glutamicum KCTC 1852 were used because they are capable of high production of L-glutamate, an important precursor for GABA production [,]. C. glutamicum ATCC 13032 was also used as control host strain []. The effect of the eight repeats of Pvgb on GABA production was evaluated under high aeration conditions because we have previously demonstrated that low aeration did not increase chemical production by recombinant strains expressing DavBA under single and eight repeats of Pvgb during shake flask cultivation (Figure 3). It was observed that the average GABA production by C. glutamicum KCTC 1852-derived strains (5.31 g/L–5.58 g/L) was higher than that by recombinant C. glutamicum ATCC 13032 and C. glutamicum KCTC 1447 strains (4.82–5.22 g/L) (Figure 5, Figure S3). This is because the host strain, C. glutamicum KCTC 1852, is naturally capable of higher L-glutamate production than C. glutamicum KCTC 1447 and C. glutamicum ATCC 13032 []. Higher GABA production was attributed to the efficient production of L-glutamate, an important pre-requisite for the development of strains for GABA production [,]. The level of GABA production in all the tested strains increased as the number of Pvgb repeats increased (Figure 5). For example, GABA production in V1GD1852 (5.31 ± 0.16 g/L) and V8GD1852 (5.58 ± 0.27 g/L) strains increased when GadBmut was expressed under Pvgb and Pvgb8, respectively. The same tendency was observed in V1GD13032 (4.82 ± 0.09 g/L) and V1GD1447 (5.09 ± 0.64 g/L) strains with respect to GABA production, under the control of the Pvgb promoter, and by V8GD13032 (4.94 ± 0.27 g/L) and V8GD1447 (5.22 ± 0.60 g/L) strains under the control of the Pvgb8 promoter. Regarding GABA production using C. glutamicum ATCC 13032 strains (4.82–4.94 g/L), similar GABA production was achieved by pLGmut (4.87 ± 0.15 g/L), V1GD13032 (4.82 ± 0.09 g/L), and V8GD13032 (4.94 ± 0.27 g/L) strains, which expressed GadBmut under the synthetic promoters PL26, Pvgb, and Pvgb8, respectively (Figure 4, Table S2) []. The level of GABA production using the constructed Pvgb promoter-based expression system (4.82–5.09 g/L) was comparable to the titers achieved by recombinant strains pHGmut (5.89 ± 0.35 g/L) and pIGmut (5.32 ± 0.04 g/L). The recombinant strains expressed GadBmut under synthetic promoters of high (PH36) and intermediate (PI16) strength []. The level of GABA production by the C. glutamicum KCTC 1852-derivative strains V1GD1852 (5.31 ± 0.16) and V8GD1852 (5.58 ± 0.27), expressing GadBmut, were similar to the level of GABA production achieved when the strong synthetic promoter PH36 was used in the H36GM1852 strain (8.47 ± 0.06 g/L) (Table S2). This shows that the strength of GadBmut expression by the Pvgb promoter-based expression system is also tunable like previously used synthetic promoters (PL26, PI16, PH36) (Table S2) [,]. Based on these results, we concur that the Pvgb -based expression system was capable of tunable expression of key proteins for 5-AVA (DavBA) and GABA (GadBmut) production by increasing the number of Pvgb repeats. The addition of the newly constructed Pvgb promoter-based expression system into the repertoire of plasmids available for metabolic engineering of C. glutamicum provides an alternative method of fine-tuning protein expression levels for validating synthetic metabolic pathways and improving biochemical production in biorefineries.
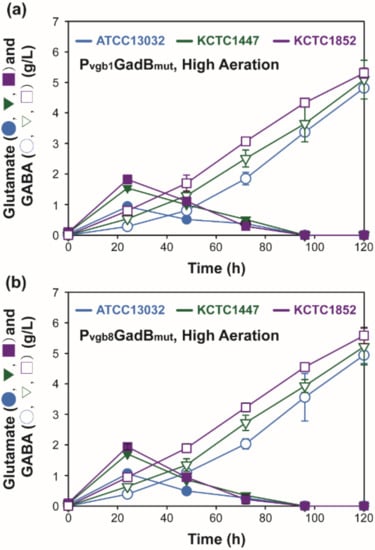
Figure 5.
Time profile of Gamma-aminobutyric acid production by recombinant C. glutamcium strains expressing GadBmut under Pvgb (a) and Pvgb8 (b) under high aeration condition.
