A Novel Method of Affinity Tag Cleavage in the Purification of a Recombinant Thermostable Lipase from Aneurinibacillus thermoaerophilus Strain HZ
Abstract
1. Introduction
2. Results
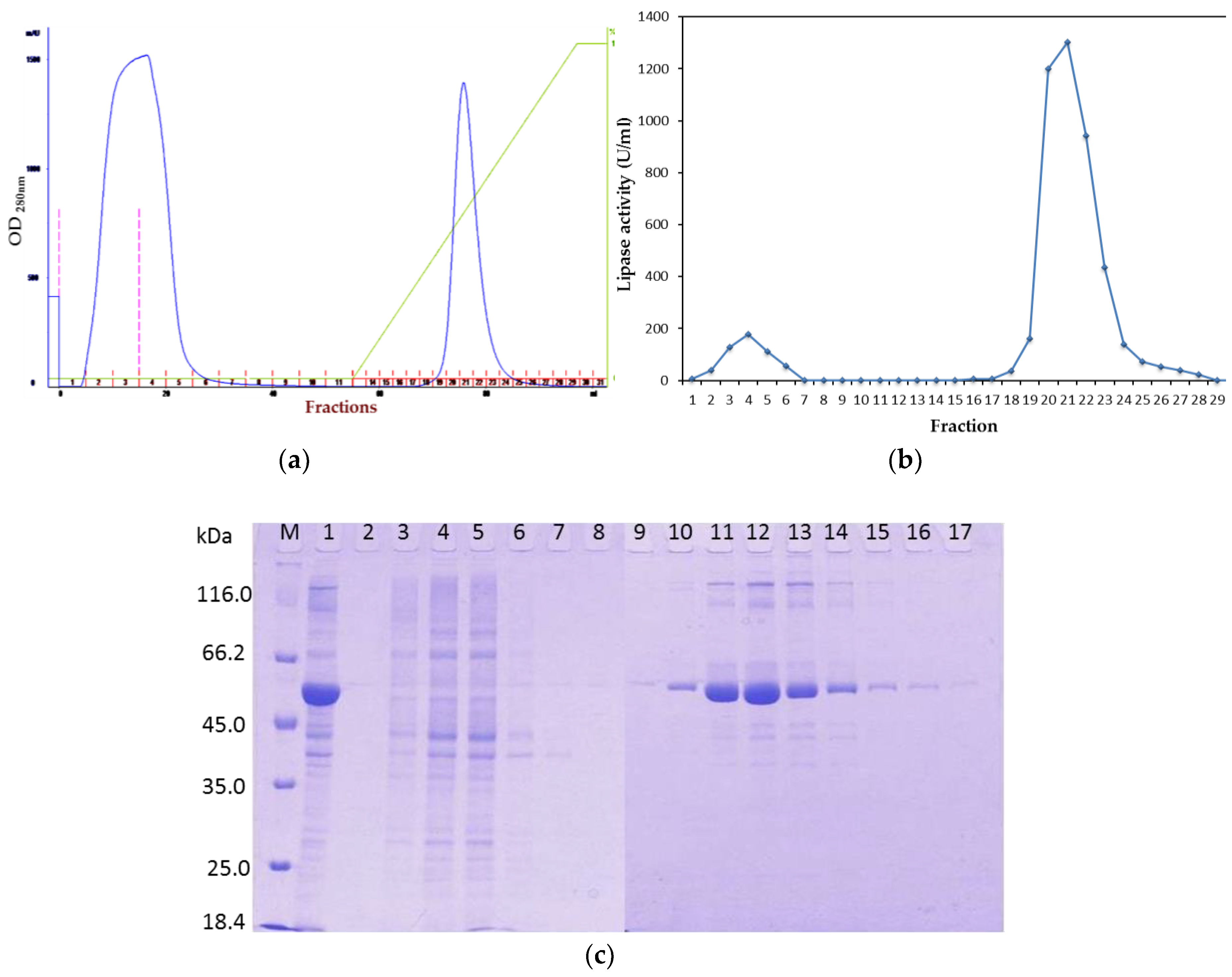
2.1. Purification of Recombinant Lipase
2.1.1. Affinity Chromatography
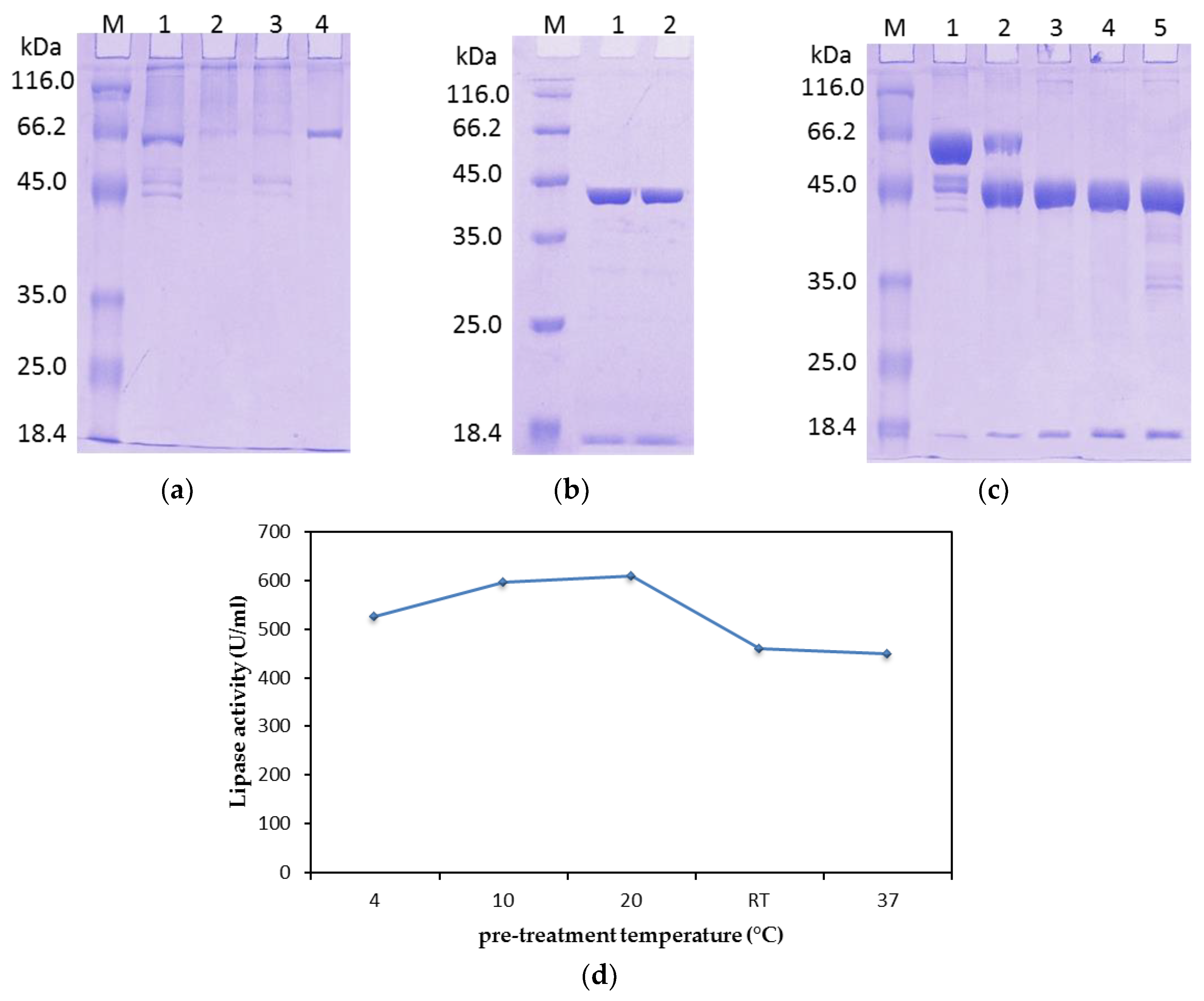
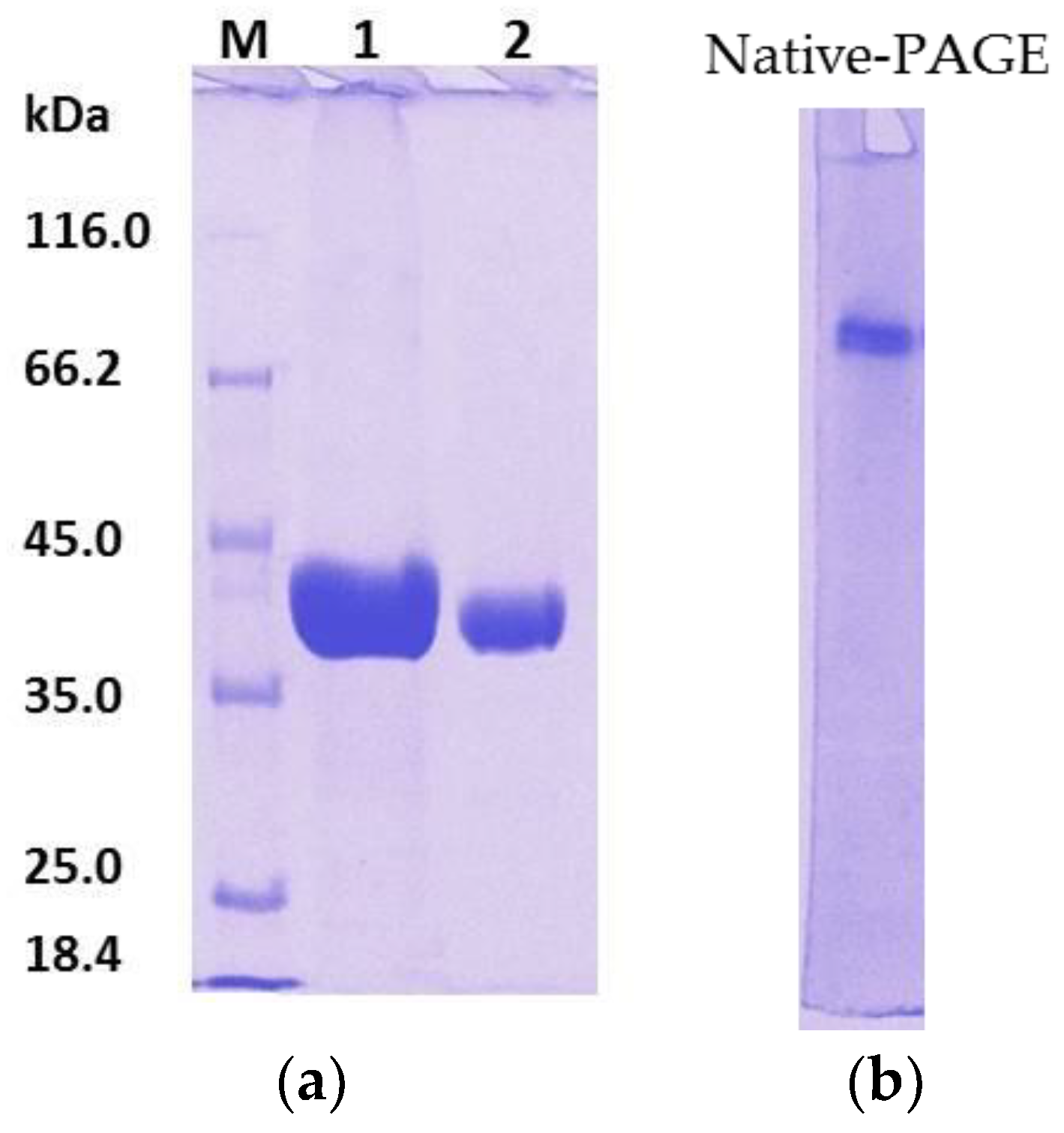
2.1.2. Tag Removal
2.1.3. N-Terminal Sequencing
2.2. Characterization of Mature rHZ Lipase
2.2.1. Effect of Temperature on the Lipase Activity and Stability
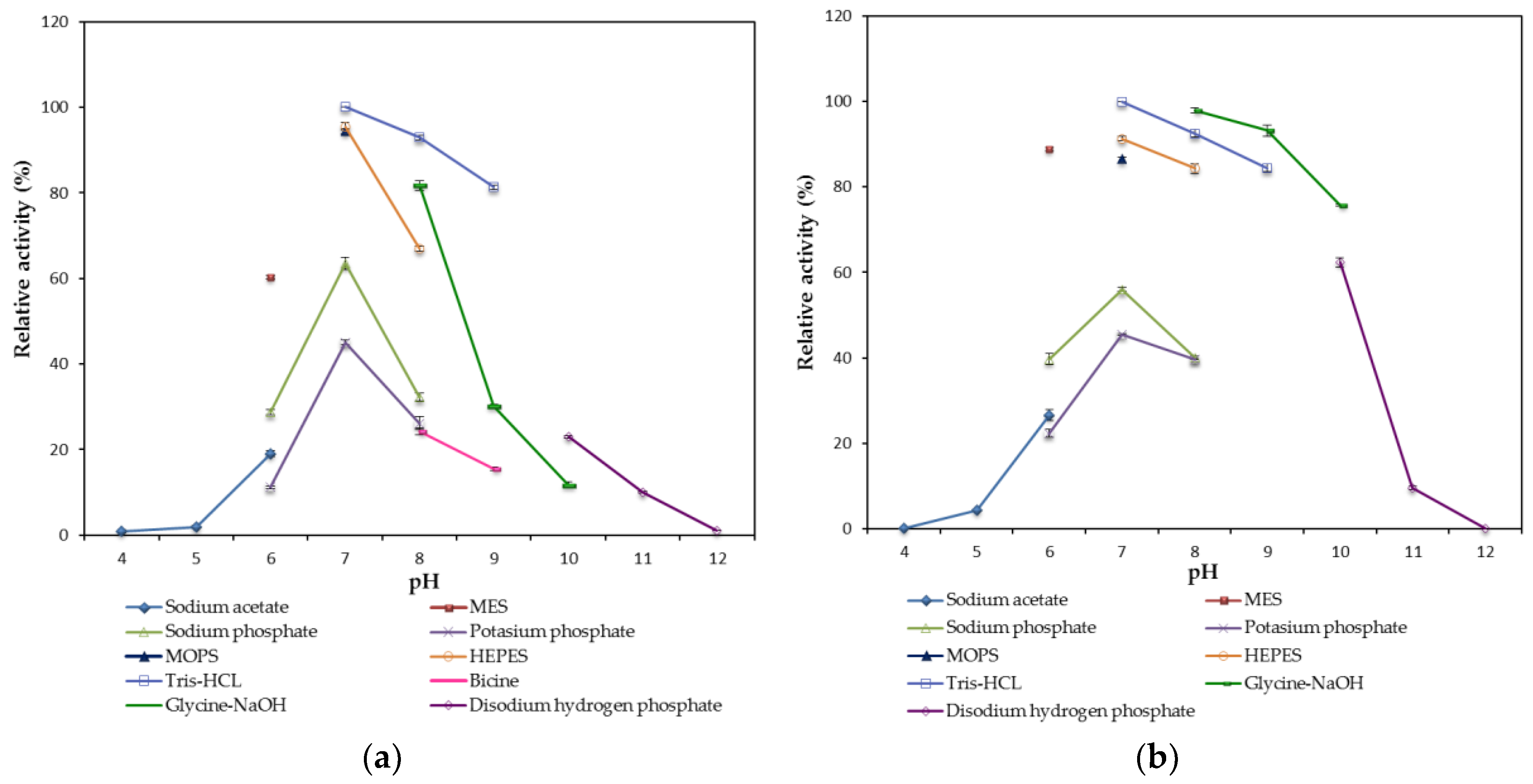
2.2.2. Effect of pH on the Lipase Activity and Stability
2.2.3. Effect of Metal Ions and Inhibitors on the Lipase Activity
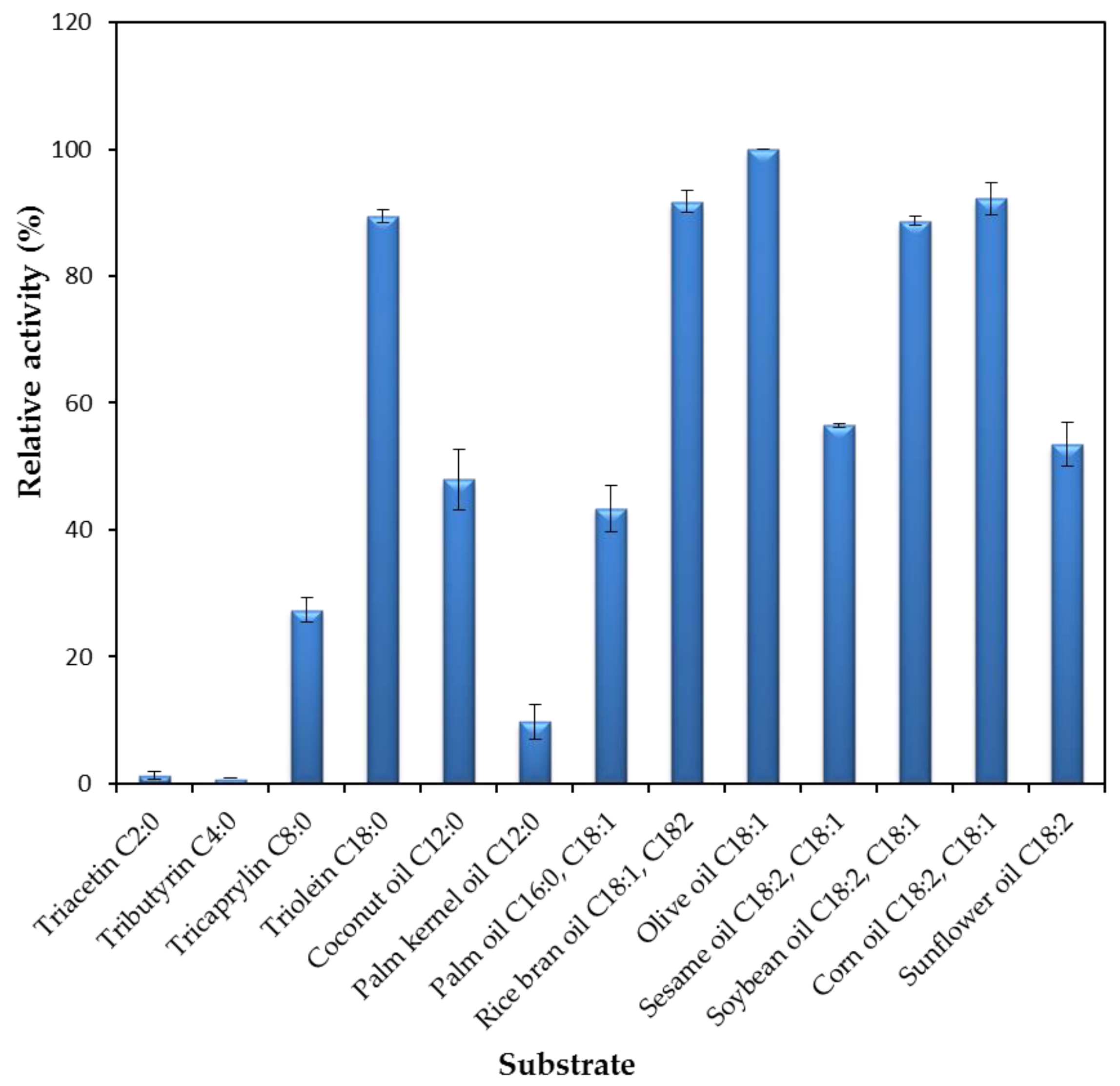
2.2.4. Substrates Specificity of the rHZ Lipase towards Triacylglycerols and Natural Oils
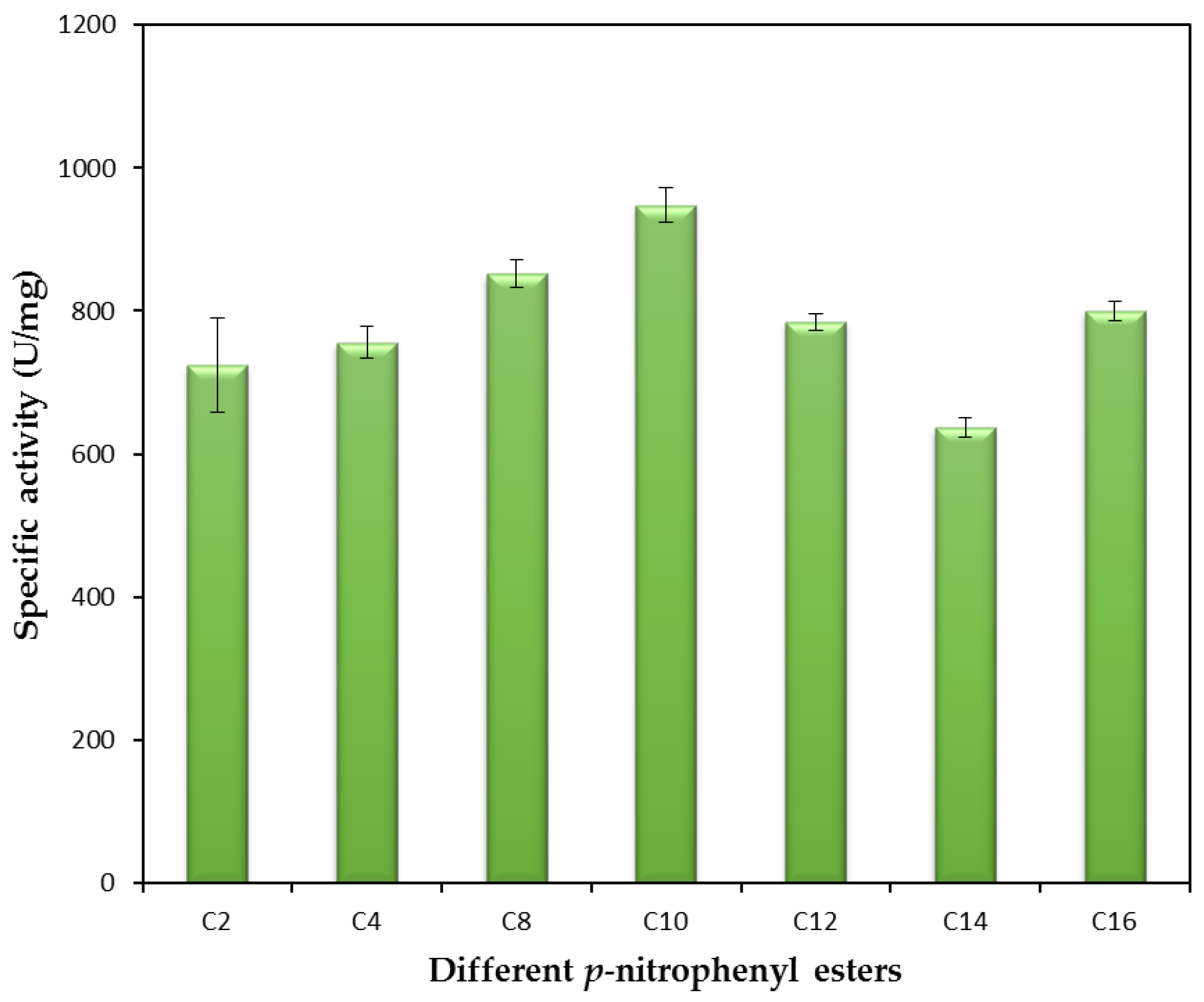
2.2.5. Substrate Specificity of the rHZ Lipase toward p-Nitrophenyl Ester Substrates and Molecular Docking
2.2.6. Determination of Km and Vmax of the rHZ Lipase
2.2.7. Effect of Organic Solvent on the Lipase Activity
3. Discussion
4. Materials and Methods
4.1. Strains and Materials
4.2. Expression of rHZ Lipase
4.3. Purification of Recombinant Lipase
4.3.1. Sample Preparation
4.3.2. Affinity Chromatography
4.3.3. Treatment of the rHZ Lipase to Remove the Tags
4.4. Lipase Assay
4.5. Determination of Protein Content
4.6. Gel Electrophoresis
4.7. N-Terminal Sequencing
4.8. Characterization of Purified rHZ Lipase
4.8.1. Effect of Temperature on the Lipase Activity and Stability
4.8.2. Effect of pH on the Lipase Activity and Stability
4.8.3. Effect of Metal Ions and Inhibitors on the Lipase Activity
4.8.4. Substrates Specificity of the Mature rHZ Lipase towards Triacylglycerols and Natural Oils
4.8.5. Substrate Specificity of the Mature rHZ Lipase Using p-NP Ester Substrates
4.8.6. Molecular Docking Study
