Preparation of a Flower-Like Immobilized D-Psicose 3-Epimerase with Enhanced Catalytic Performance
Abstract
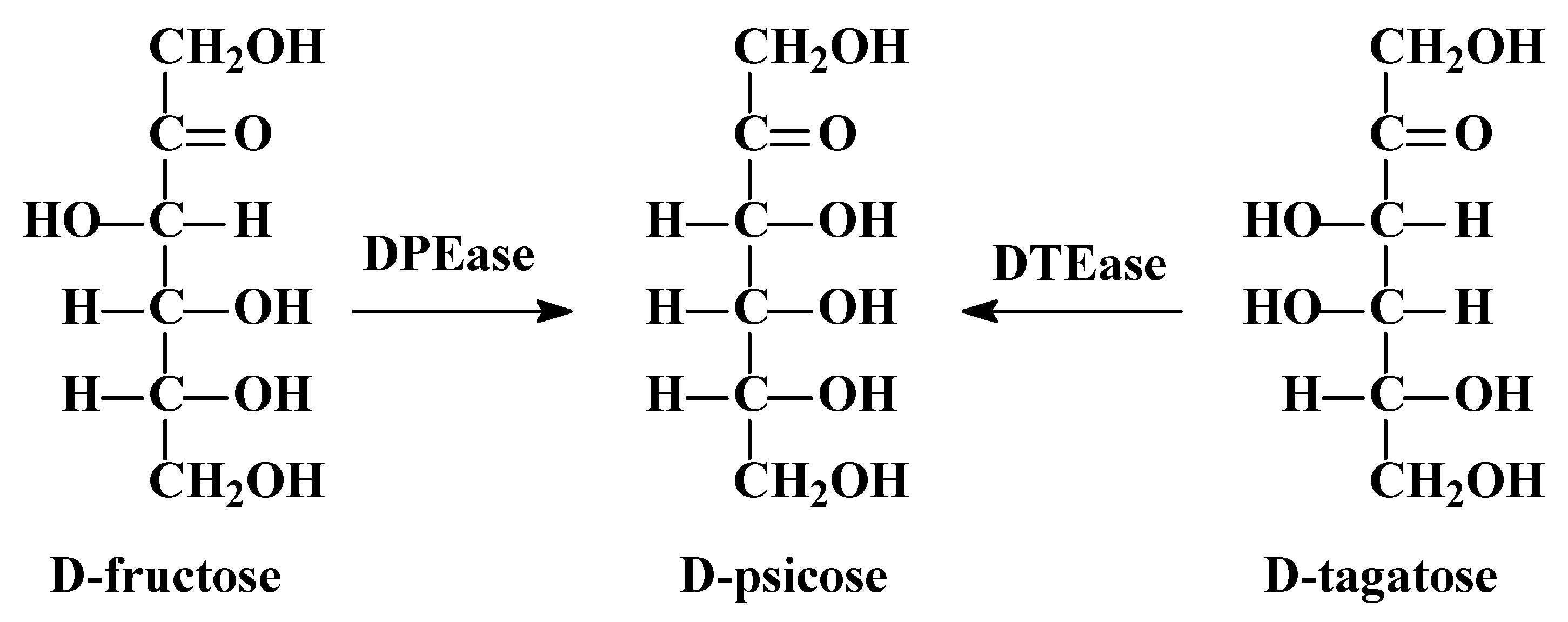
1. Introduction
2. Results and Discussion
2.1. Morphology of Nanoflower
2.2. FTIR of Nanoflowers
2.3. Comparison of Catalytic Properties of Nanoflower and Free DPEase
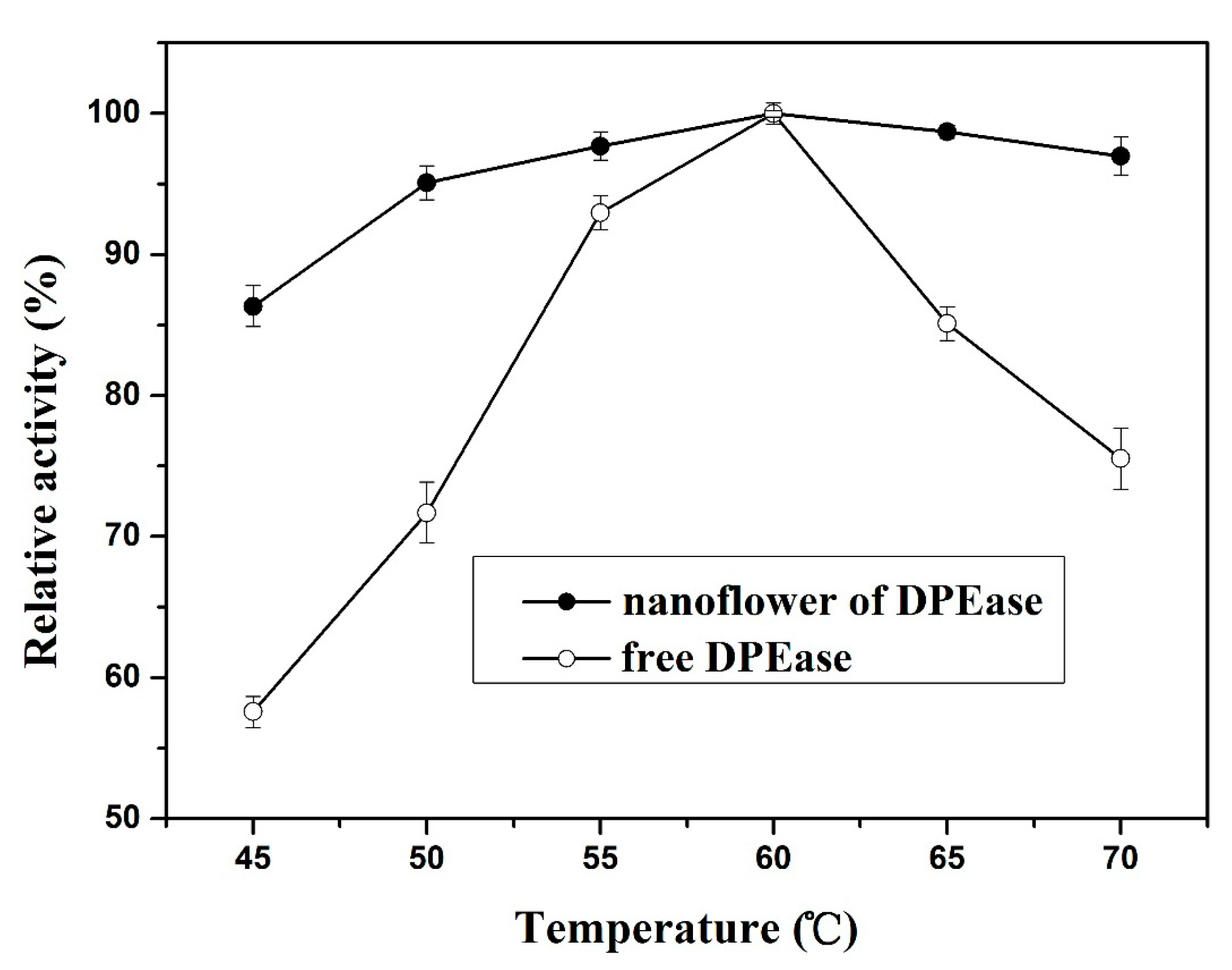
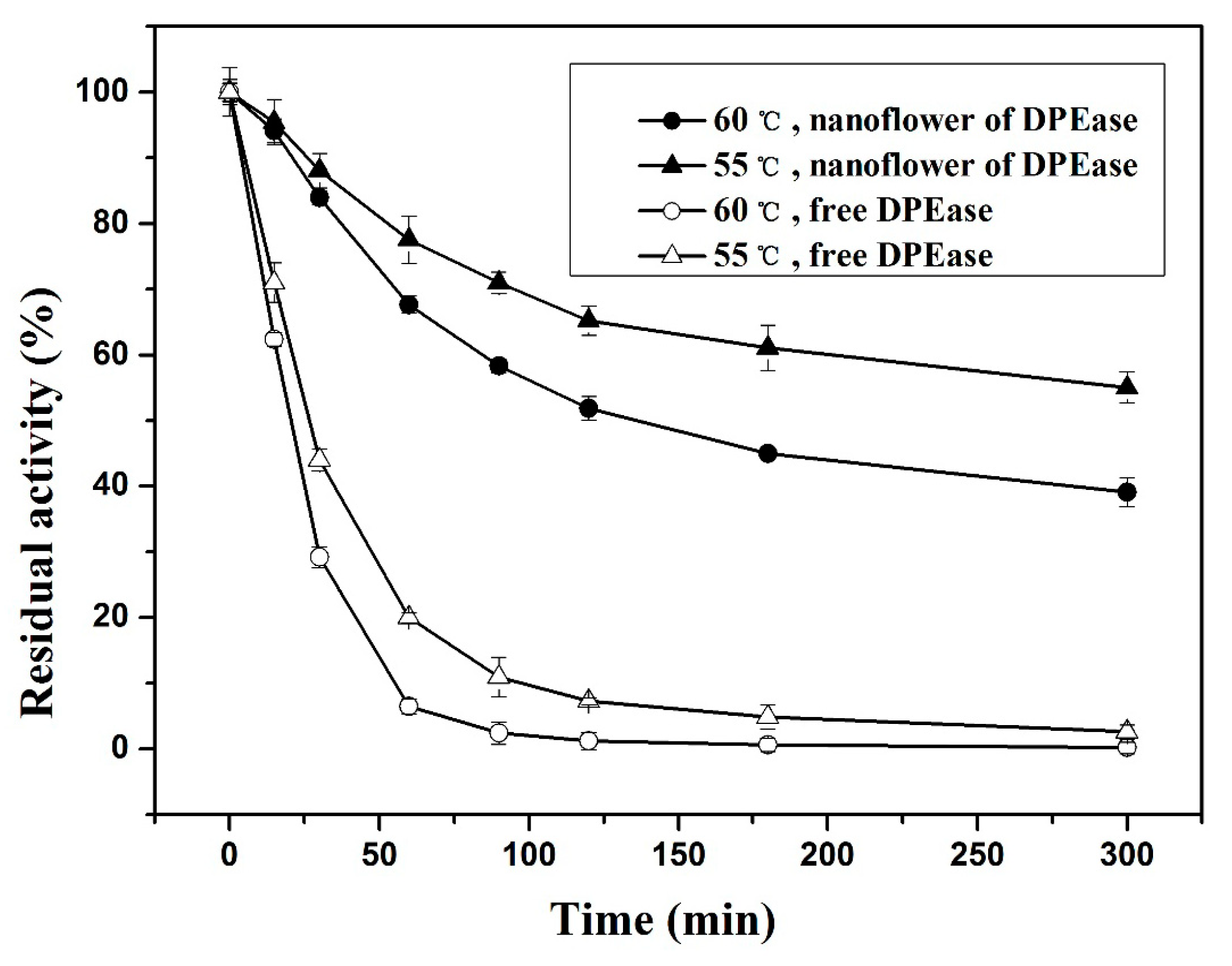
2.3.1. Effect of Temperature and Thermal Stability
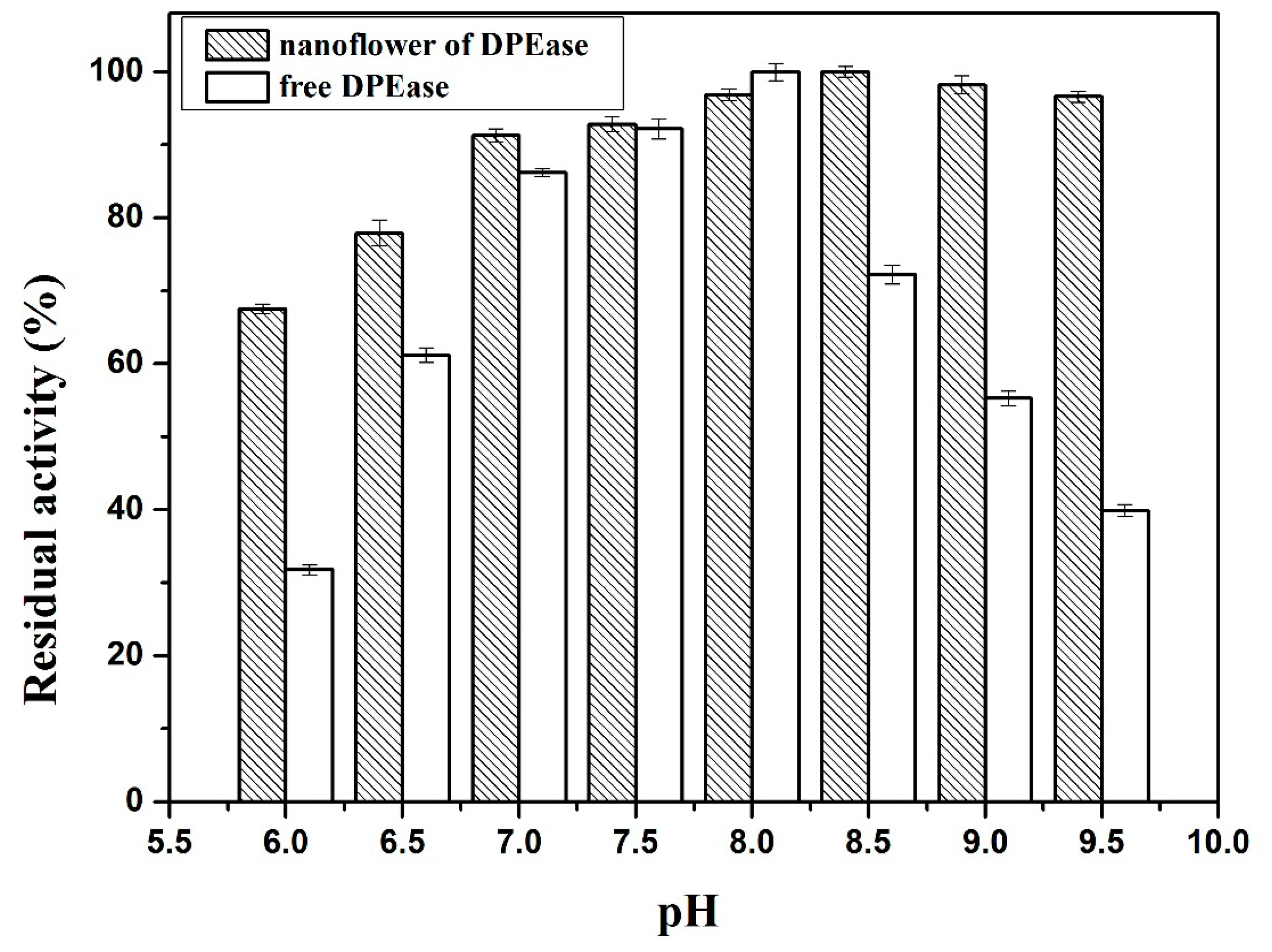
2.3.2. Effect of pH and pH Stability
2.3.3. Free DPEase vs the Prepared Nanoflower
2.3.4. Reusability
3. Materials and Methods
3.1. Materials
3.2. Preparation of the Nanoflowers
3.3. Characterization of the Prepared Nanoflowers
3.4. Standard Enzyme Activity Assay
3.5. HPLC Analysis
3.6. pH Stability
3.7. Thermal Stability
3.8. Reusability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chattopadhyay, S.; Raychaudhuri, U.; Chakraborty, R. Artificial Sweeteners—A Review. J. Food Sci. Tech. 2014, 51, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, I.; Hossain, A.; Yamaguchi, F.; Hirata, Y.; Dong, Y.; Kamitori, K.; Sui, L.; Machiko, N.; Masaki, U.; Kazuyuki, N.; et al. Intestinal Absorption, Organ Distribution, and Urinary Excretion of the Rare Sugar D-psicose. Drug Des. Dev. Ther. 2014, 8, 1955–1964. [Google Scholar]
- Mu, W.; Zhang, W.; Feng, Y.; Jiang, B.; Zhou, L. Recent advances on applications and biotechnological production of D-psicose. Appl. Microbiol. Biot. 2012, 94, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lafuente, R.; Rodrı´guez, V.; Mateo, C.; Penzol, G.; Hernández-Justiz, O.; Irazoqui, G.; Villarino, A.; Ovsejevi, K.; Batista, F.; Guisán, J.M. Stabilization of Multimeric Enzymes via Immobilization and Post-immobilization Techniques. J. Mol. Catal. B Enzym. 