3. Materials and Methods
3.1. Bacterial Strains and Plasmids
All the bacterial strains and plasmids used in this study are listed in Table 1. E. coli XL1-Blue (Stratagene, La Jolla, CA, USA) was used for the general gene cloning studies. C. glutamicum ATCC 13032, C. glutamicum KCTC 1447, C. glutamicum KCTC 1852, and C. glutamicum KCTC 1857 strains were purchased from the Korean Collection for Type Cultures (KCTC, Daejeon, Korea). The pCES208-based plasmids: pCES208H30:DavBA, pCES208H30:DavBHisA and pHGmut were constructed as previously described [,].

Table 1.
Strains and plasmids used in the present study.
3.2. Plasmid Construction
All DNA manipulations were performed following standard procedures []. Polymerase chain reaction (PCR) was performed with the C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA). The primers used in this study were synthesized at Bioneer (Daejeon, Korea). The pHGmut plasmid was cut at KpnI and BamHI sites to replace the synthetic promoter PH36 with Pvgb promoter. The subsequent repeats of the Pvgb promoter were inserted into the pCES208V:eGFP vector at BamHI/BglII sites to obtain pCES208V4:eGFP and pCES208V8:eGFP plasmids. The pCES208V:eGFP, pCES208V4:eGFP, and pCES208V8:eGFP plasmids for the expression of enhanced GFP were constructed by inserting GFP at the BamHI/NotI sites of modified pCES208-based plasmids with one, four, and eight repeats of Pvgb, respectively. The plasmids for the expression of DavBA, DavBHisA, and GadBmut were also inserted at the BamHI/NotI sites of pCES208V:eGFP and pCES208V8:eGFP (Table 1).
3.3. Culture Conditions
E. coli XL1-Blue, used for general gene cloning experiments, was cultured at 37 °C in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl). Flask cultures of the recombinant strains of C. glutamicum were obtained in triplicates by culturing at 30 °C and 250 rpm in a rotary shaker. High aeration conditions for flask cultivation were maintained by adding 20 mL of the appropriate culture medium into a 250-mL baffled flask. Low aeration conditions for flask cultivation were maintained by adding 100 mL of the culture medium into a 250-mL glass flask. Seed cultivation of C. glutamicum strains was carried out in 14-mL round-bottomed tubes containing 2 mL of Recovery Growth (RG) medium (10 g/L of glucose, 40 g/L of brain heart infusion, 10 g/L of beef extract, and 30 g/L of D-sorbitol) with incubation overnight at 30 °C and 250 rpm []. The main-flask cultures for 5-AVA production were grown in 250 mL baffled flasks containing 20 mL of CG50 medium for 120 h at 30 °C and 250 rpm. The CG50 medium for flask cultivation contained (per liter) 50 g glucose, 15 g yeast extract, 15 g (NH4)2SO4·7H2O, 0.5 g KH2PO4, 0.5 g MgSO4·7H2O, 0.01 g MnSO4·H2O, 0.01 g FeSO4·7H2O, and 20 μg/L of kanamycin (Km) for plasmid maintenance []. The GP1 medium optimized in our previous study [] was used for GABA production. The GP1 medium for flask cultivation contained (per liter) 50 g (NH4)2SO4·7H2O, 1 g K2HPO4, 3 g urea, 0.4 g MgSO4· 7H2O, 50 g peptone, 0.02 g FeSO4·7H2O, 0.007 g MnSO4·H2O, 200 μg thiamine, and 1 mM of pyridoxal 5′-phosphate hydrate (PLP) []. PLP was added to the culture medium as it is a cofactor of glutamate decarboxylase. Moreover, 0.1 mM of PLP was the optimum concentration for prolonging GABA production using recombinant C. glutamicum strains []. Kanamycin and biotin were added to the GP1 culture medium at 25 and 50 μg/L, respectively. Only 50 μg /L of biotin was used in flask cultivation for GABA production because biotin-limited condition promotes L-glutamate accumulation []. CaCO3 was added to the culture medium at 10 g/L to minimize the pH change during cultivation.
3.4. Analysis
The concentrations of glucose and organic acids were determined by high performance liquid chromatography (HPLC). The standard and sample concentrations of 5-AVA, GABA, L-lysine, and L-glutamate were determined by HPLC using an Optimapak C18 column (RStech, DaeJeon, Korea) as previously reported [].