4.8.7. Determination of Km and Vmax of the rHZ Lipase
4.8.8. Effect of Organic Solvents on the Lipase Activity
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reis, P.; Holmberg, K.; Watzke, H.; Leser, M.E.; Miller, R. Lipases at interfaces: A review. Adv. Colloid Interface Sci. 2009, 147–148, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Gause, G.; Dudnik, Y.; Laiko, A.; Netyksa, E. Induction of mutants with altered DNA composition: Effect of ultraviolet on Bacterium paracoli 5099. Science 1967, 157, 1196–1197. [Google Scholar] [CrossRef] [PubMed]
- Jonasson, P.; Liljeqvist, S.; Nygren, P.-A.K.; Ståhl, S. Genetic design for facilitated production and recovery of recombinant proteins in Escherichia coli. Biotechnol. Appl. Biochem. 2002, 35, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.K.; Sheoran, A.; Giri, B.; Davidson, W.S. Purification strategies for microbial lipases. J. Microbiol. Methods 2003, 52, 1–18. [Google Scholar] [CrossRef]
- Zhao, X.; Li, G.; Liang, S. Several Affinity Tags Commonly Used in Chromatographic Purification. J. Anal. Methods Chem. 2013, 2013, 581093. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Junceda, E.; Garcia-Garcia, J.F.; Bastida, A.; Fernandez-Mayoralas, A. Enzymes in the synthesis of bioactive compounds: The prodigious decades. Bioorg. Med. Chem. 2004, 12, 1817–1834. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, J.M. Counterdiffusion methods for macromolecular crystallization. Meth. Enzymol. 2003, 368, 130–154. [Google Scholar] [PubMed]
- Makrides, S.C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996, 60, 512–538. [Google Scholar] [PubMed]
- Diepen, M.T.-V. Crystallographic Studies of Modified Insulin; University of York: York, UK, 1996. [Google Scholar]
- Masomian, M.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Basri, M. Analysis of comparative sequence and genomic data to verify phylogenetic relationship and explore a new subfamily of bacterial lipases. PLoS ONE 2016, 11, e0149851. [Google Scholar] [CrossRef] [PubMed]
