2′-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium
Abstract
:1. Introduction
2. Results
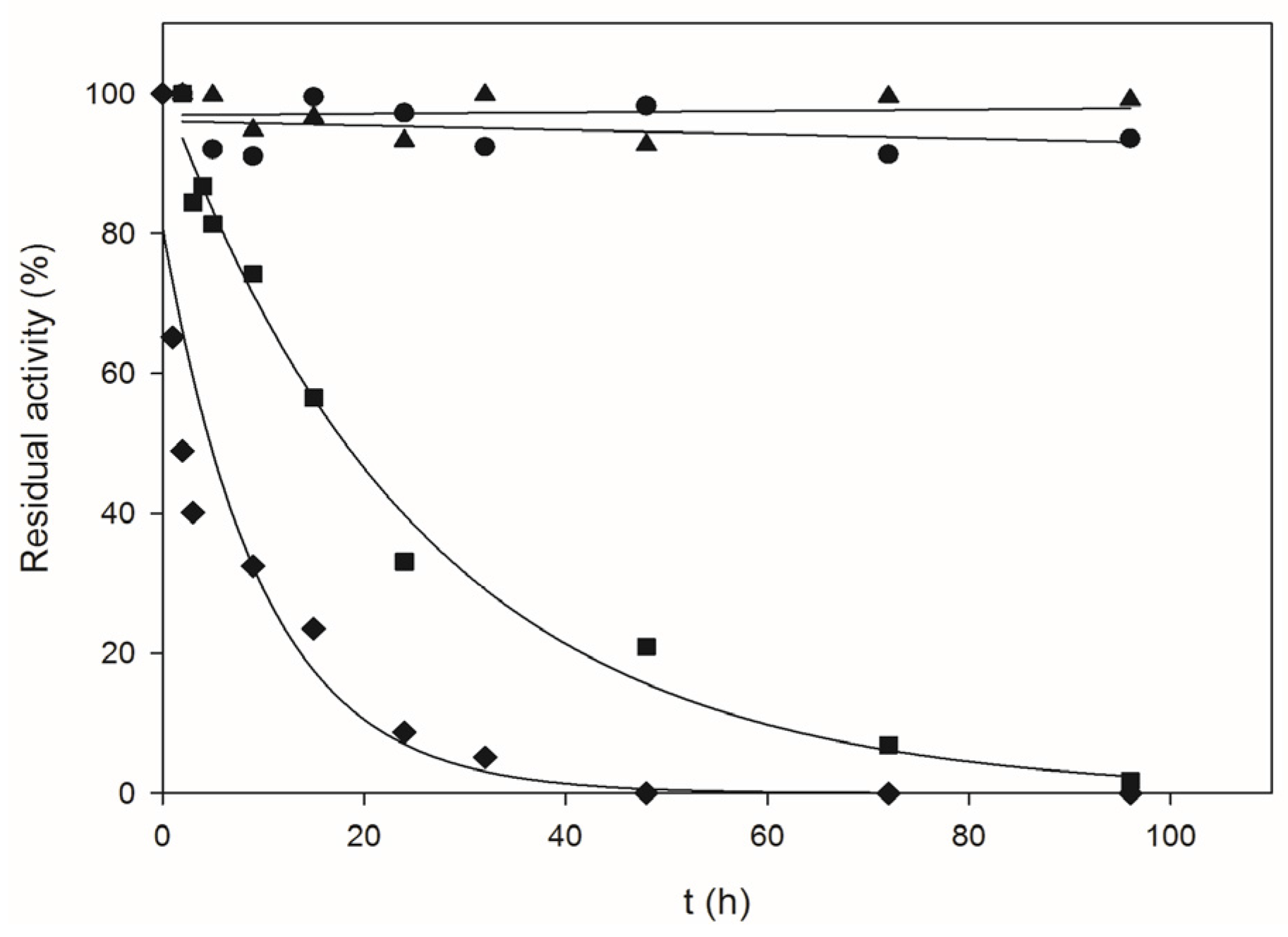
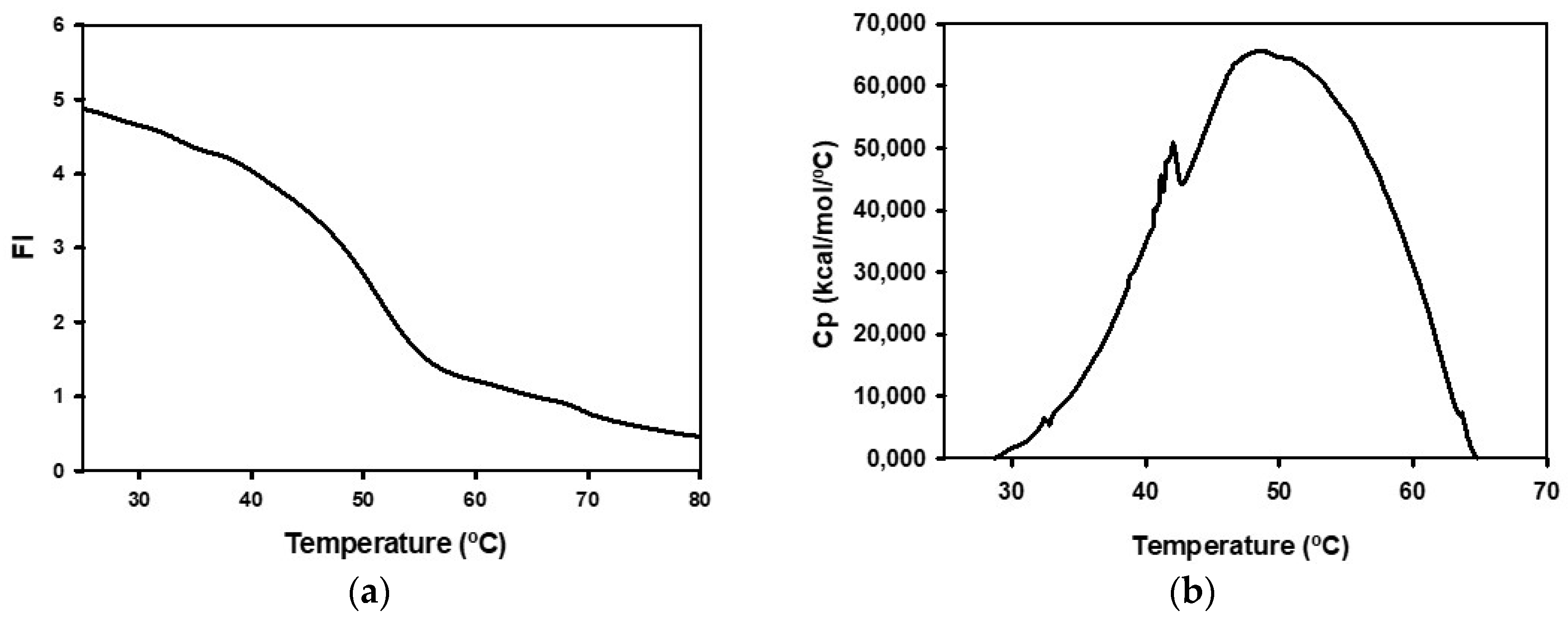
2.1. Biochemical Characterization of Recombinant BpNDT
2.2. Substrate Specificity
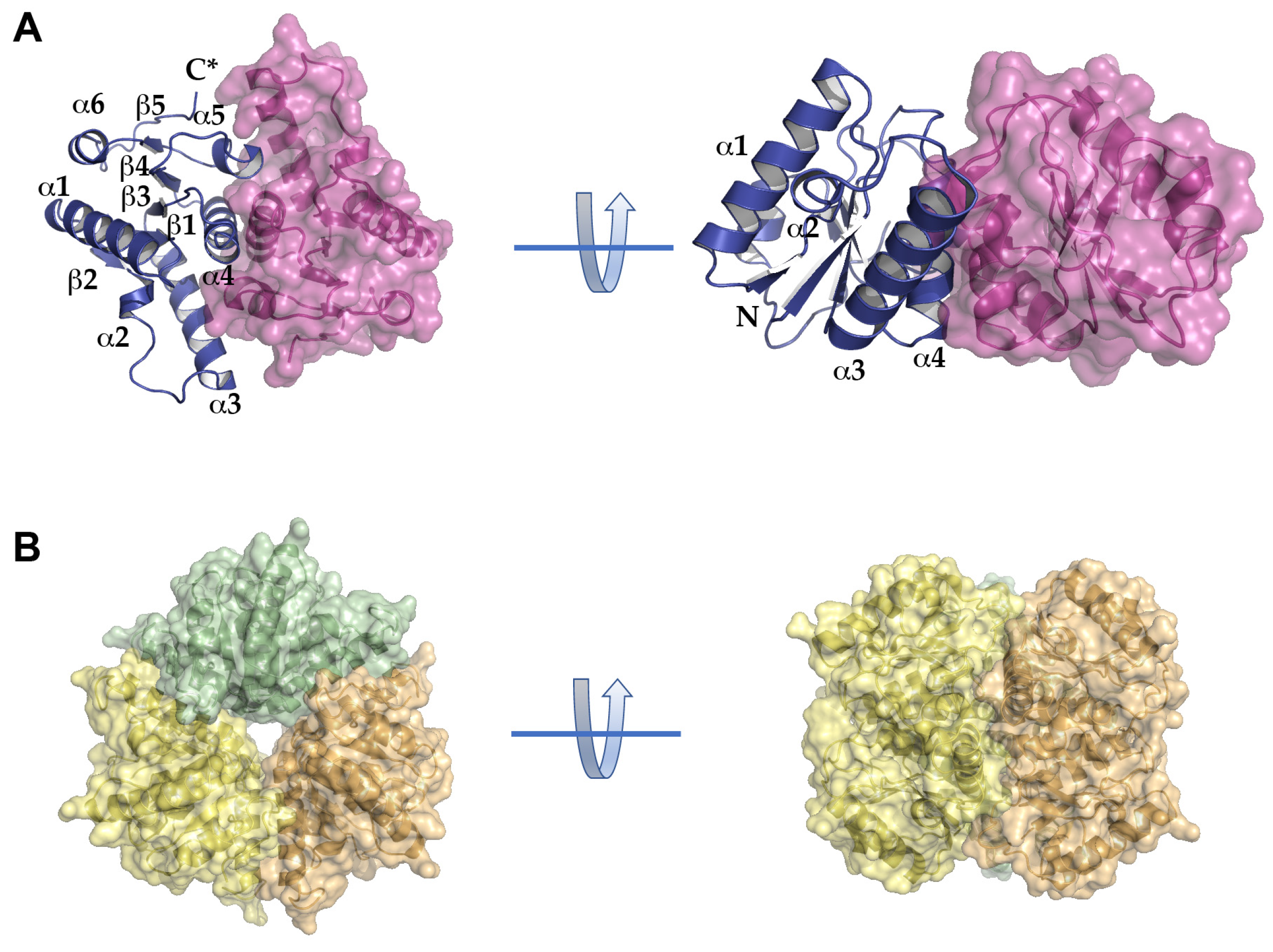
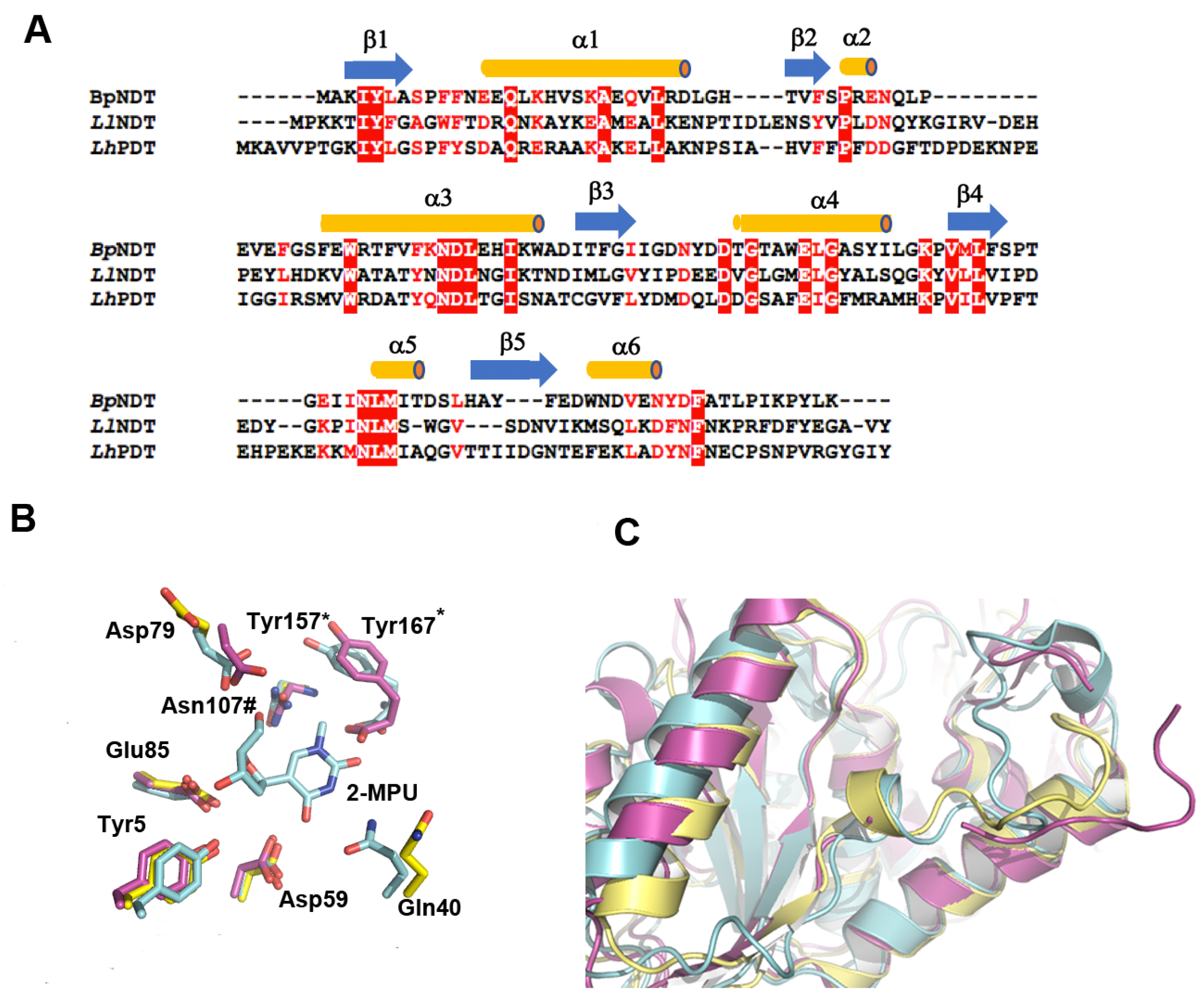
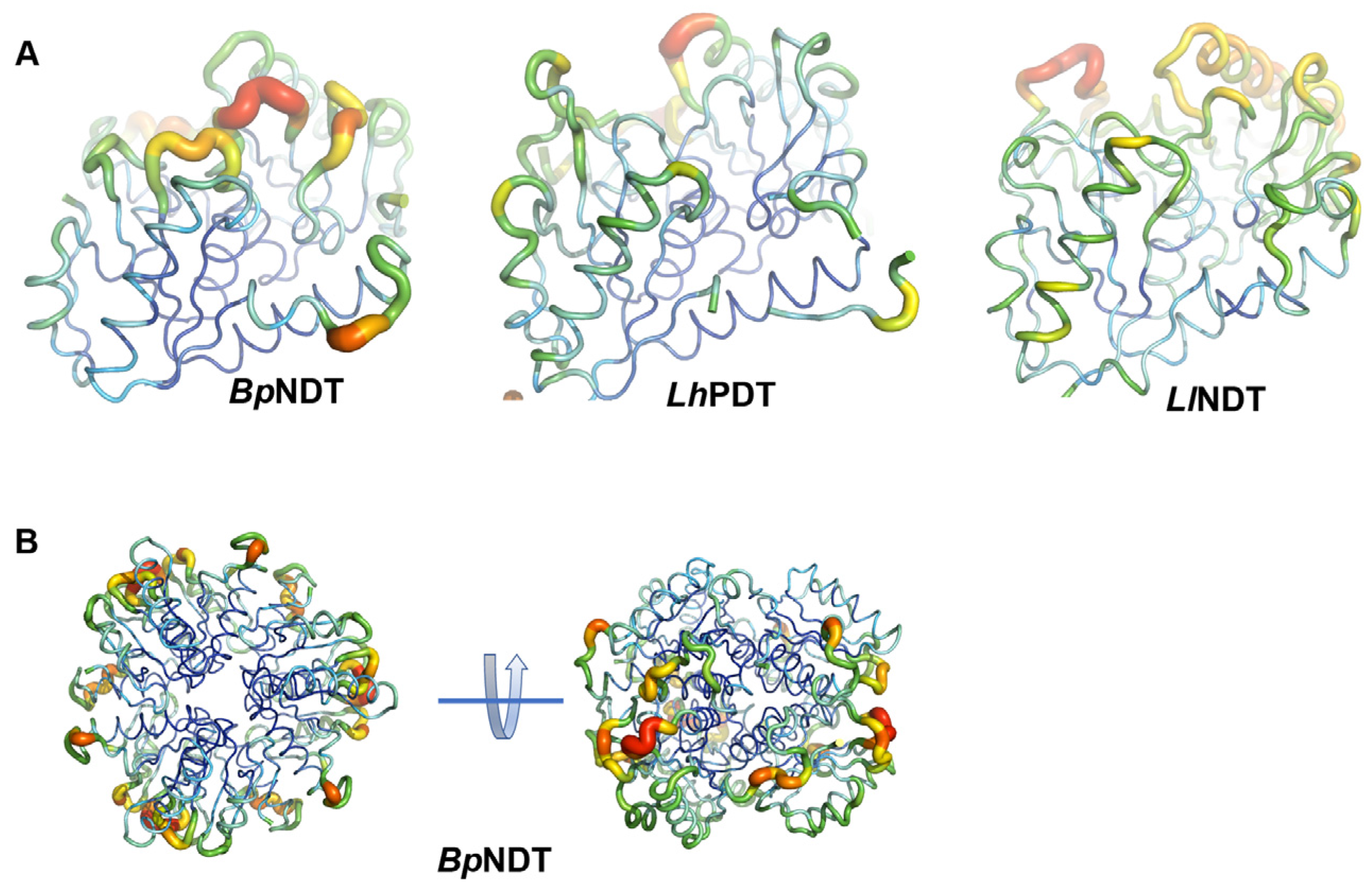
2.3. Structural Analysis of BpNDT
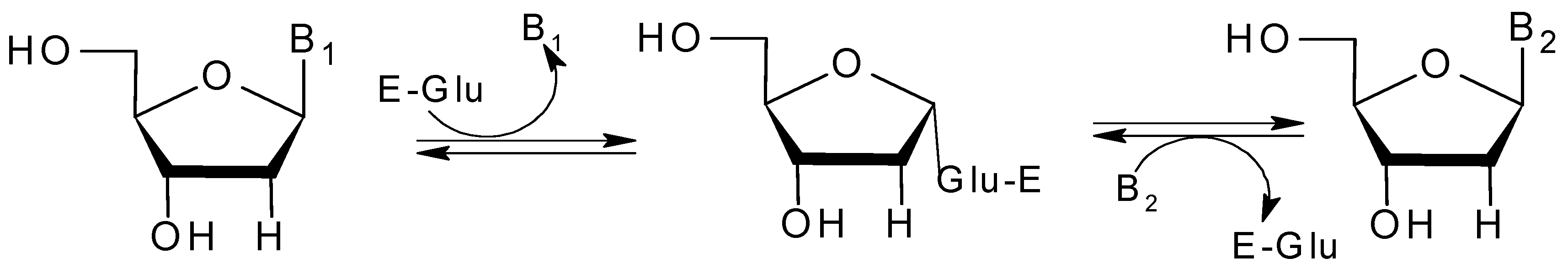
2.4. Active Site of BpNDT
2.5. Enzymatic Production of Nucleoside Analogues
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cloning, Sequencing and Tagging of BpNDT
4.3. Production and Purification of Recombinant His-Tag BpNDT
4.4. Site Directed Mutagenesis of His-tag BpNDT
4.5. N-Deoxyribosyltransferase Assay
4.6. Thermal Inactivation Studies
4.7. Effect of Ionic Strength and Cations on Enzyme Activity
4.8. Analytical Ultracentrifugation Analysis
4.9. Enzyme Crystallization and Data Collection
4.10. Structure Solution and Refinement
4.11. Spectroscopic Studies
4.12. Differential Scanning Calorimetry Studies
4.13. Kinetic Studies
4.14. Accession Number
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Omar, S.M.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D.; et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2002, 357, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [PubMed]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997, 53, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.M.; Stokes, J.L. Taxonomy of psychrophilic strains of Bacillus. J. Bacteriol. 1967, 94, 889–895. [Google Scholar] [PubMed]
- Seo, J.B.; Kim, H.S.; Jung, G.Y.; Nam, M.H.; Chung, J.H.; Kim, J.Y.; Yoo, J.S.; Kim, C.W.; Kwon, O. Psychrophilicity of Bacillus psychrosaccharolyticus: A proteomic study. Proteomics 2004, 4, 3654–3659. [Google Scholar] [CrossRef] [PubMed]
