Relationships between Substrate Promiscuity and Chiral Selectivity of Esterases from Phylogenetically and Environmentally Diverse Microorganisms
Abstract
1. Introduction
2. Results and Discussion
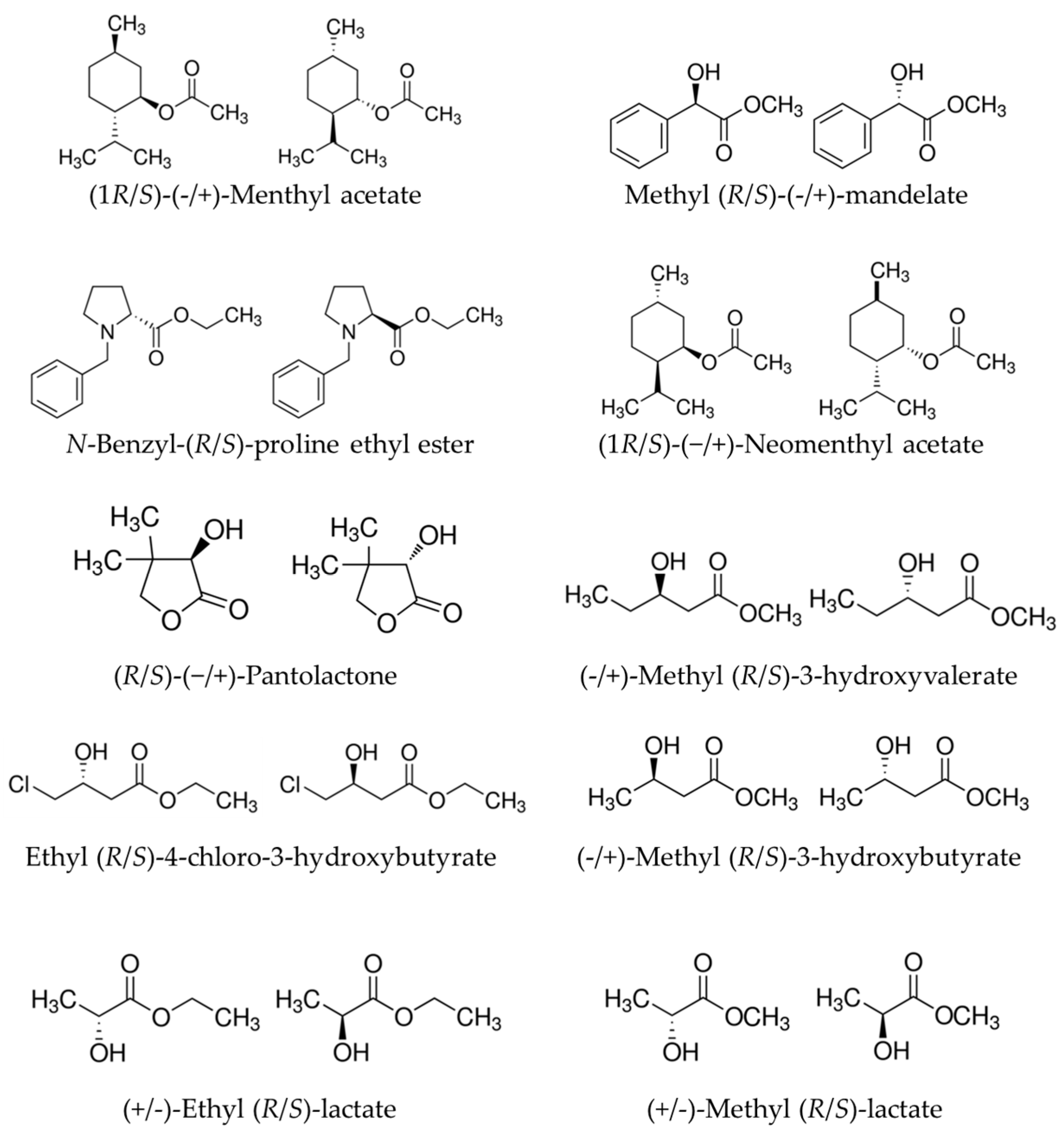
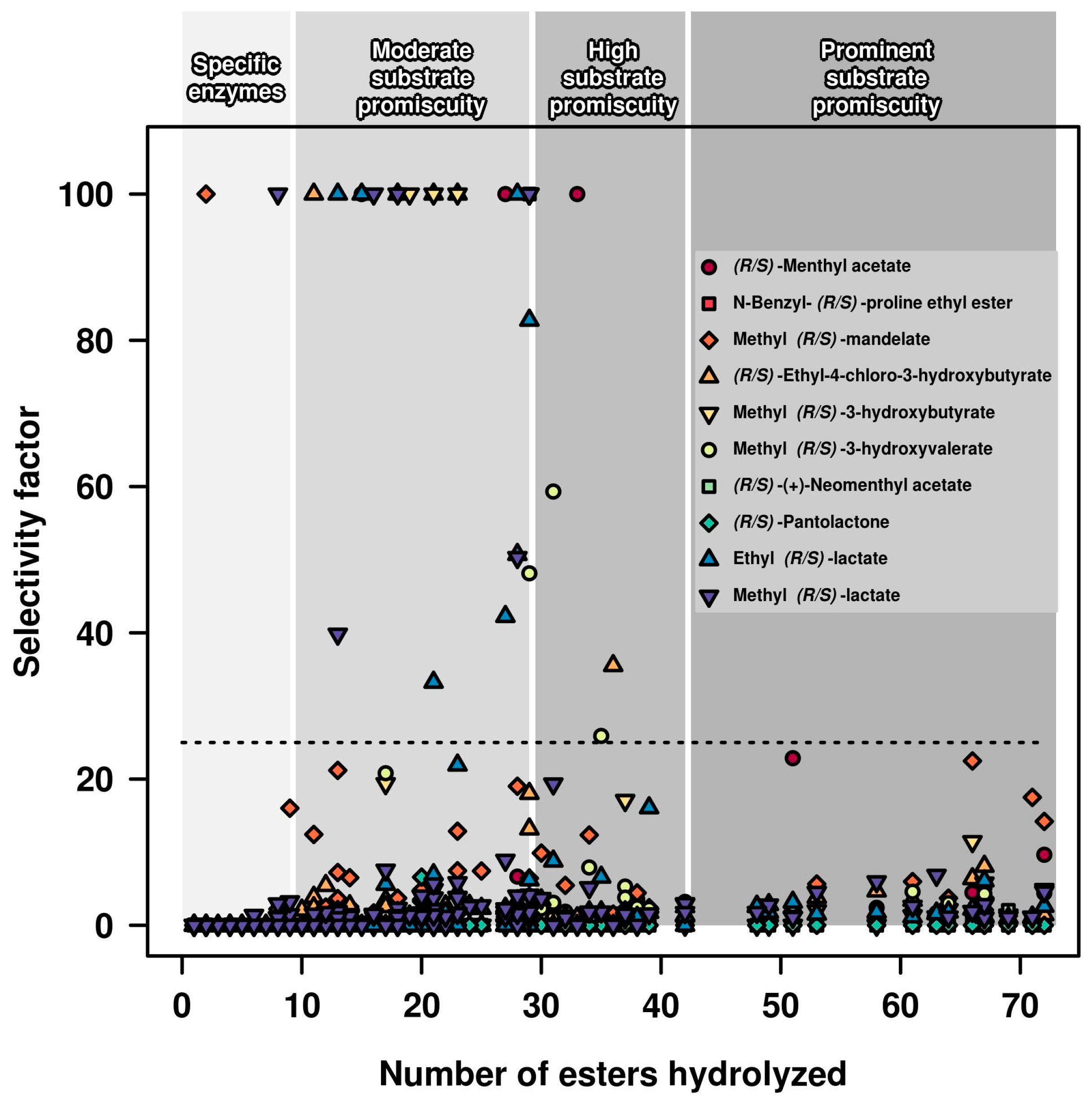
2.1. Relationships between Substrate Promiscuity and Chiral Selectivity
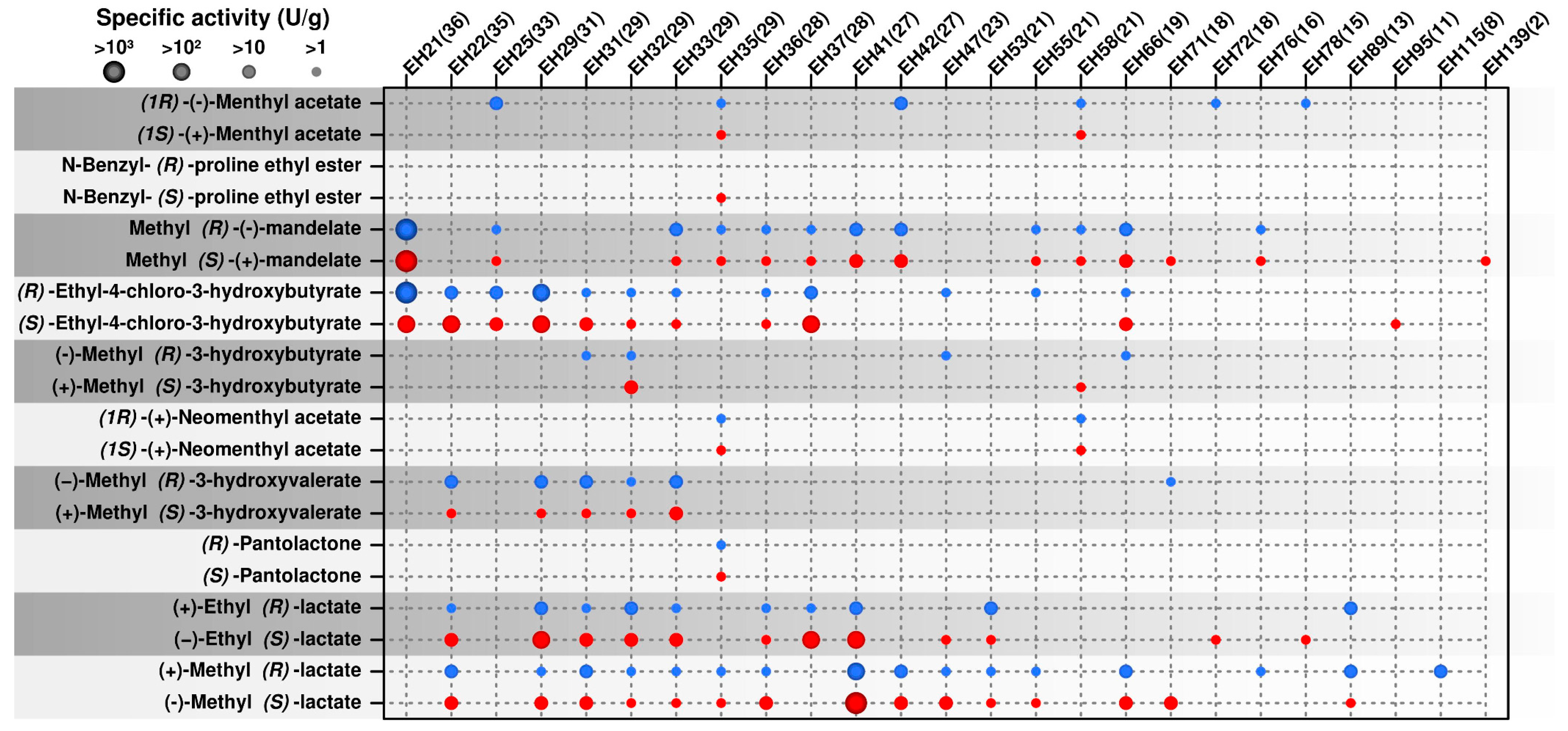
2.2. Occurrence of Multi Selective Esterases
3. Materials and Methods
3.1. Source of Chemicals, Enzymes, and Datasets
3.2. Selectivity Factor Calculation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Martínez-Martínez, M.; Bargiela, R.; Ferrer, M. Metagenomics and the search for industrial enzymes. In Biotechnology of Microbial Enzymes, 1st ed.; Brahmachari, G., Demain, A.L., Adrio, J.L., Eds.; Academic Press: Chennai, India, 2015; pp. 167–184. [Google Scholar]
- Martínez-Martínez, M.; Bargiela, R.; Coscolín, C.; Navarro-Fernández, J.; Golyshin, P.N.; Ferrer, M. Functionalization and modification of hydrocarbon-like molecules guided by metagenomics: Enzymes most requested at the industrial scale for chemical synthesis as study cases. In Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Production of Fuels and Chemicals; Lee, S.Y., Ed.; Springer International Publishing AG: Cham, Switzerland, 2016; pp. 1–26. [Google Scholar]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the success of enzyme bioprospecting through metagenomics: Current status and future trends. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, M.; Coscolín, C.; Santiago, G.; Chow, J.; Stogios, P.; Bargiela, R.; Gertler, C.; Navarro-Fernández, J.; Bollinger, A.; Thies, S.; et al. Determinants and prediction of esterase substrate promiscuity patterns. ACS Chem. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.; Dordick, J.S.; Hauer, B.; Kiener, A.; Wubbolts, M.; Witholt, B. Industrial biocatalysis today and tomorrow. Nature 2001, 409, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Bargiela, R.; Martínez-Martínez, M.; Mir, J.; Koch, R.; Golyshina, O.V.; Golyshin, P.N. Biodiversity for Biocatalysis: A review for α/β-hydrolases of the esterase-lipase superfamily as case of Study. Biocatal. Biotransform. 2015, 33, 235–249. [Google Scholar] [CrossRef]