- Galhardo, R.; Rocha, R.; Marques, M.; Menck, C. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005, 33, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Skala, W.; Goettig, P.; Brandstetter, H. Do-it-yourself histidine-tagged bovine enterokinase: A handy member of the protein engineer’s toolbox. J. Biotechnol. 2013, 168, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, S.; Sathishkuma, T. Enzyme Technology; I.K. International Publishing House Pvt. Ltd.: New Delhi, India, 2009. [Google Scholar]
- Amersham Biosciences. Affinity Chromatography: Principles and Methods; RAK Design AB: Uppsala, Sweden, 2002. [Google Scholar]
- Mutsuda, M.; Michel, K.P.; Zhang, X.; Montgomery, B.L.; Golden, S.S. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongates PCC 7942. J. Biol. Chem. 2003, 278, 19102–19110. [Google Scholar] [CrossRef] [PubMed]
- Leow, T.; Rahman, R.; Basri, M.; Salleh, A. A thermoalkaliphilic lipase of Geobacillus sp. T1. Extremophiles 2007, 11, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Gavira, J.A.; Hernandez-Hernandez, M.; Gonzalez-Ramirez, L.A.; Briggs, R.A.; Kole, S.A.; Shaw Stewart, P.D. Combining counter-diffusion and microseeding to increase the success rate in protein crystallization. Cryst. Growth Des. 2011, 11, 2122–2126. [Google Scholar] [CrossRef]
- Quyen, D.T.; Schmidt-Dannert, C.; Schmid, R.D. High-level expression of a lipase from Bacillus thermocatenulatus BTL2 in Pichia pastoris and some properties of the recombinant lipase. Protein Expr. Purif. 2003, 28, 102–110. [Google Scholar] [CrossRef]
