Microbial Spore-Based Biocatalysts: Properties, Applications and New Trends
Abstract
1. Introduction
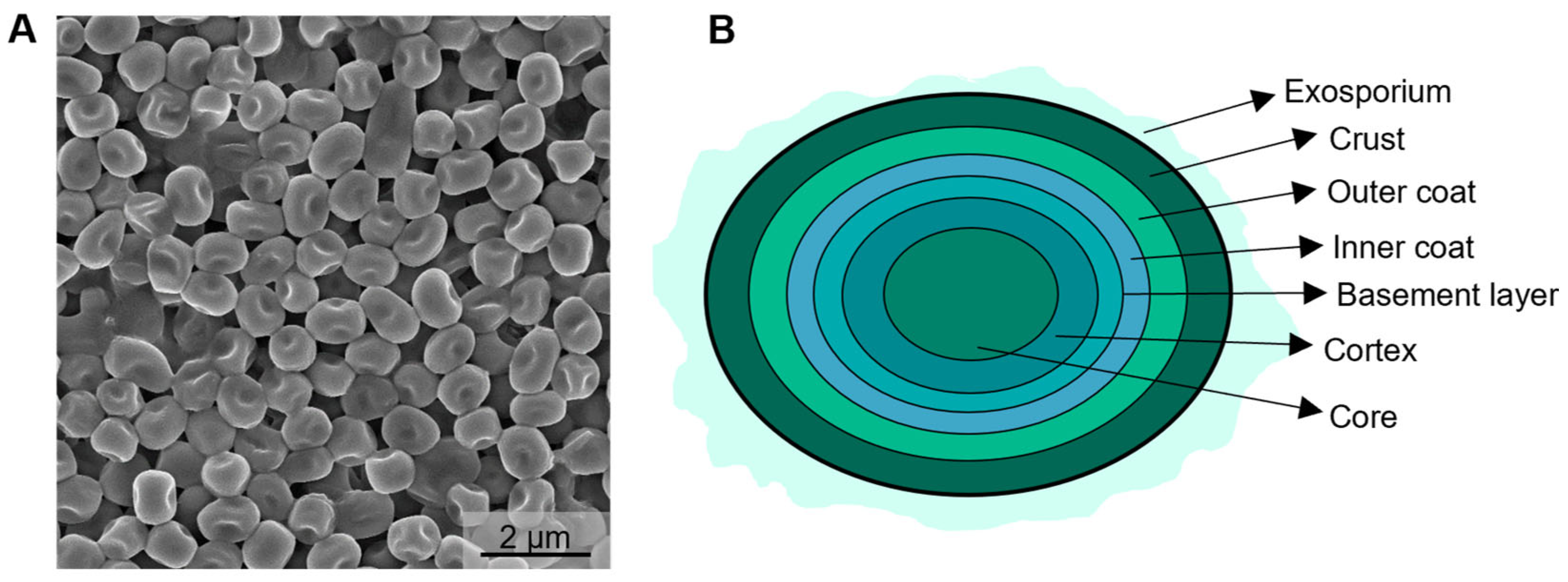
2. Naturally Occurring Biocatalytically Active Spores
3. Spore Immobilization/Display Systems for Biotechnologically Relevant Enzymes
3.1. Adsorption of Enzymes onto Spores
3.2. Covalent Immobilization of Enzymes onto Spores
3.3. Genetic Display Systems for Immobilization of Enzymes onto Spores
4. Biocatalyst Stability Improvements Through Immobilization onto Spores
| Enzyme | Source Organism | Temperature | pH | Organic Solvents | Protease | Reusability | References |
|---|---|---|---|---|---|---|---|
| Alcohol dehydrogenase | Bombyx mori Acetobacter pasteurianus |  |  |  |  | [76,86] | |
| Cellobiose dehydrogenase | Trametes sanguinea |  |  |  | [68] | ||
| Laccase | Streptomyces coelicolor |  |  | [87] | |||
| Manganese peroxidase | Irpex lacteus |  |  |  | [88] | ||
| Lignin peroxidase | Phanerochaete chrysosporium |  |  | [77,89] | |||
| Peroxiredoxin (Bcp1) | Sulfolobus solfataricus |  | [38] | ||||
| Tyrosinase | B. megaterium |  |  | [78,90] | |||
| Trehalose synthase | Pimelobacter sp. R48 |  |  |  | [79] | ||
| ω-transaminase | Vibrio fluvialis |  | [85] | ||||
| Lipoyl synthase | P. polymyxa |  |  | [41] | |||
| Esterase | B. subtilis P. polymyxa |  |  |  |  | [41,70] | |
| Lipase | Thermotoga maritima |  |  |  |  |  | [41,71,74] |
| Phytase | E.coli |  |  |  | [39,72,91] | ||
| Organophosphorus hydrolase | Pseudomonas diminuta Flavobacterium sp. |  |  |  |  |  | [56,75] |
| Peptidoglycan hydrolase | Lactobacillus rhamnosus |  |  | [92] | |||
| α-amylase | B. licheniformis |  |  |  | [55] | ||
| β-Galactosidase | Alicyclobacillus acidocaldarius B. subtilis B. stearothermophilus E. coli |  |  |  |  |  | [51,65,67,80,84,93,94,95] |