1999, 7, 181–189. [Google Scholar] [CrossRef]
- Palomo, J.M.; Muñoz, G.; Fernández-Lorente, G.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Interfacial Adsorption of Lipases on Very Hydrophobic Support (octadecyl–Sepabeads): Immobilization, Hyperactivation and Stabilization of the Open Form of Lipases. J. Mol. Catal. B Enzym. 2002, 19, 279–286. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of Enzyme Activity, Stability and Selectivity via Immobilization Techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Guisán, J.M.; Fernández-Lafuente, R. Specificity Enhancement towards Hydrophobic Substrates by Immobilization of Lipases by Interfacial Activation on Hydrophobic Supports. Enzyme Microb. Technol. 2007, 41, 565–569. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Cabrera, Z.; Godoy, C.; Fernandez-Lafuente, R.; Palomo, J.M.; Guisan, J.M. Interfacially Activated Lipases against Hydrophobic Supports: Effect of the Support Nature on the Biocatalytic Properties. Process Biochem. 2008, 43, 1061–1067. [Google Scholar] [CrossRef]
- Suescun, A.; Rueda, N.; dos Santos, J.C.; Castillo, J.J.; Ortiz, C.; Torres, R.; Barbosa, O.; Fernandez-Lafuente, R. Immobilization of Lipases on Glyoxyl–octyl Supports: Improved Stability and Reactivation Strategies. Process Biochem. 2015, 50, 1211–1217. [Google Scholar] [CrossRef]