- Fresco-Taboada, A.; del Cerro, C.; Fernández-Lucas, J.; Arroyo, M.; Acebal, C.; García, J.L.; de la Mata, I. Genome of the psychrophilic bacterium Bacillus psychrosaccharolyticus, a potential source of 2′-deoxyribosyltransferase for industrial nucleoside synthesis. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Lewkowicz, E.S.; Iribarren, A.M. Nucleoside phosphorylases. Curr. Org. Chem. 2006, 10, 1197–1215. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A. Biotechnology of nucleic acid constituents—State of the art and perspectives. Curr. Org. Chem. 2007, 11, 317–333. [Google Scholar] [CrossRef]
- Fresco-Taboada, A.; de la Mata, I.; Arroyo, M.; Fernández-Lucas, J. New insights on nucleoside 2′-deoxyribosyltransferases: A versatile biocatalyst for one-pot one-step synthesis of nucleoside analogs. Appl. Microbiol. Biotechnol. 2013, 97, 3773–3785. [Google Scholar]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002, 3, 415–424. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Brandon, M.L.; Mi, L.; Chaung, W.; Teebor, G.; Boorstein, R.J. 5-Chloro-2′-deoxyuridine cytotoxicity results from base excision repair of uracil subsequent to thymidylate synthase inhibition. Mutat. Res. 2000, 459, 161–169. [Google Scholar] [CrossRef]
- Sato, A.; Hiramoto, A.; Uchikubo, Y.; Miyazaki, E.; Satake, A.; Naito, T.; Hiraoka, O.; Miyake, T.; Kim, H.S.; Wataya, Y. Gene expression profiles of necrosis and apoptosis induced by 5-fluoro-2′-deoxyuridine. Genomics 2008, 92, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Médici, R.; Lewkowicz, E.S.; Iribarren, A.M. Microbial synthesis of 2,6-diaminopurine nucleosides. J. Mol. Catal. B Enzym. 2006, 39, 40–44. [Google Scholar] [CrossRef]