| Exochitinase | Paenibacillus barengoltzii |  |  |  | [73] | ||
| Bromelain | Ananas comosus |  | [40] | ||||
| Keratinase | B. tequilensis |  |  | [96,97] | |||
| Nitrilase | Clostridium thermocellum Thermotoga maritima |  |  |  | [98,99,100] | ||
| Hydrolase | E. coli Burkholderia cepacia |  |  | [81] | |||
| Haloalkane dehalogenase | Rhodococcus rhodochrous |  |  |  |  | [83] | |
| Photodecarboxylase | Chlorella variabilis |  | [101] | ||||
| N-acetyl-D-neuraminic acid aldolase | E. coli |  | [69] | ||||
| Cellobiose 2-epimerase | Caldicellulosiruptor saccharolyticus |  |  | [102] | |||
| D-psicose 3-epimerase | Clostridium scindens |  | [103] | ||||
| L-arabinose isomerase | Lactobacillus brevis |  | [82] |
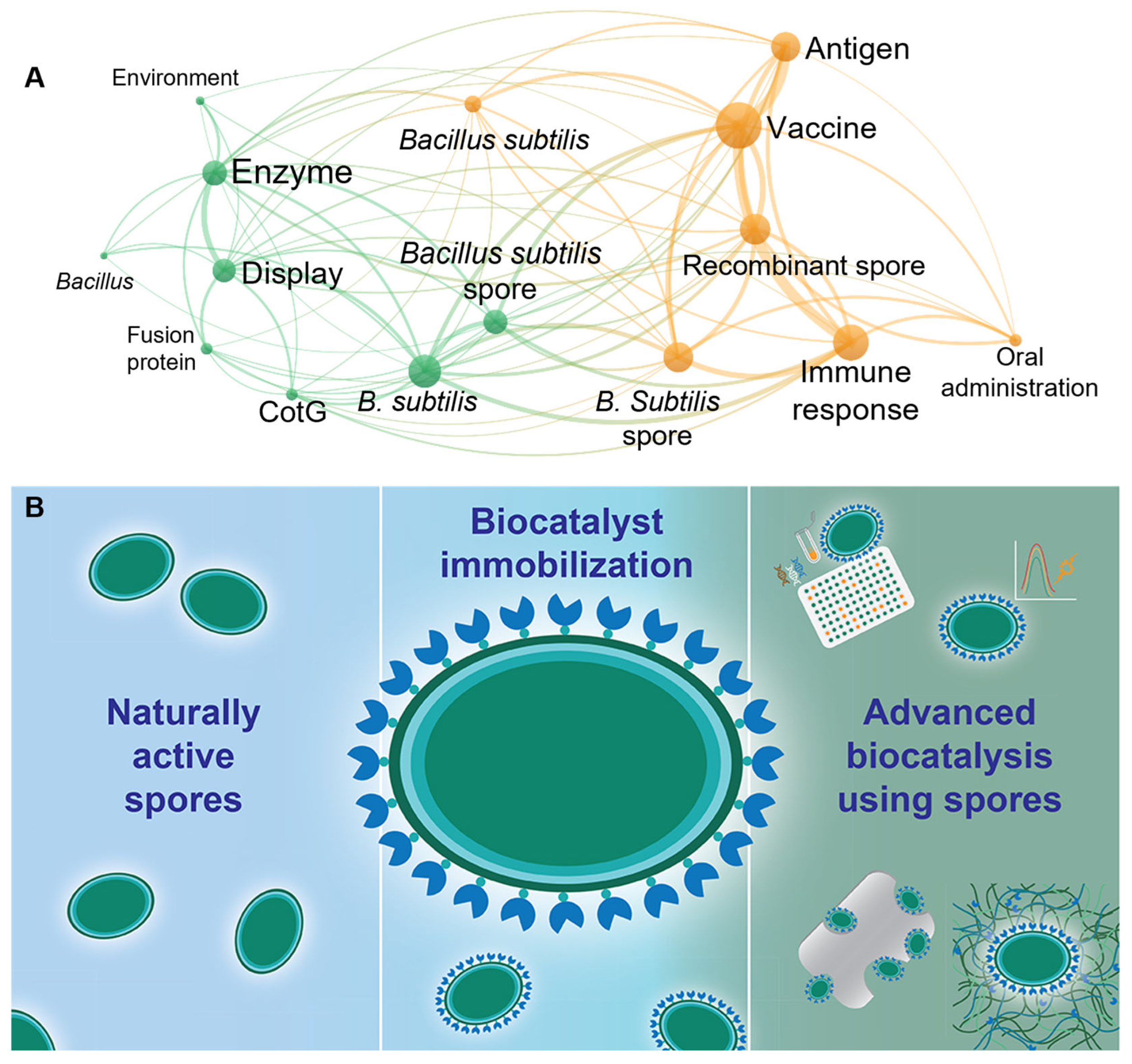
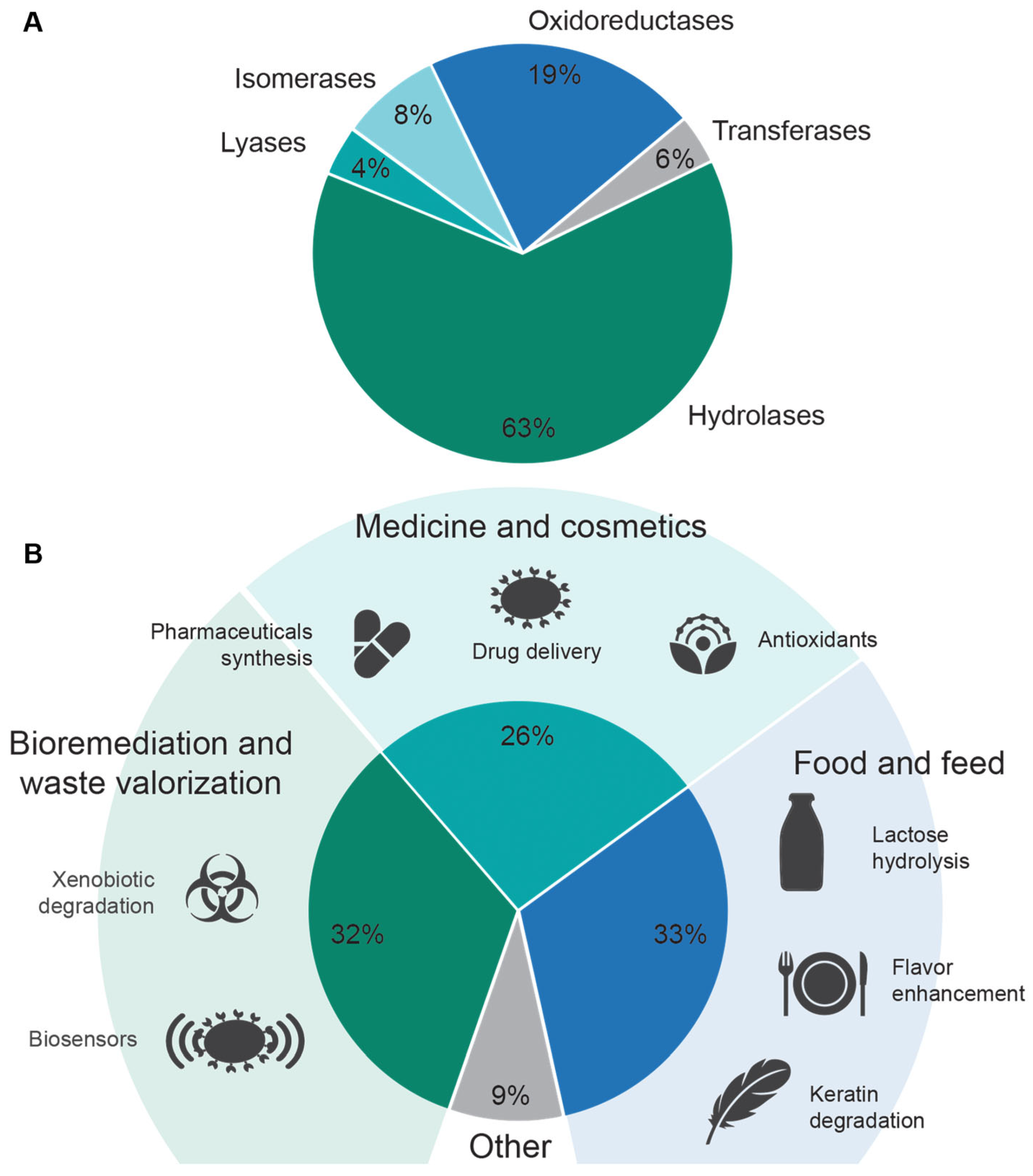
5. Application of Bacterial Spores in Traditional Biocatalysis
6. Expanding the Utility of Spores in Biocatalysis
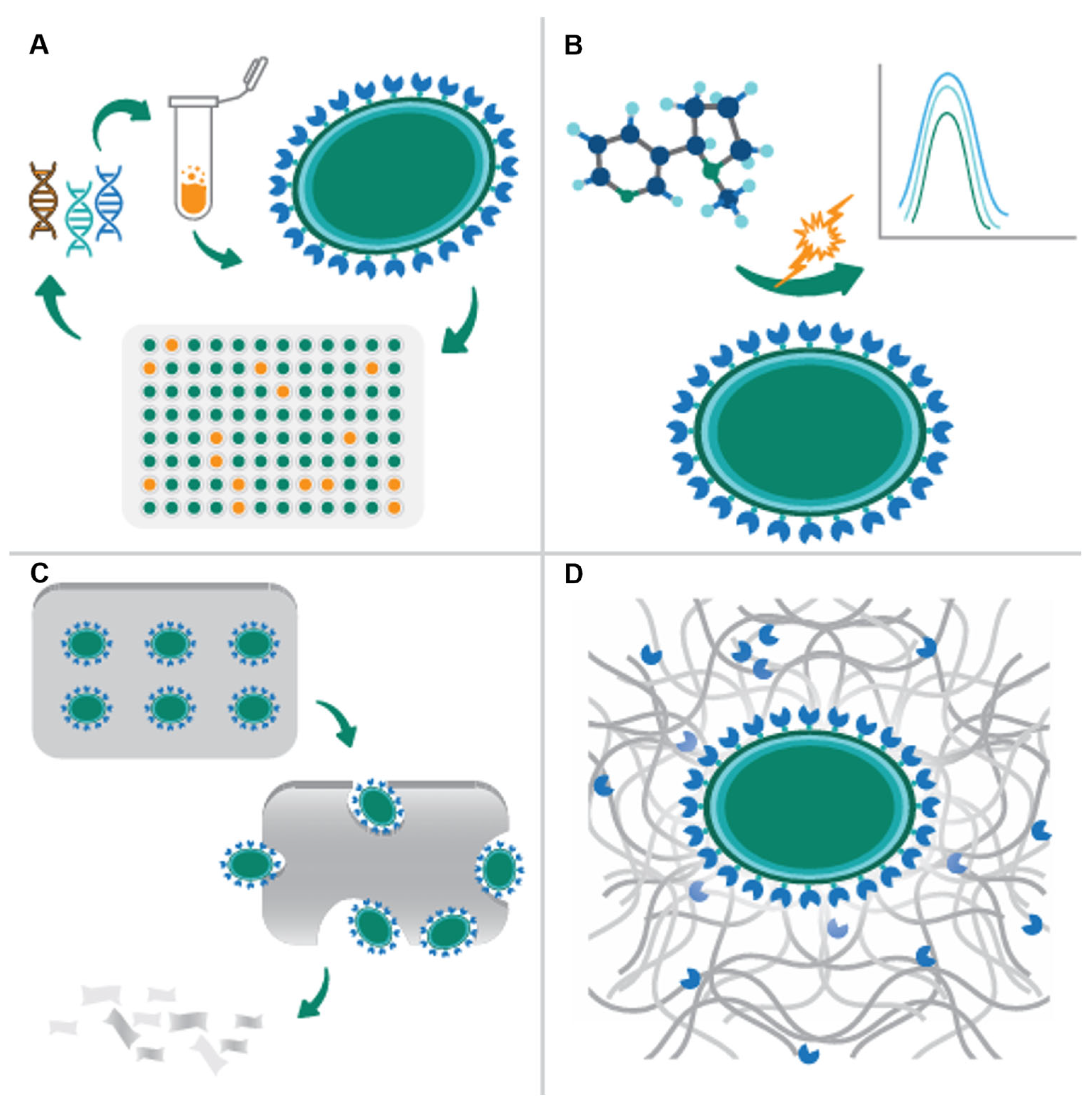
7. Artificial Spores
8. Limitations of Spore-Based Biocatalysts
9. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AdhA | Alcohol dehydrogenase A |
| BAL | Benzaldehyde lyase |
| Bcp1 | Peroxiredoxin |
| CALB | Candida antartica lipase B |
| DEAE | N,N-Diethylethanolamine |
| DMSO | Dimethyl sulfoxide |
| EC | Enzyme Commission |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl) carboniimide hydrochloride |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| LacA | β-galactosidase from B. subtilis |
| L-AI | L-arabinose isomerase |
| Neu5Ac | N-acetyl-D-neuraminic acid |
| NHS | N-hydroxysulfosuccinimide |
| OPH | Organophosphorus hydrolase |
| PCL | Poly(caprolactone) |
| PLA | Poly(lactic acid) |
| PTDH | Phosphite dehydrogenase |
| PU | Polyurethane |
| SLAC | Small laccase from Streptomyces coelicolor |