3.5. Fluorescence-Activated cell Sorting Analysis (FACS) for Measuring GFP Expression by Recombinant Strains
FACS analysis was used to investigate the GFP expression by using the constructed Pvgb system. The recombinant EGFPV1, EGFPV4 and EGFPV8 strains were grown in Brain Heart Infusion (BHI) media for 24 h at 30 °C. The cells were then collected and diluted using PBS buffer. FACS analysis (BD Biosciences, San Jose, CA, USA) was performed for 100,000 clones of each samples using argon ion laser (blue, 488 nm) and band-pass filter (530 nm ± 15 nm) [].
4. Conclusions
The Pvgb promoter-based expression system constructed in this study was capable of tunable expression of green fluorescent protein in recombinant C. glutamcium strains, when the repeats of Pvgb promoter increased from one (Pvgb) to four (Pvgb4) to eight (Pvgb8). Furthermore, GABA and 5-AVA production by recombinant C. glutamicum strains also increased when the expression of DavBA and GadBmut in the Pvgb promoter-based expression system increased from single to eight repeats of the Pvgb promoter. This shows that the strength of protein expression using the Pvgb promoter-based tunable system was comparable to that of previously established synthetic promoters (PL26, PI16, PH30 PH36) [,,,]. Based on the different levels of 5-AVA and GABA production by all the tested strains, the Pvgb promoter-based expression system was capable of tunable expression of DavBA and GadBmut by mere manipulation in the number of Pvgb repeats. The newly constructed Pvgb promoter-based expression system developed in this study expands the repertoire of plasmids available for metabolic engineering of C. glutamicum and provides another method for fine-tuning levels of protein expression for convenient and rapid validation of synthetic metabolic pathways, ultimately improving biochemical production in biorefineries.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4344/8/11/561/s1. Table S1: 5-AVA production using recombinant C. glutamicum strains expressing DavBA and DavBHisDavA under different synthetic promoters after 120 h of flask cultivation under high aeration. Table S2: GABA production by recombinant C. glutamicum strains expressing GadBmut under different synthetic promoters after 120 h of flask cultivation under high aeration.
Author Contributions
The study was conceptualized by P.S.J. The study was validated, and formal analysis was carried out by K.H.K., K.B., T.U.K., S.Y.J, B.L., J.J.S., D.H.L., B.K.S., and J.H.C. The original draft was prepared by K.B. The paper was edited and reviewed by H.T.K. and M.N.R. The resources for completion of this study were provided by P.S.J., H.T.K. and J.C.J.
Funding
Funding sources are declared in the acknowledgement section.
Acknowledgments
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea (NRF-2015M1A2A2035810), the Bio & Medical Technology Development Program MSIT through the NRF of Korea (NRF-2018M3A9H3020459) and the Lignin Biorefinery from MSIT through the NRF of Korea (NRF-2017M1A2A2087634).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Becker, J.; Wittmann, C. Advanced Biotechnology: Metabolically Engineered Cells for the Bio-Based Production of Chemicals and Fuels, Materials, and Health-Care Products. Angew. Chem. Int. Ed. 2015, 54, 3328–3350. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.H.; Eom, I.Y.; Joo, J.C.; Yu, J.H.; Song, B.G.; Lee, S.H.; Hong, S.H.; Park, S.J. Recent advances in development of biomass pretreatment technologies used in biorefinery for the production of bio-based fuels, chemicals and polymers. Korean J. Chem. Eng. 2015, 32, 1945–1959. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, H.U.; Choi, S.; Yi, J.H.; Lee, S.Y. Microbial production of building block chemicals and polymers. Curr. Opin. Biotechnol. 2011, 22, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Baritugo, K.; Kim, H.T.; David, Y.; Choi, J.H.; Choi, J.; Kim, T.W.; Park, C.; Hong, S.H.; Na, J.G.; Jeong, K.J.; et al. Recent advances in metabolic engineering of Corynebacterium glutamicum strains as potential platform microorganisms for biorefinery. Biofuel Bioprod. Biorefin. 2018, 12, 899–925. [Google Scholar] [CrossRef]