- Fernández-Lucas, J.; Acebal, C.; Sinisterra, J.V.; Arroyo, M.; de la Mata, I. Lactobacillus reuteri 2′-deoxyribosyltransferase, a novel biocatalyst for tailoring of nucleosides. Appl. Environ. Microbiol. 2010, 76, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, P.A. Functional cloning, heterologous expression, and purification of two different N-deoxyribosyltransferases from Lactobacillus helveticus. J. Biol. Chem. 2002, 277, 14400–14407. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Kaminski, P.A.; Ealick, S.E. Structures of purine 2′-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 Å resolution. Biochemistry 2004, 43, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, K.A.; Jewett, M.W.; Rosa, P.A.; Gherardini, F.C. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 2009, 72, 1517–1529. [Google Scholar] [CrossRef] [PubMed]
- Bosch, J.; Robien, M.A.; Mehlin, C.; Boni, E.; Riechers, A.; Buckner, F.S.; Van Voorhis, W.C.; Myler, P.J.; Worthey, E.A.; DeTitta, G.; et al. Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase. J. Med. Chem. 2006, 49, 5939–5946. [Google Scholar] [CrossRef] [PubMed]
- Crespo, N.; Sánchez-Murcia, P.A.; Gago, F.; Cejudo-Sanches, J.; Galmes, M.A.; Fernández-Lucas, J.; Mancheño, J.M. 2′-Deoxyribosyltransferase from Leishmania mexicana, an efficient biocatalyst for one-pot, one-step synthesis of nucleosides from poorly soluble purine bases. Appl. Microbiol. Biotechnol. 2017, 101, 7187–7200. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lucas, J.; Condezo, L.A.; Martínez-Lagos, F.; Sinisterra, J.V. Synthesis of 2′-deoxyribosylnucleosides using new 2′-deoxyribosyltransferase microorganism producers. Enzym. Microb. Technol. 2007, 40, 1147–1155. [Google Scholar] [CrossRef]