| TIED | T7 RNA polymerase enabled displayed system |
| XR | Xylose reductase |
References
- Intasian, P.; Prakinee, K.; Phintha, A.; Trisrivirat, D.; Weeranoppanant, N.; Wongnate, T.; Chaiyen, P. Enzymes, in vivo biocatalysis, and metabolic engineering for enabling a circular economy and sustainability. Chem. Rev. 2021, 121, 10367–10451. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Bayer, T.; Badenhorst, C.P.; Wu, S.; Doerr, M.; Höhne, M.; Bornscheuer, U.T. Recent trends in biocatalysis. Chem. Soc. Rev. 2021, 50, 8003–8049. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.-S. Enzyme immobilization technologies and industrial applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Huisman, G.; Kazlauskas, R.; Lutz, S.; Moore, J.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Fasim, A.; More, V.S.; More, S.S. Large-scale production of enzymes for biotechnology uses. Curr. Opin. Biotechnol. 2021, 69, 68–76. [Google Scholar] [CrossRef]
- Sharma, V.; Tsai, M.-L.; Nargotra, P.; Chen, C.-W.; Kuo, C.-H.; Sun, P.-P.; Dong, C.-D. Agro-industrial food waste as a low-cost substrate for sustainable production of industrial enzymes: A critical review. Catalysts 2022, 12, 1373. [Google Scholar] [CrossRef]
- Lin, P.; Yuan, H.; Du, J.; Liu, K.; Liu, H.; Wang, T. Progress in research and application development of surface display technology using Bacillus subtilis spores. Appl. Microbiol. Biotechnol. 2020, 104, 2319–2331. [Google Scholar] [CrossRef]
- Bahrulolum, H.; Ahmadian, G. Bacillus subtilis surface display technology: Applications in bioprocessing and sustainable manufacturing. Biotechnol. Biofuels Bioprod. 2025, 18, 34. [Google Scholar] [CrossRef]
