Catalytic Characteristics of New Antibacterials Based on Hexahistidine-Containing Organophosphorus Hydrolase
Abstract
1. Introduction
2. Results
2.1. Studies of Catalytic Properties of His6-OPH and Its Polyelectrolyte Complexes in the Presence of Antibiotics
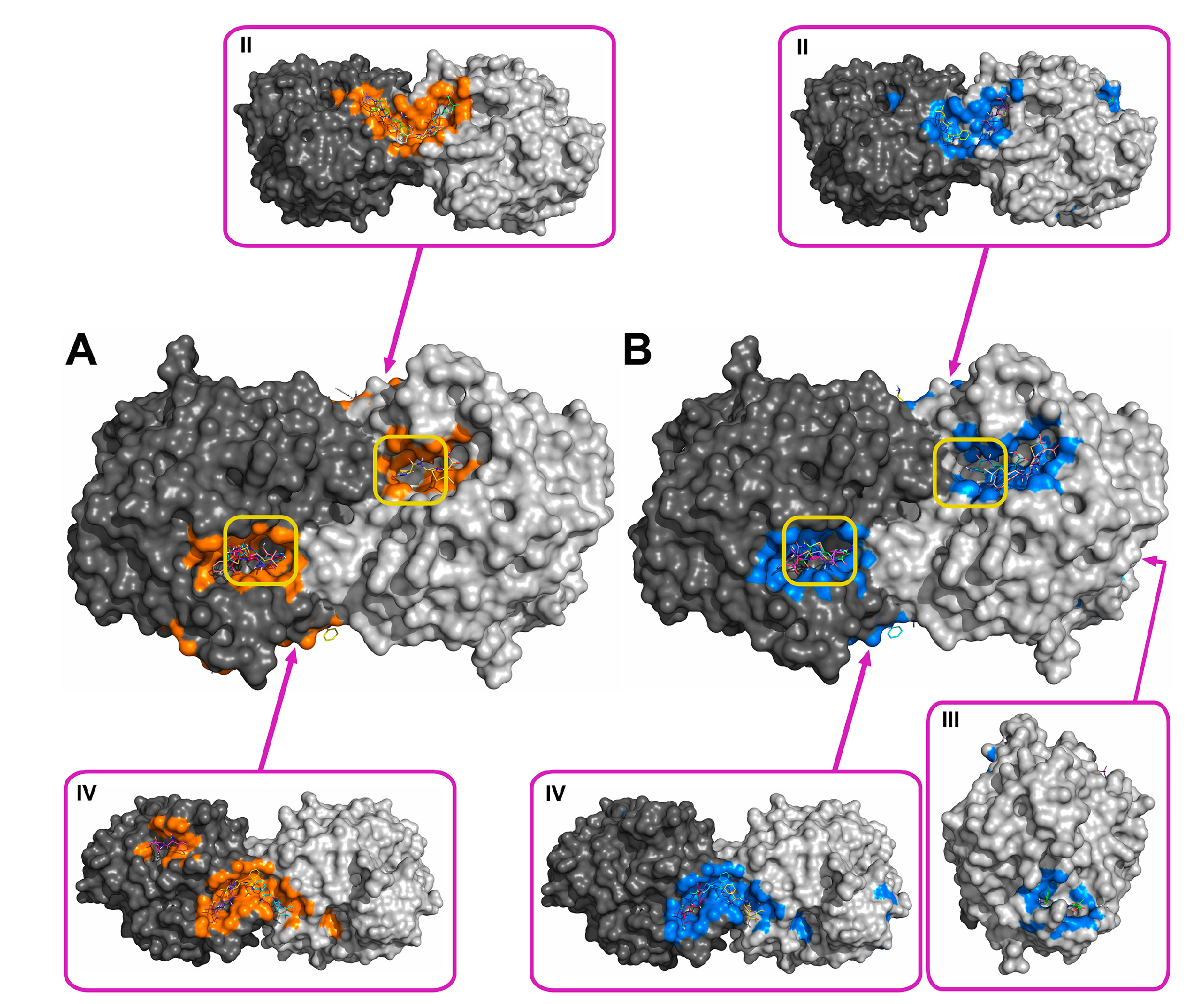
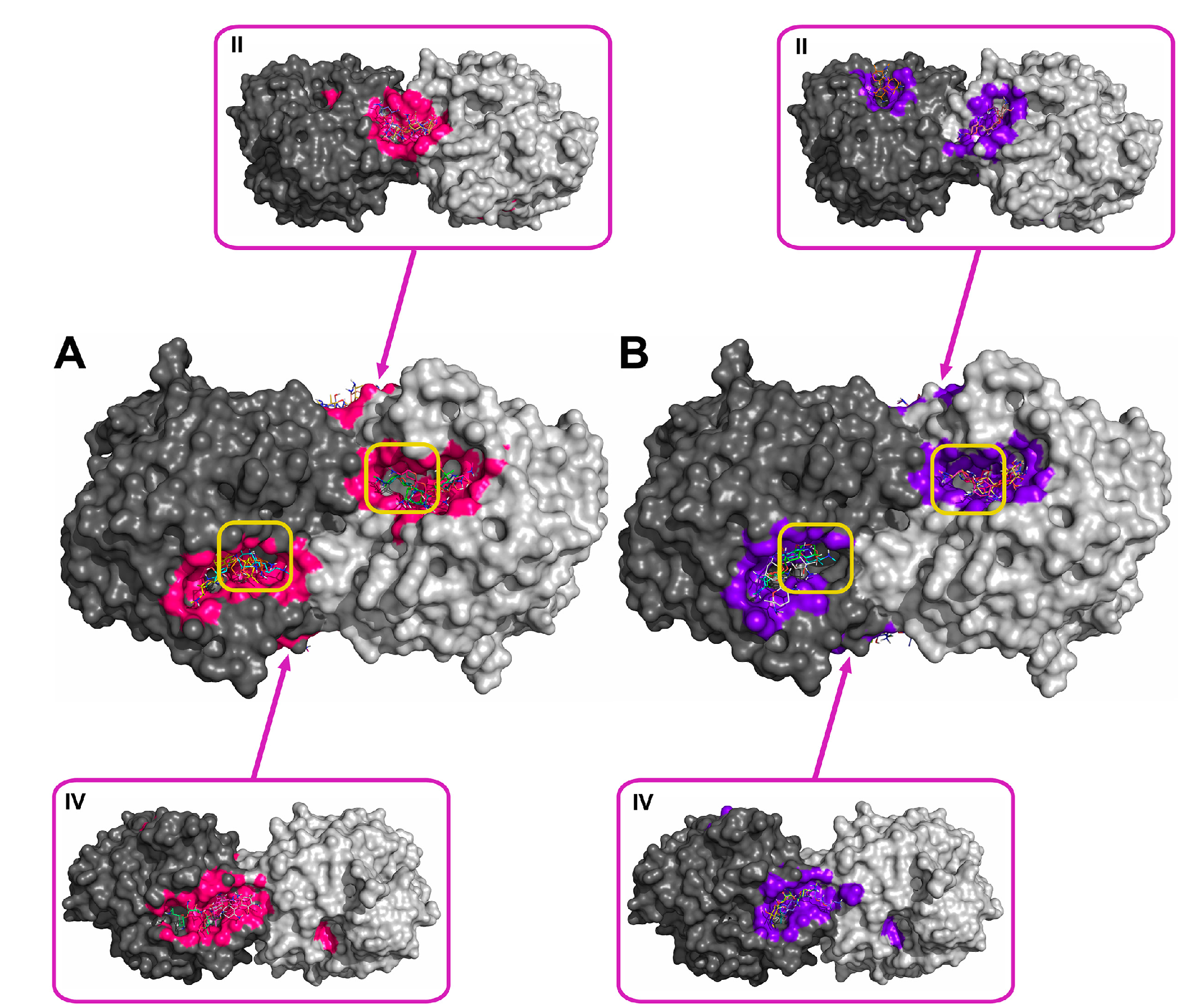
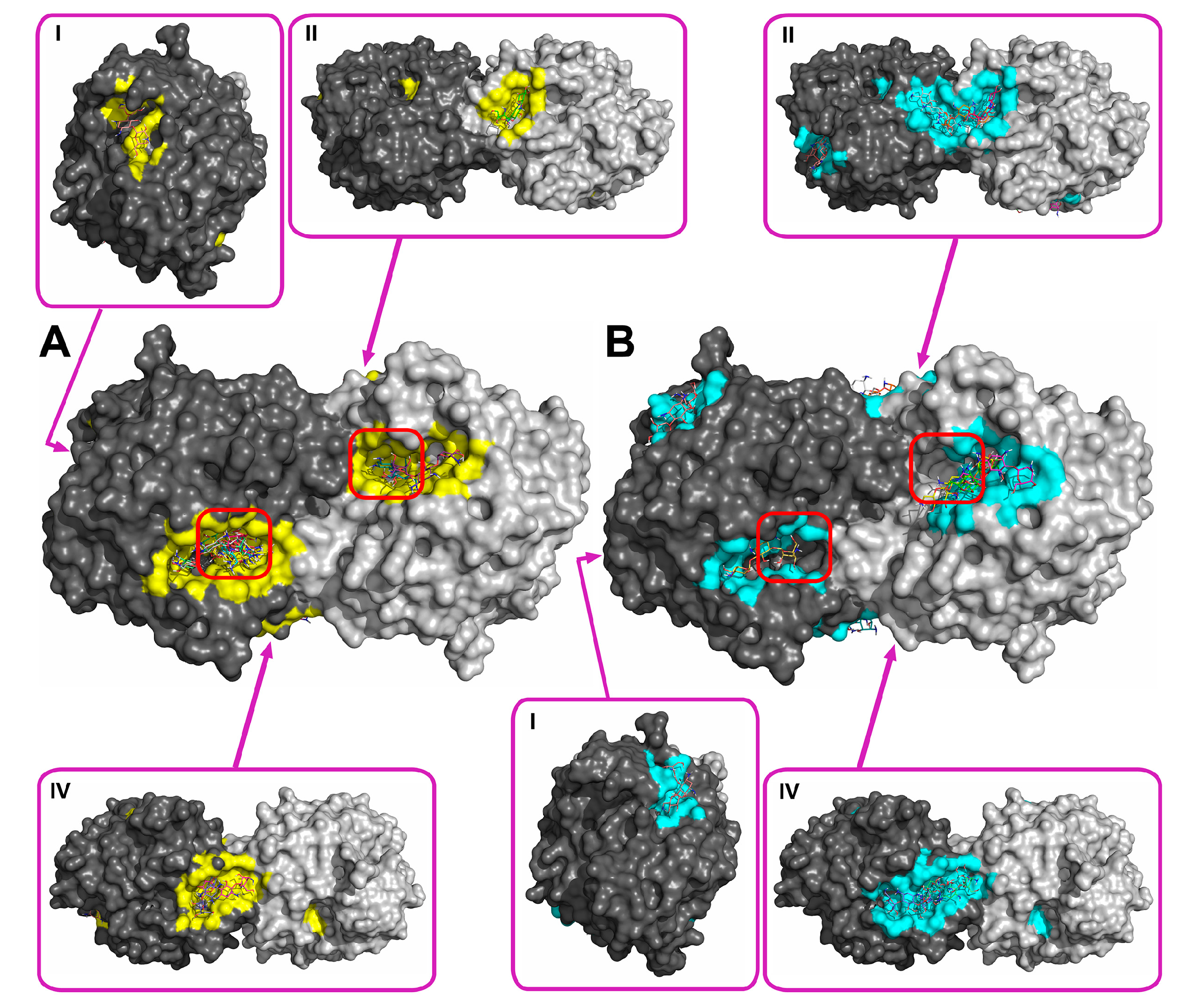
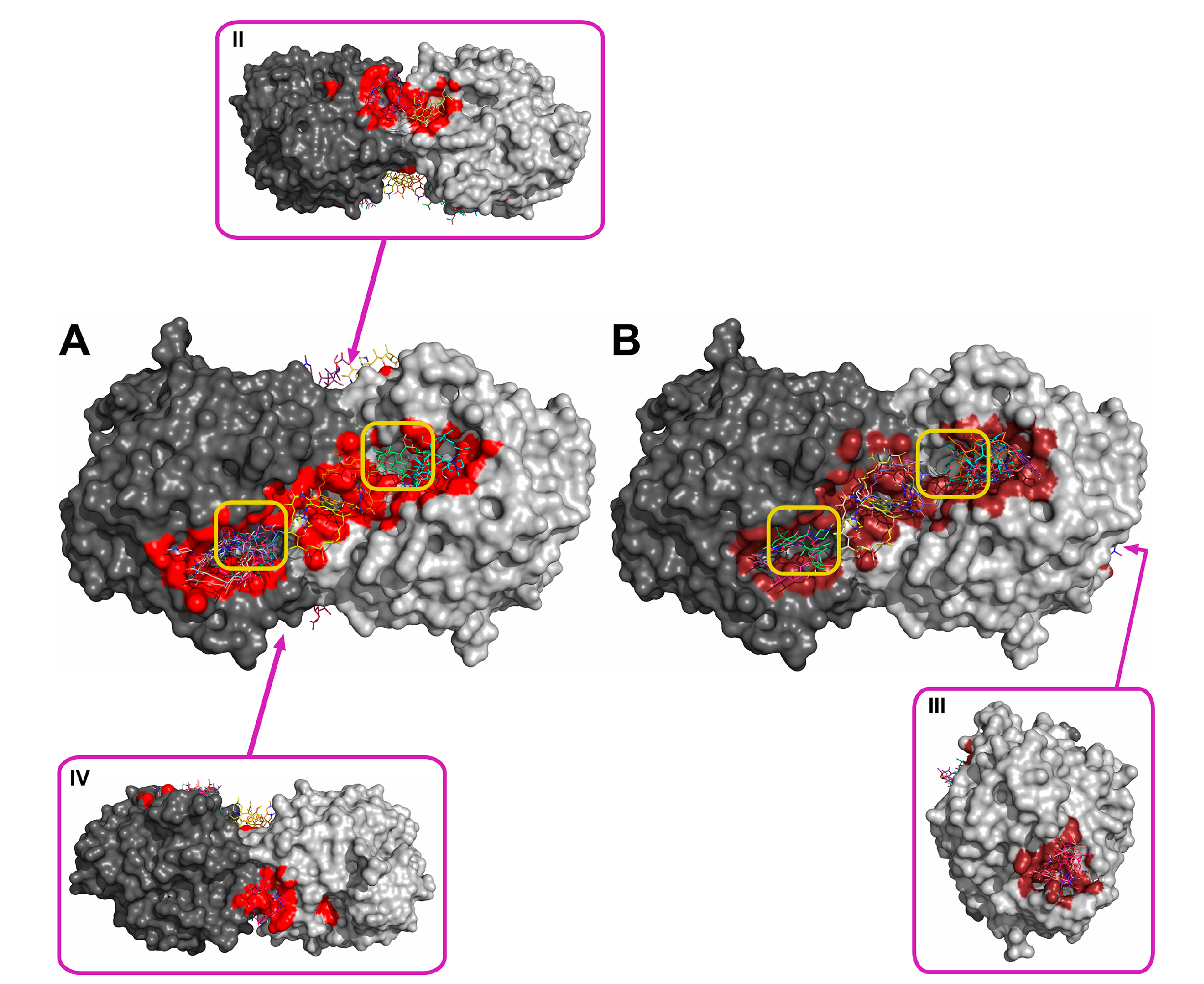
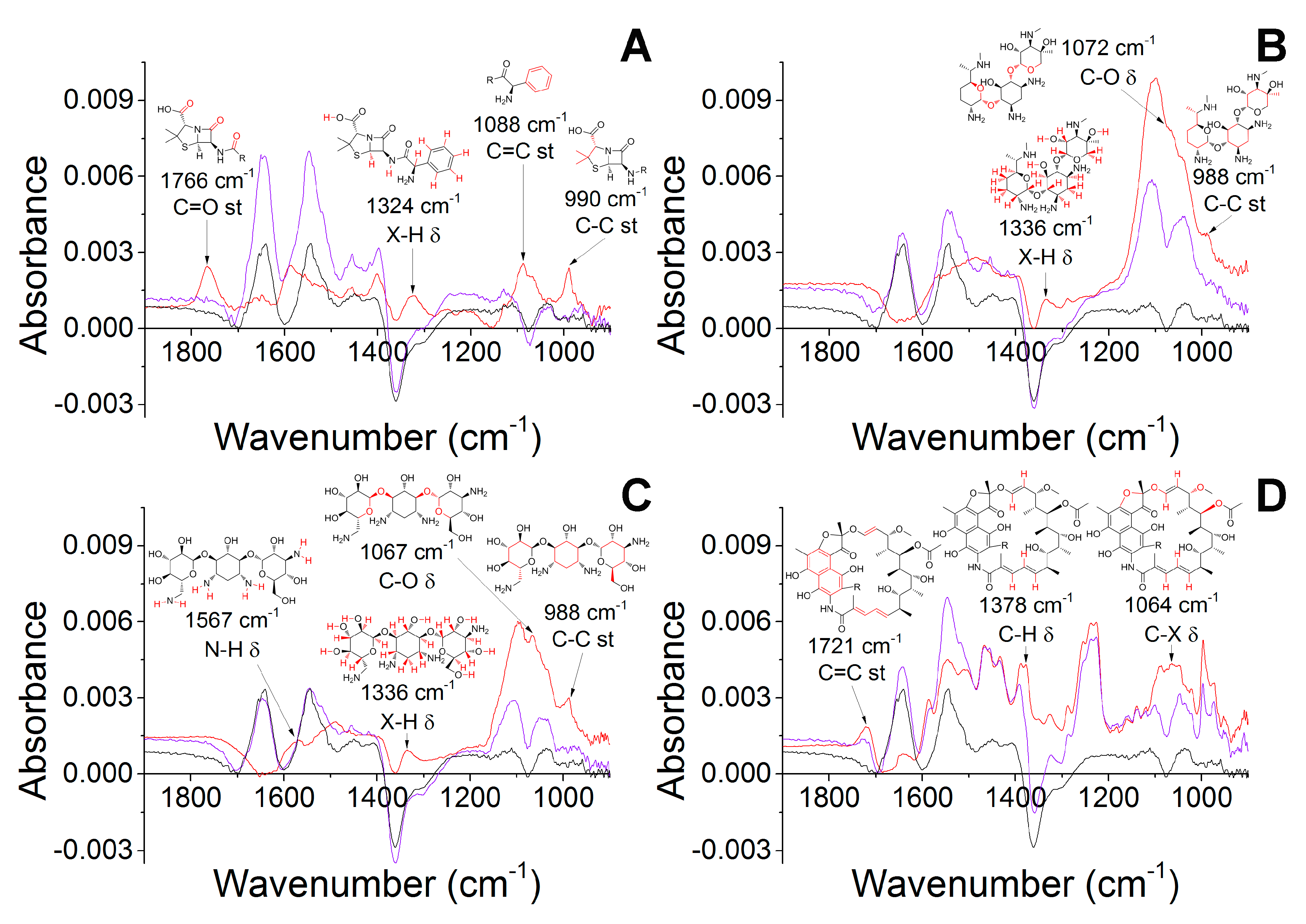
2.2. Molecular Docking of Antibiotics to the His6-OPH Dimer and Fourier-Transform Infrared (FTIR) Spectroscopy
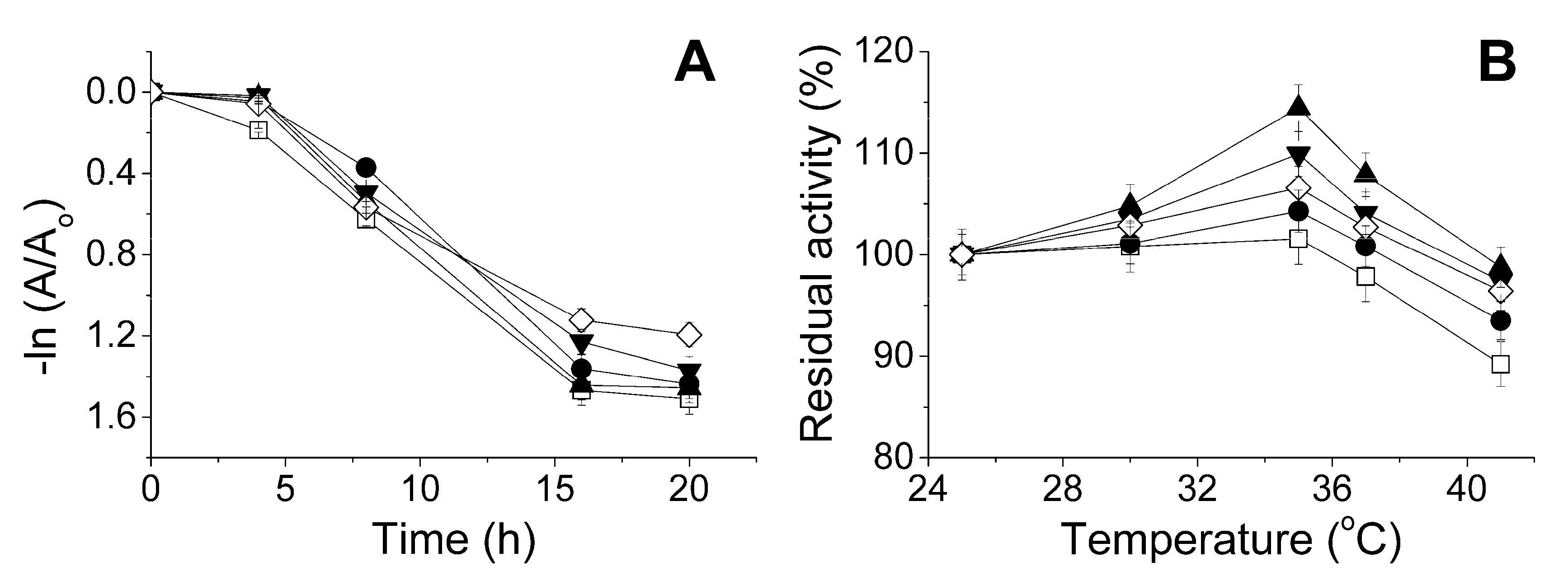
2.3. The Influence of the Physico-Chemical Factors on the Lactonase Activity of His6-OPH in the Presence of Antibiotics
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Enzyme Samples
4.3. Preparation of Enzyme-Polyelectrolyte Complexes
4.4. Measurement of Enzyme Activity
4.5. FTIR Spectroscopy
4.6. Computer Simulations
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Wyk, H. Antibiotic resistance. S. Afr. Pharm. J. 2015, 82, 20–23. [Google Scholar]
- Worthington, R.J.; Melander, C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013, 31, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V. Quorum sensing in biofilms—How to destroy the bacterial citadels or their cohesion/power? Anaerobe 2011, 17, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Algburi, A.; Comito, N.; Kashtanov, D.; Dicks, L.M.; Chikindas, M.L. Control of biofilm formation: Antibiotics and beyond. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Hong, K.W.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Quorum quenching revisited—From signal decays to signalling confusion. Sensors 2012, 12, 4661–4696. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Wang, C.; Mulchandani, A.; Ge, X. Engineering soluble human paraoxonase 2 for quorum quenching. ACS Chem. Biol. 2016, 11, 3122–3131. [Google Scholar] [CrossRef] [PubMed]
- Teplitski, M.; Mathesius, U.; Rumbaugh, K.P. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem. Rev. 2010, 111, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Kiran, S.; Sharma, P.; Harjai, K.; Capalash, N. Enzymatic quorum quenching increases antibiotic susceptibility of multidrug resistant Pseudomonas aeruginosa. Iran. J. Microbiol. 2011, 3, 1–12. [Google Scholar] [PubMed]
- Amara, N.; Krom, B.P.; Kaufmann, G.F.; Meijler, M.M. Macromolecular inhibition of quorum sensing: Enzymes, antibodies, and beyond. Chem. Rev. 2010, 111, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Raju, S.C.; Purohit, H.J. Genomic analysis reveals versatile organisms for quorum quenching enzymes: Acyl-homoserine lactone-acylase and -lactonase. Open Microbiol. J. 2011, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hiblot, J.; Bzdrenga, J.; Champion, C.; Chabriere, E.; Elias, M. Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia. Sci. Rep. 2015, 5, 8372. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, R.; Thomas, P.W.; Wu, C.X.; Nocek, B.P.; Hoang, Q.Q.; Liu, D.; Fast, W. Structural and biochemical characterization of AidC, a quorum-quenching lactonase with atypical selectivity. Biochemistry 2015, 54, 4342–4353. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Oger, P.M.; Chapelle, E.; Adeline, M.T.; Faure, D.; Dessaux, Y. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 2008, 74, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Weng, L.X.; Dong, Y.H.; Zhang, L.H. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 2004, 279, 13645–13651. [Google Scholar] [CrossRef] [PubMed]
- Sirotkina, M.; Efremenko, E.N. Rhodococcus lactonase with organophosphate hydrolase (OPH) activity and His6-tagged OPH with lactonase activity: evolutionary proximity of the enzymes and new possibilities in their application. Appl. Microbiol. Biotechnol. 2014, 98, 2647–2656. [Google Scholar] [CrossRef] [PubMed]
- Votchitseva, Y.A.; Efremenko, E.N.; Aliev, T.K.; Varfolomeyev, S.D. Properties of hexahistidine-tagged organophosphate hydrolase. Biochemistry (Moscow) 2006, 71, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Lyagin, I.V.; Andrianova, M.S.; Efremenko, E.N. Extensive hydrolysis of phosphonates as unexpected behaviour of the known His6-organophosphorus hydrolase. Appl. Microbiol. Biotechnol. 