- Aranda, J.; Cerqueira, N.M.; Fernandes, P.A.; Roca, M.; Tuñon, I.; Ramos, M.J. The Catalytic Mechanism of Carboxylesterases: A Computational Study. Biochemistry 2014, 53, 5820–5829. [Google Scholar] [PubMed]
- Elend, C.; Schmeisser, C.; Hoebenreich, H.; Steele, H.L.; Streit, W.R. Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J. Biotechnol. 2007, 130, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.; Kovacic, F.; Dall Antonia, Y.; Krauss, U.; Fersini, F.; Schmeisser, C.; Lauinger, B.; Bongen, P.; Pietruszka, J.; Schmidt, M.; et al. The Metagenome-derived enzymes LipS and LipT increase the diversity of known lipases. PLoS ONE 2012, 7, e47665. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Choi, G.S.; Kim, S.B.; Yoon, G.S.; Kim, Y.S.; Ryu, Y.W. Screening and characterization of a novel esterase from a metagenomic library. Protein Expr. Purif. 2006, 45, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Kim, S.; Ryu, Y.; Kim, T.D. Identification and characterization of a novel (S)-ketoprofen-specific esterase. Int. J. Biol. Macromol. 2007, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.D.; Ryu, B.H.; Ju, H.; Jang, E.J.; Kim, K.K.; Kim, T.D. Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/beta-lactamase homologue from a metagenomic library. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2455–2466. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Seok, S.H.; Hong, E.; Yoo, T.H.; Seo, M.D.; Ryu, Y. Crystal structure and characterization of esterase Est25 mutants reveal improved enantioselectivity toward (S)-ketoprofen ethyl ester. Appl. Microbiol. Biotechnol. 2017, 101, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Golyshina, O.V.; Chernikova, T.N.; Khachane, A.N.; Martins dos Santos, V.A.P.; Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 2005, 12, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Kourist, R.; Hari Krishna, S.; Patel, J.S.; Bartnek, F.; Hitchman, T.S.; Weiner, D.P.; Bornscheuer, U.T. Identification of a metagenome-derived esterase with high enantioselectivity in the kinetic resolution of arylaliphatic tertiary alcohols. Org. Biomol. Chem. 2007, 5, 3310–3313. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Álvaro, E.; Kourist, R.; Winter, J.; Böttcher, D.; Liebeton, K.; Naumer, C.; Eck, J.; Leggewie, C.; Jaeger, K.E.; Streit, W.; et al. Enantioselective kinetic resolution of phenylalkyl carboxylic acids using metagenome-derived esterases. Microb. Biotechnol. 2010, 3, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.M.; Liu, J.Y.; Qiao, M.; Xu, J.H. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Biochem. Biotechnol. 2013, 169, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Martini, V.; Glogauer, A.; Muller-Santos, M.; Lulek, J.; de Souza, E.; Mitchell, D.; Pedrosa, F.; Krieger, N. First co-expression of a lipase and its specific foldase obtained by metagenomics. Microb. Cell Factories 2014, 13, 171. [Google Scholar] [CrossRef] [PubMed]
- Alnoch, R.C.; Martini, V.P.; Glogauer, A.; Costa, A.C.; Piovan, L.; Muller-Santos, M.; de Souza, E.M.; de Oliveira Pedrosa, F.; Mitchell, D.A.; Krieger, N. Immobilization and characterization of a new regioselective and enantioselective lipase obtained from a metagenomic library. PLoS ONE 2015, 10, e0114945. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Kim, J.T.; Kang, S.G.; Lee, J.H.; Kim, S.J. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar. Biotechnol. 2009, 11, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Elend, C.; Schmeisser, C.; Leggewie, C.; Babiak, P.; Carballeira, J.D.; Steele, H.L.; Reymond, J.L.; Jaeger, K.E.; Streit, W.R. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Environ. Microbiol. 2006, 72, 3637–3645. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, M.; Alcaide, M.; Tchigvintsev, A.; Reva, O.; Polaina, J.; Bargiela, R.; Guazzaroni, M.-E.; Chicote, Á.; Canet, A.; Valero, F.; et al. Biochemical diversity of carboxyl esterases and lipases from Lake Arreo (Spain): A metagenomic approach. Appl. Environ. Microbiol. 2013, 79, 3553–3562. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Tchigvintsev, A.; Martinez-Martinez, M.; Popovic, A.; Reva, O.N.; Lafraya, A.; Bargiela, R.; Nechitaylo, T.Y.; Matesanz, R.; Cambon-Bonavita, M.A.; et al. Identification and characterization of carboxyl esterases of gill chamber-associated microbiota in the deep-sea shrimp Rimicaris exoculata by using functional metagenomics. Appl. Environ. Microbiol. 2015, 81, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Placido, A.; Hai, T.; Ferrer, M.; Chernikova, T.N.; Distaso, M.; Armstrong, D.; Yakunin, A.F.; Toshchakov, S.V.; Yakimov, M.M.; Kublanov, I.V.; et al. Diversity of hydrolases from hydrothermal vent sediments of the Levante Bay.; Vulcano Island (Aeolian archipelago) identified by activity-based metagenomics and biochemical characterization of new esterases and an arabinopyranosidase. Appl. Microbiol. Biotechnol. 2015, 99, 10031–10046. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Jung, W.K.; Kim, Y.H.; Ryu, B.H.; Kim, T.D.; Kim, J.; Kim, H. Characterization of a novel alkaline family VIII esterase with S-enantiomer preference from a compost metagenomic library. J. Microbiol. Biotechnol. 2016, 26, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Fan, H.; Chen, L.; Wang, H.; Wei, D. Efficient kinetic resolution of secondary alcohols using an organic solvent-tolerant esterase in non-aqueous medium. Biotechnol. Lett. 2016, 38, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Banoth, L.; Banerjee, U.C.; Kaur, J. Enantiomeric separation of pharmaceutically important drug intermediates using a metagenomic lipase and optimization of its large scale production. Int. J. Biol. Macromol. 2017, 95, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, D.; Zägel, P.; Schmidt, M.; Bornscheuer, U.T. A microtiter plate-based assay to screen for active and stereoselective hydrolytic enzymes in enzyme libraries. Methods Mol. Biol. 2017, 1539, 197–204. [Google Scholar] [PubMed]
- Gawley, R.E. Do the terms “% ee” and “% de” make sense as expressions of stereoisomer composition or stereoselectivity? J. Org. Chem. 2006, 71, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. (Ed.) Introduction to directed evolution. In Directed Evolution of Selective Enzymes: Catalysts for Organic Chemistry and Biotechnology; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; pp. 1–16. [Google Scholar]