- Baritugo, K.; Kim, H.T.; David, Y.; Choi, J.; Hong, S.H.; Jeong, K.J.; Joo, J.C.; Park, S.J. Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 2018, 102, 3915–3937. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.C.; Oh, Y.H.; Yu, J.H.; Hyun, S.M.; Khang, T.U.; Kang, K.H.; Song, B.K.; Park, K.; Oh, M.K.; Lee, S.Y.; et al. Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process. Bioresour. Technol. 2017, 244, 1692–1700. [Google Scholar] [CrossRef] [PubMed]
- David, Y.; Joo, J.C.; Yang, J.E.; Oh, Y.H.; Lee, S.Y.; Park, S.J. Biosynthesis of 2-hydroxyacid-containing polyhydroxyalkanoates by employing butyryl-CoA transferases in metabolically engineered Escherichia coli. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Parks, S.J.; Kim, W.J.; Yang, J.E.; Lee, H.; Shin, J.; Lee, S.Y. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat. Biotechnol. 2016, 34, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Kind, S.; Neubauer, S.; Becker, J.; Yamamoto, M.; Völkert, M.; Abendroth, G.; Zelder, O.; Wittmann, C. From zero to hero—Production of bio-based nylon from renewable re-sources using engineered Corynebacterium glutamicum. Metab. Eng. 2014, 25, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Chae, C.G.; Kim, Y.J.; Lee, S.J.; Oh, Y.H.; Yang, J.E.; Joo, J.C.; Kang, K.H.; Jang, Y.A.; Lee, H.; Park, A.R.; et al. Biosynthesis of poly(2-hydroxybutyrate-co-lactate) in metabolically engineered Escherichia coli. Biotechnol. Bioprocess Eng. 2016, 21, 169–174. [Google Scholar] [CrossRef]
- David, Y.; Baylon, M.G.; Sudheer, P.D.V.N.; Baritugo, K.; Chae, C.G.; Kim, Y.J.; Kim, T.W.; Kim, M.; Na, J.G.; Park, S.J. Screening of microorganisms able to degrade low-rank coal in aerobic conditions: Potential coal bio-solubilization mediators from coal to biochemical. Biotechnol. Bioprocess Eng. 2017, 22, 178–185. [Google Scholar] [CrossRef]
- Baylon, G.; David, Y.; Pamidimarri, S.; Baritugo, K.; Chae, C.G.; Kim, Y.J.; Wan, T.W.; Kim, M.S.; Na, J.G.; Park, S.J. Bio-solubilization of the untreated low rank coal by alkali-producing bacteria isolated from soil. Korean J. Chem. Eng. 2016, 34, 105–109. [Google Scholar] [CrossRef]
- Sudheer, P.D.V.N.; David, Y.; Chae, C.; Kim, Y.J.; Baylon, M.G.; Baritugo, K.; Kim, T.W.; Kim, M.; Na, J.G.; Park, S.J. Advances in the biological treatment of coal for synthetic natural gas and chemicals. Korean J. Chem. Eng. 2016, 10, 2788–2801. [Google Scholar] [CrossRef]
- Yang, J.E.; Park, S.J.; Kim, W.J.; Kim, H.J.; Kim, B.; Lee, H.; Shin, J.; Lee, S.Y. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 2018, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.C.; Khusnutdinova, A.N.; Flick, R.; Kim, T.H.; Bornscheuer, U.T.; Yakunin, A.F.; Mahadevan, R. Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem. Sci. 2017, 8, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lama, S.; Kim, J.R.; Park, S.H. Production of 1,3-Propanediol from Glucose by Recombinant Escherichia coli BL21(DE3). Biotechnol. Bioprocess Eng. 2017, 23, 250–258. [Google Scholar] [CrossRef]
- Li, J.; Feng, R.; Wen, Z.; Zhang, A. Overexpression of ARO10 in pdc5Δmutant resulted in higher isobutanol titers in Saccharomyces cerevisiae. Biotechnol. Bioprocess Eng. 2017, 22, 382–389. [Google Scholar] [CrossRef]
- Zong, H.; Liu, X.; Chen, W.; Zhuge, B.; Sun, J. Construction of glycerol synthesis pathway in Klebsiella pneumoniae for bioconversion of glucose into 1,3-propanediol. Biotechnol. Bioprocess Eng. 2017, 22, 549–555. [Google Scholar] [CrossRef]