2016, 100, 5829–5838. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, E.N.; Lyagin, I.V.; Klyachko, N.L.; Bronich, T.; Zavyalova, N.V.; Jiang, Y.; Kabanov, A.V. A simple and highly effective catalytic nanozyme scavenger for organophosphorus neurotoxins. J. Control. Release 2017, 247, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Pretsch, E.; Bühlman, P.; Badertscher, M. Structure Determination of Organic Compounds: Tables of Spectral Data, 4th ed.; Springer: Berlin, Germany, 2009; pp. 269–335. ISBN 978-3-540-93809-5. [Google Scholar]
- Hooper, D.C. Mechanisms of action of antimicrobials: Focus on fluoroquinolones. Clin. Infect. Dis. 2001, 32, S9–S15. [Google Scholar] [CrossRef] [PubMed]
- Floss, H.G.; Yu, T.W. Rifamycin mode of action, resistance, and biosynthesis. Chem. Rev. 2005, 105, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef] [PubMed]
- Van Bambeke, F.; Barcia-Macay, M.; Lemaire, S.; Tulkens, P.M. Cellular pharmacodynamics and pharmacokinetics of antibiotics: Current views and perspectives. Curr. Opin. Drug Discov. Dev. 2006, 9, 218–230. [Google Scholar]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, V.S.; Efremenko, E.N.; Kazankov, G.M.; Varfolomeyev, S.D. Double effect of organic amines (activation and inhibition) on the phosphotriesterase. J. Mol. Catal. B Enzym. 2000, 10, 571–576. [Google Scholar] [CrossRef]