- Wilson, L.; Palomo, J.M.; Fernández-Lorente, G.; Illanes, A.; Guisán, J.M.; Fernández-Lafuente, R. Improvement of the Functional Properties of a Thermostable Lipase from Alcaligenes sp. via Strong Adsorption on Hydrophobic Supports. Enzyme Microb. Technol. 2006, 38, 975–980. [Google Scholar] [CrossRef]
- Takeshita, K.; Suga, A.; Takada, G.; Izumori, K. Mass Production of D-psicose from D-fructose by a Continuous Bioreactor System Using Immobilized D-Tagatose 3-Epimerase. J. Biosci. Bioeng. 2000, 90, 453–455. [Google Scholar] [CrossRef]
- Lim, B.C.; Kim, H.J.; Oh, D.K. A Stable Immobilized D-Psicose 3-Epimerase for the Production of D-Psicose in the Presence of Borate. Process Biochem. 2009, 44, 822–828. [Google Scholar] [CrossRef]
- Dedania, S.R.; Patel, M.J.; Patel, D.J.; Patel, D.M.; Akhani, R.C.; Patel, D.H. Immobilization on Graphene Oxide Improves the Thermal Stability and Bioconversion Efficiency of D-Psicose 3-Epimerase for Rare Sugar Production. Enzyme Microb. Technol. 2017, 107, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Lei, J.; Zare, R.N. Protein–inorganic Hybrid Nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; He, R.; Zhuang, A.; Wang, X.; Zeng, J.; Hou, J.G. A New Nanobiocatalytic System Based on Allosteric Effect with Dramatically Enhanced Enzymatic Performance. J. Am. Chem. Soc. 2013, 135, 1272–1275. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Fan, H.; Wu, Z.; Zheng, L.; Wang, L.; Wang, Z.; Chen, G. Ultrasound-assisted enantioselective esterification of ibuprofen catalyzed by a flower-like nanobioreactor. Molecules 2016, 21, 565. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, H.J.; Oh, D.K.; Cha, S.S.; Rhee, S. Crystal Structure of D-psicose 3-epimerase from Agrobacterium Tumefaciens and Its Complex with True Substrate D-fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and Its Conformational Changes. J. Mol. Biol. 2006, 361, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Jeong, J.M.; Lee, S.J.; Choi, B.G.; Lee, K.G. Protein-directed Assembly of Cobalt Phosphate Hybrid Nanoflowers. J. Colloid Interf. Sci. 2016, 484, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Gao, J.; He, Y.; Zhou, L.; Ma, L.; Huang, Z.; Jiang, Y. Preparation of a Flowerlike Nanobiocatalyst System via Biomimetic Mineralization of Cobalt Phosphate with Enzyme. Ind. Eng. Chem. Res. 2017, 56, 14923–14930. [Google Scholar] [CrossRef]
- Cornishbowden, A. Enthalpy-entropy Compensation and the Isokinetic Temperature in Enzyme Catalysis. J. Biosci. 2017, 42, 665–670. [Google Scholar] [CrossRef]
- Daniel, R.M.; Danson, M.J.; Eisenthal, R.; Lee, C.K.; Peterson, M.E. The Effect of Temperature on Enzyme Activity: New Insights and Their Implications. Extremophiles 2008, 12, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.M.; Peterson, M.E.; Danson, M.J.; Price, N.C.; Kelly, S.M.; Monk, C.R.; Weinberg, C.S.; Oudshoorn, M.L.; Lee, C.K. The Molecular Basis of the Effect of Temperature on Enzyme Activity. Biochem. J. 2010, 425, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.I.; Chiba, M.; Tsunoda, T.; Aritomo, Y. Enzyme Immobilization in Mesoporous Silica for Enhancement of Thermostability. J. Nanosci. Nanotechno. 