- Paul, C.; Filippidou, S.; Jamil, I.; Kooli, W.; House, G.L.; Estoppey, A.; Hayoz, M.; Junier, T.; Palmieri, F.; Wunderlin, T. Bacterial spores, from ecology to biotechnology. Adv. Appl. Microbiol. 2019, 106, 79–111. [Google Scholar]
- Horneck, G.; Rettberg, P.; Reitz, G.; Wehner, J.; Eschweiler, U.; Strauch, K.; Panitz, C.; Starke, V.; Baumstark-Khan, C. Protection of bacterial spores in space, a contribution to the discussion on panspermia. Orig. Life Evol. Biosph. 2001, 31, 527–547. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, E.A.; Miller, D.A.; Angert, E.R. Sporulation in bacteria: Beyond the standard model. In The Bacterial Spore: From Molecules to Systems; ASM Press: Washington, DC, USA, 2016; pp. 87–102. [Google Scholar]
- Brbić, M.; Piškorec, M.; Vidulin, V.; Kriško, A.; Šmuc, T.; Supek, F. The landscape of microbial phenotypic traits and associated genes. Nucleic Acids Res. 2016, 44, 10074–10090. [Google Scholar] [CrossRef]
- Tocheva, E.I.; Ortega, D.R.; Jensen, G.J. Sporulation, bacterial cell envelopes and the origin of life. Nat. Rev. Microbiol. 2016, 14, 535–542. [Google Scholar] [CrossRef]
- Mahmoodi, A.; Farinas, E.T. Applications of Bacillus subtilis protein display for medicine, catalysis, environmental remediation, and protein engineering. Microorganisms 2024, 12, 97. [Google Scholar] [CrossRef]
- McKenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013, 11, 33–44. [Google Scholar] [CrossRef]
- Bauda, E.; Gallet, B.; Moravcova, J.; Effantin, G.; Chan, H.; Novacek, J.; Jouneau, P.-H.; Rodrigues, C.D.; Schoehn, G.; Moriscot, C. Ultrastructure of macromolecular assemblies contributing to bacterial spore resistance revealed by in situ cryo-electron tomography. Nat. Commun. 2024, 15, 1376. [Google Scholar] [CrossRef] [PubMed]