- Chaskar, P.; Zoete, V.; Röhrig, U.F. Toward on-the-fly quantum mechanical/molecular mechanical (QM/MM) docking: Development and benchmark of a scoring function. J. Chem. Inf. Model. 2014, 54, 3137–3152. [Google Scholar] [CrossRef] [PubMed]
- Nowosielski, M.; Hoffmann, M.; Kuron, A.; Korycka-Machala, M.; Dziadek, J. The MM2QM tool for combining docking, molecular dynamics, molecular mechanics, and quantum mechanics. J. Comput. Chem. 2013, 34, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Samra, Z.Q.; Shabir, S.; Rehmat, Z.; Zaman, M.; Nazir, A.; Dar, N.; Athar, M.A. Synthesis of cholesterol-conjugated magnetic nanoparticles for purification of human paraoxonase 1. Appl. Biochem. Biotechnol. 2010, 162, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, E.N.; Votchitseva, Y.A.; Aliev, T.K.; Varfolomeev, S.D. Recombinant Plasmid DNA pTES-HIS-OPH and Producer of Oligohistidine-Containing Organophosphate Hydrolase. RU Patent 2255975 C 1, 10 July 2005. [Google Scholar]
- Efremenko, E.; Votchitseva, Y.; Plieva, F.; Galaev, I.; Mattiasson, B. Purification of His6-organophosphate hydrolase using monolithic supermacroporous polyacrylamide cryogels developed for immobilized metal affinity chromatography. Appl. Microbiol. Biotechnol. 2006, 70, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Freeman, L.; Buisson, M.; Tarbouriech, N.; Van der Heyden, A.; Labbé, P.; Burmeister, W.P. The flexible motif V of Epstein-Barr virus deoxyuridine 5′-triphosphate pyrophosphatase is essential for catalysis. J. Biol. Chem. 2009, 284, 25280–25289. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, E.N.; Lyagin, I.V.; Le, H.C.; Le, M.H. Antioxidants as stabilizers for His6-OPH: Is this an unusual or regular role for them with enzymes? J. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Sadovnichy, V.; Tikhonravov, A.; Voevodin, V.; Opanasenko, V. “Lomonosov”: Supercomputing at Moscow State University. In Contemporary High Performance Computing: From Petascale toward Exascale; Vetter, J.S., Ed.; Chapman & Hall/CRC Computational Science Series; CRC Press: Boca Raton, FL, USA, 2013; pp. 283–307. ISBN 9781466568341. [Google Scholar]
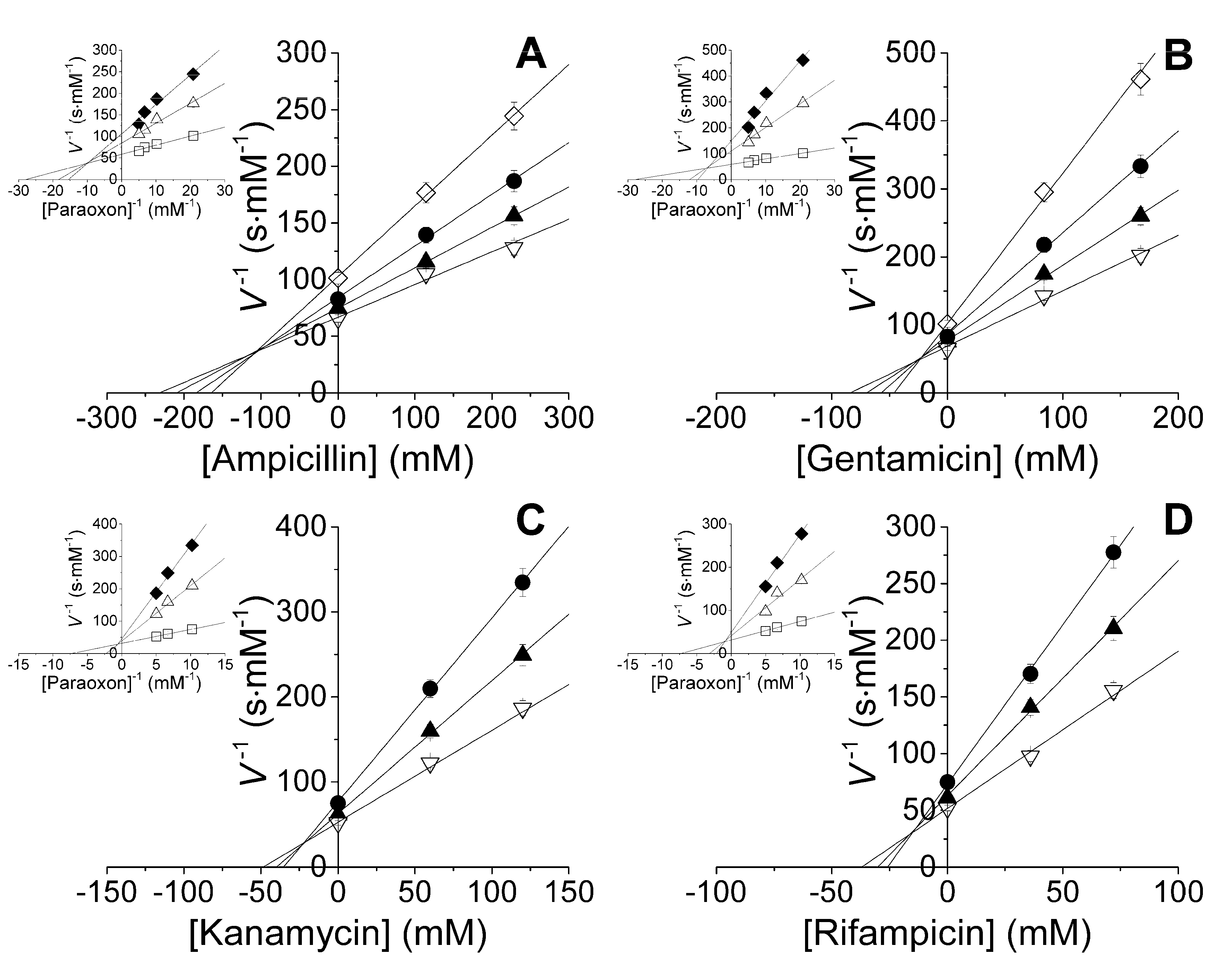

| Inhibitor | Enzyme or EPC | Kic (mM) | Kiu (mM) | Km (µM) | Vmax·E0−1 (s−1) | Vmax·E0−1·Km−1 (106 s−1·M−1) |
|---|---|---|---|---|---|---|
| – | His6-OPH | – | – | 10.5 ± 2.2 | 5040 ± 340 | 480 ± 133 |
| His6-OPH/PLD50 | – | – | 12.1 ± 1.0 | 5240 ± 140 | 433 ± 47 | |
| His6-OPH/PLE50 | – | – | 15.0 ± 2.1 | 4880 ± 240 | 325 ± 62 | |