- Sinchaikul, S.; Sookkheo, B.; Phutrakul, S.; Pan, F.-M.; Chen, S.-T. Optimization of a thermostable lipase from Bacillus stearothermophilus P1: Overexpression, purification, and characterization. Protein Expr. Purif. 2001, 22, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Shariff, F.M.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. A newly isolated thermostable lipase from Bacillus sp. Int. J. Mol. Sci. 2011, 12, 2917–2934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cremer, P.S. Interactions between macromolecules and ions: The Hofmeister series. Curr. Opin. Chem. Biol. 2006, 10, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Guncheva, M.; Zhiryakova, D. Catalytic properties and potential applications of Bacillus lipases. J. Mol. Catal. B Enzym. 2011, 68, 1–21. [Google Scholar] [CrossRef]
- Masomian, M.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Basri, M. A new thermostable and organic solvent-tolerant lipase from Aneurinibacillus thermoaerophilus strain HZ. Process Biochem. 2013, 48, 169–175. [Google Scholar] [CrossRef]
- Kambourova, M.; Kirilova, N.; Mandeva, R.; Derekova, A. Purification and properties of thermostable lipase from a thermophilic Bacillus stearothermophilus MC 7. J. Mol. Catal. B Enzym. 2003, 22, 307–313. [Google Scholar] [CrossRef]
- Gray, C.J. Stabilization of enzymes with soluble additives. In Thermostability of Enzymes; Gupta, M.N., Ed.; Narosa: New Delhi, India, 1995; pp. 124–143. [Google Scholar]
- Klibanov, A.M. Why are enzymes less active in organic solvents than in water? Trends Biotechnol. 1997, 15, 97–101. [Google Scholar] [CrossRef]
- Kim, H.K.; Park, S.Y.; Lee, J.K.; Oh, T.K. Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci. Biotechnol. Biochem. 1998, 62, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Bradoo, S.; Saxena, R.K.; Gupta, R. Two acidothermotolerant lipases from new variants of Bacillus spp. World J. Microbiol. Biotechnol. 1999, 15, 87–91. [Google Scholar] [CrossRef]