- Fresco-Taboada, A.; Serra, I.; Fernandez-Lucas, J.; Acebal, C.; Arroyo, M.; Terreni, M.; de la Mata, I. Nucleoside 2′-deoxyribosyltransferase from psychrophilic bacterium Bacillus psychrosaccharolyticus—Preparation of an immobilized biocatalyst for the enzymatic synthesis of therapeutic nucleosides. Molecules 2014, 19, 11231–11249. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Masaki, T.; Chohnan, S. Characterization of N-deoxyribosyltransferase from Lactococcus lactis subsp lactis. BBA Proteins Proteom. 2007, 1774, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, P.A.; Dacher, P.; Dugué, L.; Pochet, S. In vivo reshaping the catalytic site of nucleoside 2′-deoxyribosyltransferase for dideoxy- and didehydronucleosides via a single amino acid substitution. J. Biol. Chem. 2008, 283, 20053–20059. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Rosenström, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Godzik, A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 2003, 19, ii246–ii255. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, S.R.; Cook, W.J.; Short, S.A.; Ealick, S.E. Crystal structures of nucleoside 2′-deoxyribosyltransferase in native and ligand-bound forms reveal architecture of the active site. Structure 1996, 4, 97–107. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold-adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Short, S.A.; Armstrong, S.R.; Ealick, S.E.; Porter, D.J. Active site amino acids that participate in the catalytic mechanism of nucleoside 2′-deoxyribosyltransferase. J. Biol. Chem. 1996, 271, 4978–4987. [Google Scholar] [PubMed]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1988, 6, 351–361. [Google Scholar] [CrossRef]
- Aghajari, N.; Van Petegem, F.; Villeret, V.; Chessa, J.P.; Gerday, C.; Haser, R.; Van Beeumen, J. Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold adapted proteases. Proteins 2003, 50, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Heo, D.; Choi, S.; Kim, K.; Shim, J.; Kim, C.; Sung, H.; Yun, C. Functional characterization of Hsp33 protein from Bacillus psychrosaccharolyticus; additional function of HSP33 on resistance to solvent stress. Biochem. Biophys. Res. Commun. 