2018, 18, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Tian, L.; Ma, D.; Wu, H.; Wang, Z.; Wang, L.; Fang, X. Microwave-assisted Fatty Acid Methyl Ester Production from Soybean Oil by Novozym 435. Green Chem. 2010, 12, 844–850. [Google Scholar] [CrossRef]
- Da Silva, T.M.; Maller, A.; de Lima Damásio, A.R.; Michelin, M.; Ward, R.J.; Hirata, I.Y.; Jorge, J.A.; Terenzi, H.F.; de Polizeli, M.L.T.M. Properties of a Purified Thermostable Glucoamylase from Aspergillus Niveus. J. Ind. Microbiol. Biot. 2009, 36, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Zamost, B.L.; Nielsen, H.K.; Starnes, R.L. Thermostable Enzymes for Industrial Applications. J. Ind. Microbiol. Biot. 1991, 8, 71–81. [Google Scholar] [CrossRef]
- Yang, C.-H.; Huang, Y.-C.; Chen, C.-Y.; Wen, C.-Y. Heterologous Expression of Thermobifida Fusca, Thermostable Alpha-amylase in Yarrowia Lipolytica, and its Application in Boiling Stable Resistant Sago Starch Preparation. J. Ind. Microbiol. Biot. 2010, 37, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Galante, Y.M.; Formantici, C. Enzyme Applications in Detergency and in Manufacturing Industries. Curr. Org. Chem. 2003, 7, 1399–1422. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An Overview of Technologies for Immobilization of Enzymes and Surface Analysis Techniques for Immobilized Enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; dos Santos, J.C.S; Ortiz, C.; Torres, R.; Barbosa, O.; Rodrigus, R.C.; Berengure-Murcia, Á.; Fernandez-Lafuente, R. Chemical Modification in the Design of Immobilized Enzyme Biocatalysts: Drawbacks and Opportunities. Chem. Rec. 2016, 16, 1436–1455. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. The Influence of Macromolecular Crowding and Macromolecular Confinement on Biochemical Reactions in Physiological Media. J. Biol. Chem. 2001, 276, 10577–10580. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; Dill, K.A. Stabilization of Proteins in Confined Spaces. Biochemistry 2001, 40, 11289–11293. [Google Scholar] [CrossRef]
- Zheng, L.; Xie, X.; Wang, Z.; Zhang, Y.; Wang, L.; Cui, X.; Huang, H.; Zhuang, H. Fabrication of a Nano-biocatalyst for Regioselective Acylation of Arbutin. Green Chem. Lett. Rev. 2018, 11, 55–61. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Li, F.; Yue, H.; He, C.; Xie, F.; Wang, Z. Enantioselective Transesterification of (R,S)-2-pentanol Catalyzed by a New Flower-like Nanobioreactor. RSC Adv. 2014, 4, 33998–34002. [Google Scholar] [CrossRef]
- Mehta, J.; Bhardwaj, N.; Bhardwaj, S.K.; Kim, K.H.; Deep, A. Recent Advances in Enzyme Immobilization Techniques: Metal-organic Frameworks as Novel Substrates. Coordin. Chem. Rev. 2016, 322, 30–40. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, B.; Zhang, Z.; Lou, D.; Tan, J.; Zhu, L. The Effects of Macromolecular Crowding and Surface Charge on the Properties of an Immobilized Enzyme: Activity, Thermal Stability, Catalytic Efficiency and Reusability. RSC Adv. 2017, 7, 38028–38036. [Google Scholar] [CrossRef]