- Ricca, E.; Baccigalupi, L.; Isticato, R. Spore-adsorption: Mechanism and applications of a non-recombinant display system. Biotechnol. Adv. 2021, 47, 107693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Al-Dossary, A.; Hussain, M.; Setlow, P.; Li, J. Applications of Bacillus subtilis spores in biotechnology and advanced materials. Appl. Environ. Microbiol. 2020, 86, e01096-20. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Cui, Z.; Sim, S. Stress-tolerant, recyclable, and renewable biocatalyst platform enabled by engineered bacterial spores. ACS Synth. Biol. 2022, 11, 2857–2868. [Google Scholar] [CrossRef]
- Mohsin, M.Z.; Omer, R.; Huang, J.; Mohsin, A.; Guo, M.; Qian, J.; Zhuang, Y. Advances in engineered Bacillus subtilis biofilms and spores, and their applications in bioremediation, biocatalysis, and biomaterials. Synth. Syst. Biotechnol. 2021, 6, 180–191. [Google Scholar] [CrossRef]
- Lozančić, M.; Sk. Hossain, A.; Mrša, V.; Teparić, R. Surface display—An alternative to classic enzyme immobilization. Catalysts 2019, 9, 728. [Google Scholar] [CrossRef]
- Bahrulolum, H.; Beyranvand, P.; Ahmadian, G. Harnessing Bacillus subtilis spore surface display (BSSD) technology for mucosal vaccines and drug delivery: Innovations in respiratory virus immunization. Drugs Drug Candidates 2024, 3, 774–795. [Google Scholar] [CrossRef]
- Oggioni, M.R.; Ciabattini, A.; Cuppone, A.M.; Pozzi, G. Bacillus spores for vaccine delivery. Vaccine 2003, 21, S96–S101. [Google Scholar] [CrossRef]
- Arruda, H.; Silva, E.R.; Lessa, M.; Proença Jr, D.; Bartholo, R. VOSviewer and bibliometrix. J. Med. Libr. Assoc. 2022, 110, 392. [Google Scholar] [CrossRef]
- Leggett, M.J.; McDonnell, G.; Denyer, S.P.; Setlow, P.; Maillard, J.Y. Bacterial spore structures and their protective role in biocide resistance. J. Appl. Microbiol. 2012, 113, 485–498. [Google Scholar] [CrossRef]