2007, 358, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Okubo, Y.; Yokoigawa, K.; Esaki, N.; Soda, K.; Misono, H. High catalytic activity of alanine racemase from psychrophilic Bacillus psychrosaccharolyticus at high temperatures in the presence of pyridoxal 5′-phosphate. FEMS Microbiol. Lett. 2000, 192, 169–173. [Google Scholar] [CrossRef]
- Nandakumar, R.; Mattiasson, B. Affinity isolation of a cold-adapted enzyme: Lactate dehydrogenase from Bacillus psychrosaccharolyticus. Bioseparation 1999, 7, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Fresco-Taboada, A.; Serra, I.; Arroyo, M.; Fernández-Lucas, J.; de la Mata, I.; Terreni, M. Development of an immobilized biocatalyst based on Bacillus psychrosaccharolyticus NDT for the preparative synthesis of trifluridine and decytabine. Catal. Today 2016, 259, 197–204. [Google Scholar] [CrossRef]
- Xu, Y.; Feller, G.; Gerday, C.; Glansdorff, N. Metabolic enzymes from psychrophilic bacteria: Challenge of adaptation to low temperatures in ornithine carbamoyltransferase from Moritella abyssi. J. Bacteriol. 2003, 185, 2161–2168. [Google Scholar] [CrossRef] [PubMed]
- Danzin, C.; Cardinaud, R. Deoxyribosyl transfer catalysis with trans-N-deoxyribosylase. Eur. J. Biochem. 1976, 62, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.J.; Short, S.A. Nucleoside 2-deoxyribosyltransferase. Pre-steady-state kinetic analysis of native enzyme and mutant enzyme with an alanyl residue replacing Glu-98. J. Biol. Chem. 1995, 270, 15557–15562. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.B. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem. Rev. 2009, 109, 2880–2893. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.; Nakamura, J. 5-Hydroxymethyl-2′-deoxyuridine, but not temozolomide, enhances the selective synthetic lethality in BRCA1 and BRCA2- deficient cells caused by PARP inhibition. Cancer Res. 2013, 73. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.J.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 72, 5463–5464. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brown, P.H.; Schuck, P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006, 90, 4651–4661. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.L. Analysis of heterogeneous interactions. Methods Enzymol. 2004, 384, 212–232. [Google Scholar] [PubMed]
- Minton, A.; Jaenicke, R.; Durchschlag, H. Alternative Strategies for the Characterization of Associations in Multicomponent Solutions via Measurement of Sedimentation Equilibrium Analytical Ultracentrifugation IV. Progress in Colloid and Polymer Science; Springer: Berlin/Heidelberg, Germany, 1997; Volume 107, pp. 11–19. [Google Scholar]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Touw, W.G.; Baakman, C.; Black, J.; te Beek, T.A.; Krieger, E.; Joosten, R.P.; Vriend, G. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015, 43, D364–D368. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Ther. 2000, 88, 349–425. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J.T.; Kang, S.; Lee, J.H.; Kim, S.J. Characterization and its potential application of two esterases derived from the Arctic sediment metagenome. Mar. Biotechnol. 2009, 11, 307–316. [Google Scholar] [CrossRef] [PubMed]
| Additive | Relative Activity (%) at 1 mM | Relative Activity (%) at 5 mM |
|---|---|---|
| None | 100 | 100 |
| K2SO4 | 105.8 | 96.1 |
| KCl | 105.1 | 104.8 |
| LiCl | 102.0 | 100 |
| Na2SO4 | 97.1 | 95.7 |
| NaCl | 96.6 | 98.2 |
| RbCl | 95.9 | 101.8 |
| BaCl2 | 98.1 | 91.8 |
| CaCl2 | 84.0 | 74.7 |
| CoCl2 | 33.0 | 21.8 |
| CuSO4 | 63.7 | 22.3 |
| MgSO4 | 86.6 | 95.7 |
| MgCl2 | 88.5 | 90.1 |
| MnCl2 | 32.6 | 8.1 |