- Romano, D.; Bonomi, F.; de Mattos, M.C.; de Sousa Fonseca, T.; de Oliveira Mda, C.; Molinari, F. Esterases as stereoselective biocatalysts. Biotechnol. Adv. 2015, 33, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Wikmark, Y.; Svedendahl Humble, M.; Bäckvall, J.E. Combinatorial library based engineering of Candida antarctica lipase A for enantioselective transacylation of sec-alcohols in organic solvent. Angew. Chem. Int. Ed. Engl. 2015, 54, 4284–4288. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coscolín, C.; Martínez-Martínez, M.; Chow, J.; Bargiela, R.; García-Moyano, A.; Bjerga, G.E.K.; Bollinger, A.; Stokke, R.; Steen, I.H.; Golyshina, O.V.; et al. Relationships between Substrate Promiscuity and Chiral Selectivity of Esterases from Phylogenetically and Environmentally Diverse Microorganisms. Catalysts 2018, 8, 10. https://doi.org/10.3390/catal8010010
Coscolín C, Martínez-Martínez M, Chow J, Bargiela R, García-Moyano A, Bjerga GEK, Bollinger A, Stokke R, Steen IH, Golyshina OV, et al. Relationships between Substrate Promiscuity and Chiral Selectivity of Esterases from Phylogenetically and Environmentally Diverse Microorganisms. Catalysts. 2018; 8(1):10. https://doi.org/10.3390/catal8010010
Chicago/Turabian StyleCoscolín, Cristina, Mónica Martínez-Martínez, Jennifer Chow, Rafael Bargiela, Antonio García-Moyano, Gro E. K. Bjerga, Alexander Bollinger, Runar Stokke, Ida H. Steen, Olga V. Golyshina, and et al. 2018. "Relationships between Substrate Promiscuity and Chiral Selectivity of Esterases from Phylogenetically and Environmentally Diverse Microorganisms" Catalysts 8, no. 1: 10. https://doi.org/10.3390/catal8010010
APA StyleCoscolín, C., Martínez-Martínez, M., Chow, J., Bargiela, R., García-Moyano, A., Bjerga, G. E. K., Bollinger, A., Stokke, R., Steen, I. H., Golyshina, O. V., Yakimov, M. M., Jaeger, K.-E., Yakunin, A. F., Streit, W. R., Golyshin, P. N., & Ferrer, M. (2018). Relationships between Substrate Promiscuity and Chiral Selectivity of Esterases from Phylogenetically and Environmentally Diverse Microorganisms. Catalysts, 8(1), 10. https://doi.org/10.3390/catal8010010