- Lu, L.; Zhao, M.; Wang, T.-N.; Zhao, L.-Y.; Du, M.-H.; Li, T.-L.; Li, D.-B. Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresour. Technol. 2012, 115, 35–40. [Google Scholar] [CrossRef]
- El-Bendary, M.A.; Ezzat, S.M.; Ewais, E.A.; Al-Zalama, M.A. Optimization of spore laccase production by Bacillus amyloliquefaciens isolated from wastewater and its potential in green biodecolorization of synthetic textile dyes. Prep. Biochem. Biotechnol. 2021, 51, 16–27. [Google Scholar] [CrossRef]
- El-Bendary, M.A.; Ewais, E.A.; Ezzat, S.M.; Al-Zalama, M.A. Process optimization of the bio-decolorization of textile dyes by spore-bound laccase of Bacillus amyloliquefaciens. Microbiology 2019, 13, 23–33. [Google Scholar]
- Peng, F.; Zheng, B.; Zhang, Y.; Faheem, A.; Chai, Y.; Jiang, T.; Chen, X.; Hu, Y. Biocatalytic oxidation of aromatic compounds by spore-based system. ACS Sustain. Chem. Eng. 2020, 8, 14159–14165. [Google Scholar] [CrossRef]
- Lu, L.; Zhao, M.; Li, G.-F.; Li, J.; Wang, T.-N.; Li, D.-B.; Xu, T.-F. Decolorization of synthetic dyes by immobilized spore from Bacillus amyloliquefaciens. Catal. Commun. 2012, 26, 58–62. [Google Scholar] [CrossRef]
- Asadi, E.; Makhdoumi, A.; Asoodeh, A. Laccase mediator system obtained from a marine spore exhibits decolorization potential in harsh environmental conditions. Ecotoxicol. Environ. Saf. 2020, 191, 110184. [Google Scholar] [CrossRef]
- Zhou, W.; Guan, Z.-B.; Cai, Y.-J.; Chen, Y.; Zhang, N.; Liao, X.-R. Preparation and characterization of immobilized spores with laccase activity from Bacillus pumilus W3 on DEAE-cellulose and their application in dye decolorization. Braz. J. Chem. Eng. 2017, 34, 41–52. [Google Scholar] [CrossRef]