| Ampicillin | His6-OPH | 229 ± 15 | 72 ± 15 | 18.2 ± 3.8 | 3360 ± 230 | 185 ± 51 |
| His6-OPH/PLD50 | 515 ± 14 | 241 ± 20 | 14.6 ± 1.2 | 4450 ± 120 | 305 ± 33 | |
| His6-OPH/PLE50 | 146 ± 7 | 48 ± 7 | 28.5 ± 4.0 | 2760 ± 140 | 97 ± 19 | |
| Gentamicin | His6-OPH | 264 ± 18 | 61 ± 13 | 18.9 ± 4.0 | 2230 ± 150 | 118 ± 33 |
| His6-OPH/PLD50 | 377 ± 10 | 109 ± 9 | 17.5 ± 1.4 | 4390 ± 120 | 251 ± 27 | |
| His6-OPH/PLE50 | 162 ± 8 | 41 ± 6 | 30.2 ± 4.2 | 3250 ± 160 | 108 ± 20 | |
| Kanamycin | His6-OPH | 72 ± 5 | 62 ± 13 | 11.3 ± 2.4 | 1340 ± 90 | 119 ± 33 |
| His6-OPH/PLD50 | 142 ± 4 | 23 ± 2 | 30.6 ± 2.5 | 4700 ± 130 | 154 ± 17 | |
| His6-OPH/PLE50 | 44 ± 2 | 26 ± 4 | 21.1 ± 3.0 | 1340 ± 70 | 64 ± 12 | |
| Rifampicin | His6-OPH | 29 ± 2 | 26 ± 5 | 11.2 ± 2.3 | 1750 ± 120 | 156 ± 43 |
| His6-OPH/PLD50 | 79 ± 2 | 10 ± 1 | 39.1 ± 3.2 | 3580 ± 100 | 92 ± 10 | |
| His6-OPH/PLE50 | 32 ± 2 | 15 ± 2 | 23.7 ± 3.3 | 3250 ± 160 | 137 ± 26 |
| Antibiotic | pH | Affinity (kJ·mol−1) | Area (%) | |||
|---|---|---|---|---|---|---|
| Mean | Median | Upper (Lower) Bounds | Near Active Sites | Total | ||
| Ampicillin | 7.5 | −28.61 | −28.68 ± 1.47 | −26.36 (−30.54) | 5.2 | 11.8 |
| 10.5 | −27.29 | −27.00 ± 1.25 | −25.94 (−29.29) | 6.0 | 12.2 | |
| Gentamicin | 7.5 | −27.32 | −27.21 ± 0.48 | −26.78 (−28.03) | 6.8 | 11.5 |
| 10.5 | −25.85 | −25.75 ± 1.16 | −23.85 (−27.62) | 6.8 | 12.0 | |