- Lima, G.M.V.; Kreiger, N.; Mitchell, A.D.; Baratti, C.J.; Filippisde, I.; Fontana, D.J. Evaluation of the potential for the use in biocatalysis of a lipase from a wild strain of Bacillus megaterium. J. Mol. Catal. B Enzym. 2004, 31, 53–61. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Strye, L. Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Simons, J.W.F.A.; Kampen, V.M.D.; Verheij, H.M. Cloning, purification and characterization of the lipase from Staphylococcus epidermidis comparison of the substrate specificity with those of other microbial lipases. Eur. J. Biochem. 1998, 253, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Snellman, E.A.; Sullivan, E.R.; Colwell, R.R. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur. J. Biochem. 2002, 269, 5771–5779. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Gao, S.; Han, S.; Cao, S. Purification and characterization of a Pseudomonas sp. lipase and its properties in non-aqueous media. Appl. Microbiol. Biotechnol. 1999, 30, 251–256. [Google Scholar]
- Lee, D.W.; Kim, H.W.; Lee, K.W.; Kim, B.C.; Choe, E.A.; Lee, H.S.; Kim, D.S.; Pyun, Y.R. Purification and characterization of two distinct thermostable lipases from the gram positive thermophilic bacterium Bacillus thermoleovorans ID-1. Enzyme Microb. Technol. 2001, 25, 363–372. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Sabri, S.; Rahman, R.N.Z.R.A.; Leow, T.C.; Basri, M.; Salleh, A.B. Secretory expression and characterization of a highly Ca2+-activated thermostable L2 lipase. Protein Expr. Purif. 2009, 68, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; He, Y.; Xu, L.; Zhang, H.; Yan, Y. A new extracellular thermo-solvent-stable lipase from Burkholderia ubonensis SL-4: Identification, characterization and application for biodiesel production. J. Mol. Catal. B Enzym. 2016, 126, 76–89. [Google Scholar] [CrossRef]