- Paredes-Sabja, D.; Shen, A.; Sorg, J.A. Clostridium difficile spore biology: Sporulation, germination, and spore structural proteins. Trends Microbiol. 2014, 22, 406–416. [Google Scholar] [CrossRef]
- Creuly, C.; Larroche, C.; Gros, J.-B. Bioconversion of fatty acids into methyl ketones by spores of Penicillium roquefortii in a water-organic solvent, two-phase system. Enzym. Microb. Technol. 1992, 14, 669–678. [Google Scholar] [CrossRef]
- Wolken, W.; Van Der Werf, M. Geraniol biotransformation-pathway in spores of Penicillium digitatum. Appl. Microbiol. Biotechnol. 2001, 57, 731–737. [Google Scholar] [CrossRef]
- Stülke, J.; Grüppen, A.; Bramkamp, M.; Pelzer, S. Bacillus subtilis, a swiss army knife in science and biotechnology. J. Bacteriol. 2023, 205, e00102-23. [Google Scholar] [CrossRef]
- Hsieh, H.-Y.; Lin, C.-H.; Hsu, S.-Y.; Stewart, G.C. A Bacillus spore-based display system for bioremediation of atrazine. Appl. Environ. Microbiol. 2020, 86, e01230-20. [Google Scholar] [CrossRef] [PubMed]
- Lanzilli, M.; Donadio, G.; Fusco, F.A.; Sarcinelli, C.; Limauro, D.; Ricca, E.; Isticato, R. Display of the peroxiredoxin Bcp1 of Sulfolobus solfataricus on probiotic spores of Bacillus megaterium. New Biotechnol. 2018, 46, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.-A.; Kim, E.-J.; Pan, J.-G. Adsorption immobilization of Escherichia coli phytase on probiotic Bacillus polyfermenticus spores. Enzym. Microb. Technol. 2011, 49, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Corona, R.; Bontà, V.; Baccigalupi, L.; Ricca, E. Probiotic spores of Shouchella clausii SF174 and displayed bromelain show beneficial additive potential. Int. J. Mol. Sci. 2025, 26, 942. [Google Scholar] [CrossRef]
- Zander, M.; Schmid, J.; Kabisch, J. Implementation of spore display in Paenibacillus polymyxa with different hydrolytic enzymes. Microorganisms 2024, 12, 1438. [Google Scholar] [CrossRef]