| ZnSO4 | 91.5 | 57.2 |
| (NH4)2SO4 | 103.8 | 98.8 |
| 2-mercaptoethanol | 98.9 | 97.5 |
| Al2(SO4)3 | 90.7 | 18.2 |
| FeCl3 | 95.6 | 114.8 |
| EDTA | 101.3 | 110.8 |
| Temperature (°C) | kd (h−1) | t1/2 (h) |
|---|---|---|
| 60 | 0.032 | 21.8 |
| 70 | 0.118 | 5.9 |
| Donor | Specific Activity (IU/mg Protein) with Acceptor | ||||
|---|---|---|---|---|---|
| Ade | Ura | Cyt | Thy | Hyp | |
| dAdo | -- | 24.4 | 36.8 | 26.1 | 43.5 |
| dUrd | 40.0 | -- | 45.4 | 24.9 | 41.7 |
| dCyd | 61.2 | 60.0 | -- | 47.8 | 84.6 |
| dThd | 51.0 | 26.1 | 31.3 | -- | 38.3 |
| dIno | 20.7 | 15.4 | 18.0 | 40.6 | -- |
| dGuo | 38.8 | 16.2 | 36.3 | 8.1 | 22.4 |
| BpNDT | |
|---|---|
| PDB code | 6EVS |
| Data collection | |
| Synchrotron source | ESRF |
| Beamline | ID29 |
| Wavelength (Å) | 0.9792 |
| Space group | R3 |
| Unit-cell parameters | a = b = 107.55, c = 61.24 α = β = 90°, γ = 120° |
| Resolution range (Å) | 37.07–1.90 |
| No. of measured reflections a | 108,568 (14,833) |
| No. of unique reflections | 20,796 (3009) |
| Mean (I/σI) | 14.8 (2.9) |
| Completeness (%) | 100 (100) |
| Multiplicity | 5.2 (4.9) |
| Rmeas (%); Rpim (%) | 5.7 (59.8); 2.5 (26.7) |
| CC1/2 | 0.988 (0.2845) |
| B-factor (Wilson plot, Å2) | 32.7 |
| Molecules/non-H atoms | |
| Protein | 2/2257 |
| Water | 104/104 |
| Refinement statistics | |
| Rwork(%)/Rfree(%) | 17.7/21.6 |
| Average B-factors (Å2) | |
| protein | 47.3 |
| water | 47.2 |
| Rms deviation bond length (Å) | 0.009 |
| Rms deviation angles (°) | 0.927 |
| Ramachandran | |
| Favoured (%) | 95.6 |
| Disallowed (%) | 1.48 |
| Mutation | None | Tyr5Phe | Tyr5His | Gln40Glu | Gln40Lys | Asp59Asn | Asp59His | Asp79Asn |
|---|---|---|---|---|---|---|---|---|
| Relative activity (%) | 100 | 1.17 | 1.38 | 0 | 5.91 | 5.8 | 0 | 0 |
| Mutation | Asp79His | Glu85Asp | Glu85Gln | Glu85His | Asn107Asp | Asn107His | Lys142Tyr | ΔK142 |