- Sharma, R.; Soni, S.K.; Vohra, R.M.; Gupta, L.K.; Gupta, J.K. Purification and characterization of a thermostable alkaline lipase from a new thermophilic Bacillus sp. RSJ-1. Process Biochem. 2002, 37, 1075–1084. [Google Scholar] [CrossRef]
- Massadeh, M.; Sabra, F.; Dajani, R.; Arafat, A. Purification of lipase enzyme produced by Bacillus Stearothermophilus HU1. In Proceedings of the International Conference on Eco-Systems and Biological Sciences, Penang, Malaysia, 19–20 May 2012; pp. 34–37. [Google Scholar]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Castro-Ochoa, L.D.; Rodríguez-Gómez, C.; Valerio-Alfaro, G.; Oliart Ros, R. Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme Microb. Technol. 2005, 37, 648–654. [Google Scholar] [CrossRef]
- Ghatorae, A.S.; Bell, G.; Halling, P.J. Inactivation of enzymes by organic solvents: New technique with well-defined interfacial area. Biotechnol. Bioeng. 1994, 43, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.Y.; Rhee, J.S. A simple and rapid colorimetric method for determination of free fatty acid for lipase assay. J. Am. Oil Chem. Soc. 1986, 63, 69–92. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microorganism quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavge of structure protein during assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef]
- Edman, P.; Begg, G. A protein sequenator. Eur. J. Biochem. 1967, 1, 80–91. [Google Scholar] [CrossRef] [PubMed]



| Step | Total Activity (U) | Total Protein (mg) | Specific Activity (U mg−1) | Recovery (%) | Purification Fold |
|---|---|---|---|---|---|
| Crude | 16,776.00 | 36.38 | 461.20 | 100.0 | 1.0 |
| Affinity | 13,787.81 | 26.25 | 525.25 | 82.2 | 1.1 |
| Dialysis | 13,338.65 | 23.70 | 562.80 | 79.5 | 1.2 |
| Treatment at 20 °C | 13,235.02 | 21.78 | 607.58 | 78.9 | 1.3 |
| Cycle | Major Sequence | Minor Sequence |
|---|---|---|
| 1 | D | M |
| 2 | M | Q, G |
| 3 | Q | K, N |
| 4 | K | E |
| 5 | E | R, G |
| 6 | R | - |
| 7 | K | N, A, T |