- Mu, W.; Chu, F.; Xing, Q.; Yu, S.; Zhou, L.; Jiang, B. Cloning, Expression, and Characterization of a D-psicose 3-epimerase from Clostridium Cellulolyticum H10. J. Agric. Food Chem. 2011, 59, 7785–7792. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Liese, A.; Hilterhaus, L. Evaluation of Immobilized Enzymes for Industrial Applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef] [PubMed]




| Samples | C Element (%) | H Element (%) | N Element (%) |
|---|---|---|---|
| DPEase | 40.997 | 6.031 | 7.102 |
| Co3(PO4)2 | 0.130 | 1.865 | 0.007 |
| Nanoflower | 3.154 | 2.095 | 0.880 |
| Sample | Enzyme Activity (U/mg) |
|---|---|
| Free DPEase a | 5.0 ± 0.2 |
| Nanoflower b | 36.2 ± 0.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, L.; Sun, Y.; Wang, J.; Huang, H.; Geng, X.; Tong, Y.; Wang, Z. Preparation of a Flower-Like Immobilized D-Psicose 3-Epimerase with Enhanced Catalytic Performance. Catalysts 2018, 8, 468. https://doi.org/10.3390/catal8100468
Zheng L, Sun Y, Wang J, Huang H, Geng X, Tong Y, Wang Z. Preparation of a Flower-Like Immobilized D-Psicose 3-Epimerase with Enhanced Catalytic Performance. Catalysts. 2018; 8(10):468. https://doi.org/10.3390/catal8100468
Chicago/Turabian StyleZheng, Lu, Yining Sun, Jing Wang, He Huang, Xin Geng, Yi Tong, and Zhi Wang. 2018. "Preparation of a Flower-Like Immobilized D-Psicose 3-Epimerase with Enhanced Catalytic Performance" Catalysts 8, no. 10: 468. https://doi.org/10.3390/catal8100468
APA StyleZheng, L., Sun, Y., Wang, J., Huang, H., Geng, X., Tong, Y., & Wang, Z. (2018). Preparation of a Flower-Like Immobilized D-Psicose 3-Epimerase with Enhanced Catalytic Performance. Catalysts, 8(10), 468. https://doi.org/10.3390/catal8100468