| Relative activity (%) | 0 | 0 | 0 | 0 | 0 | 2.12 | 97.72 | 21.5 |
| Acceptor | Specific Activity, IU/mg Protein (Conversion, %) with Donor: | |
|---|---|---|
| dUrd | dCyd | |
| 5-Azacytosine (5-Acyt) | 4.2 (42) | ND |
| Benzimidazole (B) | 7 (75) | 6.2 (68) |
| 5-Ethyluracil (5-Eura) | 3.6 (40) | 3.5 (40) |
| 2-Fluoroadenine (2-FAde) | 7.5 (86) | 6.2 (72) |
| 5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (TFThy) | 4.2 (42) | 3.5 (35.5) |
| 2,6-Diaminopurine (DAP) | 8.9 (89) | 7.4 (82) |
| 6-Mercaptopurine (6-M) | 5.5 (55) | 4 (40) |
| 5-Chlorouracil (5-ClUra) | 3.8 (38) | 3.5 (35) |
| 5-Fluorocytosine (5-FCyt) | ND | 5 (50) |
| 5-Fluoro-2-methoxy-4(1H)pyrimidinone (FMP) | 1.15 (18) | 0.4 (5) |
| 5-Fluorouracil (5-FUra) | 4.0 (40) | 4.0 (40) |
| 5-Bromouracil (5-BrUra) | 0.5 (3.6) | 1.0 (3.5) |
| 5-Iodouracil (5-IUra) | 4.1 (41) | 3.9 (39) |
| 7-Deaza-6-hydroxypurine (DHP) | 1.6 (15) | 1.6 (16) |
| 5-Hydroxymethyluracil (5-HMUra) | ND | 4 (42) |
| 5-Methylcytosine (5-MCyt) | 0.9 | 0.7 |
| Theophylline (Teo) | 4.5 (45) | 4.2 (42) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fresco-Taboada, A.; Fernández-Lucas, J.; Acebal, C.; Arroyo, M.; Ramón, F.; De la Mata, I.; Mancheño, J.M. 2′-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium. Catalysts 2018, 8, 8. https://doi.org/10.3390/catal8010008
Fresco-Taboada A, Fernández-Lucas J, Acebal C, Arroyo M, Ramón F, De la Mata I, Mancheño JM. 2′-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium. Catalysts. 2018; 8(1):8. https://doi.org/10.3390/catal8010008
Chicago/Turabian StyleFresco-Taboada, Alba, Jesús Fernández-Lucas, Carmen Acebal, Miguel Arroyo, Fernando Ramón, Isabel De la Mata, and José Miguel Mancheño. 2018. "2′-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium" Catalysts 8, no. 1: 8. https://doi.org/10.3390/catal8010008