| Metal Ions/Inhibitors | Concentration (mM) | Relative Activity (%) ± SE |
|---|---|---|
| Control | - | 100 |
| Li+ | 1 | 82.7 ± 0.8 |
| 5 | 118.4 ± 1.1 | |
| Na+ | 1 | 100.1 ± 1.9 |
| 5 | 108.3 ± 1.2 | |
| K+ | 1 | 85.2 ± 3.0 |
| 5 | 112.3 ± 1.2 | |
| Rb+ | 1 | 100.3 ± 1.1 |
| 5 | 112.6 ± 2.0 | |
| Cs+ | 1 | 95.3 ± 1.1 |
| 5 | 115.7 ± 3.8 | |
| Mg2+ | 1 | 115.7 ± 1.4 |
| 5 | 113.0 ± 2.4 | |
| Ca2+ | 1 | 127.9 ± 0.9 |
| 5 | 137.3 ± 1.3 | |
| Sr2+ | 1 | 111.6 ± 1.2 |
| 5 | 127.8 ± 2.6 | |
| Mn2+ | 1 | 105.2 ± 4.3 |
| 5 | 112.2 ± 1.2 | |
| Fe2+ | 1 | 47.2 ± 3.5 |
| 5 | 1.5 ± 0.3 | |
| Co2+ | 1 | 100.4 ± 1.2 |
| 5 | 79.3 ± 4.3 | |
| Ni2+ | 1 | 51.4 ± 1.8 |
| 5 | 57.9 ± 2.9 | |
| Cu2+ | 1 | 10.5 ± 1.0 |
| 5 | 9.8 ± 0.4 | |
| Zn2+ | 1 | 8.3 ± 1.0 |
| 5 | 6.9 ± 1.5 | |
| PMSF | 1 | 70.7 ± 2.9 |
| 5 | 67.8 ± 2.1 | |
| DTT | 1 | 121.4 ± 1.1 |
| 5 | 118.2 ± 1.3 | |
| β-mercaptoethanol | 1 | 113.0 ± 1.7 |
| 5 | 106.6 ± 3.5 | |
| EDTA | 1 | 0.5 ± 0 |
| 5 | 0.3 ± 0 | |
| pepstatin | 1 | 81.3 ± 1.0 |
| 5 | 60.8 ± 1.8 |
| p-NP Substrates | Distance between Oγ-Ser113 and Carbonyl Carbon of the Substrate [Å] | Binding Energy [kcal/mol] | Dissociation Constant [pM] |
|---|---|---|---|
| 4-nitrophenyl acetate (C2) | 3.485 | 3.232 | 4274768384.000 |
| 4-nitrophenyl butyrate (C4) | 3.352 | 3.201 | 4504389120.000 |
| 4-nitrophenyl hexanoate (C6) | 3.565 | 3.925 | 1327201408.000 |
| 4-nitrophenyl octanoate (C8) | 3.644 | 4.257 | 757840192.000 |
| 4-nitrophenyl decanoate (C10) | 3.576 | 4.444 | 552719552.000 |
| 4-nitrophenyl laurate (C12) | 3.932 | 3.867 | 1463697792.000 |
| 4-nitrophenyl myristate (C14) | 3.965 | 3.697 | 1950127744.000 |
| 4-nitrophenyl palmitate (C16) | 3.706 | 4.068 | 1042596992.000 |
| Plot | Vmax (μmole min−1) | Km (mM) |
|---|---|---|
| Michaelis-Menten | 87.42 | 0.212 |
| Lineweaver-Burk | 89.29 | 0.223 |
| Hanes-Woolf | 88.50 | 0.212 |