| Kanamycin | 7.5 | −26.83 | −26.59 ± 1.93 | −23.85 (−29.71) | 7.0 | 11.8 |
| 10.5 | −25.36 | −24.91 ± 1.71 | −23.85 (−29.71) | 6.1 | 13.8 | |
| Rifampicin | 7.5 | −29.90 | −29.52 ± 1.63 | −28.03 (−33.05) | 8.7 | 12.6 |
| 10.5 | −30.11 | −29.73 ± 1.44 | −28.45 (−33.05) | 9.0 | 11.1 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maslova, O.; Aslanli, A.; Stepanov, N.; Lyagin, I.; Efremenko, E. Catalytic Characteristics of New Antibacterials Based on Hexahistidine-Containing Organophosphorus Hydrolase. Catalysts 2017, 7, 271. https://doi.org/10.3390/catal7090271
Maslova O, Aslanli A, Stepanov N, Lyagin I, Efremenko E. Catalytic Characteristics of New Antibacterials Based on Hexahistidine-Containing Organophosphorus Hydrolase. Catalysts. 2017; 7(9):271. https://doi.org/10.3390/catal7090271
Chicago/Turabian StyleMaslova, Olga, Aysel Aslanli, Nikolay Stepanov, Ilya Lyagin, and Elena Efremenko. 2017. "Catalytic Characteristics of New Antibacterials Based on Hexahistidine-Containing Organophosphorus Hydrolase" Catalysts 7, no. 9: 271. https://doi.org/10.3390/catal7090271
APA StyleMaslova, O., Aslanli, A., Stepanov, N., Lyagin, I., & Efremenko, E. (2017). Catalytic Characteristics of New Antibacterials Based on Hexahistidine-Containing Organophosphorus Hydrolase. Catalysts, 7(9), 271. https://doi.org/10.3390/catal7090271