- Kim, H.S.; Oh, Y.H.; Jang, Y.; Kang, K.H.; David, Y.; Yu, J.H.; Song, B.K.; Choi, J.; Chang, Y.K.; Joo, J.C.; et al. Recombinant Ralstonia eutropha engineered to utilize xylose and its use for the production of poly(3-hydroxybutyrate) from sunflower stalk hydrolysate solution. Microb. Cell Factories 2016, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jang, Y.A.; Noh, W.; Oh, Y.H.; Lee, H.; David, Y.; Baylon, M.G.; Shin, J.; Yang, J.E.; Choi, S.Y.; et al. Metabolic engineering of Ralstonia eutropha for the production of polyhydroxyalkanoates from sucrose. Biotechnol. Bioeng. 2015, 112, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Zhuang, X.; Tang, Z.; Chen, X. Polylactic acid (PLA): Research, development and industrialization. Biotechnol. J. 2010, 5, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, B.H. Poly (butylene succinate) and its copolymers: Research, development and industrialization. Biotechnol. J. 2010, 5, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Kind, S.; Wittmann, C. Bio-based production of the platform chemical 1, 5-diaminopentane. Appl. Microbiol. Biotechnol. 2011, 91, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, E.Y.; Noh, W.; Park, H.M.; Oh, Y.H.; Lee, S.H.; Song, B.K.; Jegal, J.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab. Eng. 2013, 16, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, S.H.; Oh, Y.H.; Choi, J.W.; Lee, M.H.; Cho, J.S.; Jeong, K.J.; Joo, J.C.; Yu, J.; Park, S.J.; et al. Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb. Cell Factories 2016, 15, 174. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Yim, S.S.; Lee, S.H.; Kang, T.J.; Park, S.J.; Jeong, K.J. Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell Factories 2015, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, E.Y.; Won, N.; Oh, Y.H.; Kim, H.Y.; Song, B.K.; Cho, K.M.; Hong, S.H.; Lee, S.H.; Jegal, J. Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst. Eng. 2013, 36, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Baritugo, K.A.; Oh, Y.H.; Hyun, S.M.; Khang, T.U.; Kang, K.H.; Jung, S.H.; Song, B.K.; Park, K.; Kim, I.K.; et al. Metabolic engineering of Corynebacterium glutamicum for the high-level production of cadaverine that can be used for synthesis of biopolyamide 510. ACS Sustain. Chem. Eng. 2018, 6, 5296–5305. [Google Scholar] [CrossRef]
- Oh, Y.H.; Choi, J.W.; Kim, E.Y.; Song, B.K.; Jeong, K.J.; Park, K.; Kim, I.; Woo, H.M.; Lee, S.H.; Park, S.J. Construction of Synthetic Promoter-Based Expression Cassettes for the production of Cadaverine in Recombinant Corynebacterium glutamicum. Appl. Biochem. Biotech. 2015, 176, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kang, T.U.; Baritugo, K.; Hyun, S.M.; Kang, K.H.; Jung, S.H.; Song, B.K.; Park, K.; Oh, M.; Kim, G.B.; et al. Metabolic engineering of Corynebacterium glutamicum for the production of glutaric acid, a C5 dicarboxylic acid platform chemical. Metab. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Baritugo, K.; Kim, H.T.; David, Y.; Khang, T.U.; Hyun, S.M.; Kang, K.H.; Yu, J.H.; Choi, J.H.; Song, J.J.; Joo, J.C.; et al. Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum strains from empty fruit bunch biosugar solution. Microb. Cell Factories 2018, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, J.; Geng, Y.; Li, Y.; Wang, Q.; Liang, Q.; Qi, Q. A strategy of gene overexpression based on tandem repetitive promoters in Escherichia coli. Microb. Cell Factories 2012, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, H.; Chen, J.; Chen, G.Q. Effects of cascaded vgb promoters on poly(hydroxybutyrate) (PHB) synthesis by recombinant Escherichia coli grown micro-aerobically. Appl. Microbiol. Biotechnol. 2014, 98, 10013–10021. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, J.; Wei, X.X.; Ouyang, S.P.; Wu, Q.; Chen, G.Q. Microbial production of L-glutamate and L-glutamine by recombinant Corynebacterium glutamicum harboring Vitreoscilla hemoglobin gene vgb. Appl. Microbiol. Biotechnol. 2008, 77, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.; Lee, B.; Cheong, D.; Han, Y.; Choi, J.H.; Song, J.J. Bacterial Cell Surface Display of a Multifunctional Cellulolytic Enzyme Screened from a Bovine Rumen Metagenomic Resource. J. Microbiol. Biotechnol. 2015, 25, 1835–1841. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3. [Google Scholar]
- Cabezudo, M.D.; Hermosín, I.; Chicón, R.M. Free amino acid composition and botanical origin of honey. Food Chem. 2003, 83, 263–268. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).