| Solvents | Log P | Relative Activity (%) ±SE |
|---|---|---|
| Control | - | 100 |
| Glycerol | −3.0 | 109.35 ± 2.61 |
| DMSO | −1.3 | 128.25 ± 3.36 |
| Methanol | −0.76 | 67.05 ± 1.37 |
| Ethanol | −0.18 | 15.49 ± 1.23 |
| 1-Propanol | 0.28 | 1.44 ± 0.00 |
| Pyridine | 0.77 | 2.74 ± 0.10 |
| 1-Butanol | 0.84 | 1.00 ± 0.01 |
| Propyl acetate | 1.2 | 2.45 ± 0.53 |
| Isoamyl alcohol | 1.36 | 0.62 ± 0.02 |
| Benzene | 2.0 | 107.85 ± 2.67 |
| Toluene | 2.5 | 105.82 ± 4.79 |
| Xylene | 3.15 | 95.73 ± 2.18 |
| Hexane | 3.9 | 100.31 ± 3.71 |
| Isooctane | 4.7 | 104.85 ± 2.97 |
| Octane | 5.2 | 95.66 ± 2.55 |
| Decane | 5.8 | 100.19 ± 2.86 |
| n-Tridecane | 7.33 | 95.70 ± 2.65 |
| n-Tetradecane | 7.6 | 102.11 ± 2.24 |
| n-Hexadecane | 8.86 | 102.04 ± 0.34 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masomian, M.; Rahman, R.N.Z.R.A.; Salleh, A.B. A Novel Method of Affinity Tag Cleavage in the Purification of a Recombinant Thermostable Lipase from Aneurinibacillus thermoaerophilus Strain HZ. Catalysts 2018, 8, 479. https://doi.org/10.3390/catal8100479
Masomian M, Rahman RNZRA, Salleh AB. A Novel Method of Affinity Tag Cleavage in the Purification of a Recombinant Thermostable Lipase from Aneurinibacillus thermoaerophilus Strain HZ. Catalysts. 2018; 8(10):479. https://doi.org/10.3390/catal8100479
Chicago/Turabian StyleMasomian, Malihe, Raja Noor Zaliha Raja Abd Rahman, and Abu Bakar Salleh. 2018. "A Novel Method of Affinity Tag Cleavage in the Purification of a Recombinant Thermostable Lipase from Aneurinibacillus thermoaerophilus Strain HZ" Catalysts 8, no. 10: 479. https://doi.org/10.3390/catal8100479
APA StyleMasomian, M., Rahman, R. N. Z. R. A., & Salleh, A. B. (2018). A Novel Method of Affinity Tag Cleavage in the Purification of a Recombinant Thermostable Lipase from Aneurinibacillus thermoaerophilus Strain HZ. Catalysts, 8(10), 479. https://doi.org/10.3390/catal8100479