- Brena, B.; González-Pombo, P.; Batista-Viera, F. Immobilization of enzymes: A literature survey. In Immobilization of Enzymes and Cells, 3rd ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 15–31. [Google Scholar]
- Datta, R.; Anand, S.; Moulick, A.; Baraniya, D.; Imran Pathan, S.; Rejsek, K.; Vranova, V.; Sharma, M.; Sharma, D.; Formanek, P. How enzymes are adsorbed on soil solid phase and factors limiting its activity: A Review. Int. Agrophysics 2017, 31, 287–302. [Google Scholar] [CrossRef]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Kim, M. An overview of techniques in enzyme immobilization. Appl. Sci. Converg. Technol. 2017, 26, 157–163. [Google Scholar] [CrossRef]
- Górecka, E.; Jastrzębska, M. Immobilization techniques and biopolymer carriers. Biotechnol. Food Sci. 2011, 75, 65–86. [Google Scholar]
- Kummetha, L.R.; Oh, J.-J.; van der Linden, F.H.; Aubin-Tam, M.-E. Leveraging the versatile properties of bacterial spores in materials. Trends Biotechnol. 2025, 43, 812–825. [Google Scholar] [CrossRef]
- Huang, J.-M.; Hong, H.A.; Van Tong, H.; Hoang, T.H.; Brisson, A.; Cutting, S.M. Mucosal delivery of antigens using adsorption to bacterial spores. Vaccine 2010, 28, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wan, Q.; Krajcikova, D.; Tang, J.; Tzokov, S.B.; Barak, I.; Bullough, P.A. Diverse supramolecular structures formed by self-assembling proteins of the Bacillus subtilis spore coat. Mol. Microbiol. 2015, 97, 347–359. [Google Scholar] [CrossRef]
- Donadio, G.; Lanzilli, M.; Sirec, T.; Ricca, E.; Isticato, R. Localization of a red fluorescence protein adsorbed on wild type and mutant spores of Bacillus subtilis. Microb. Cell Factories 2016, 15, 153. [Google Scholar] [CrossRef]
- Sirec, T.; Strazzulli, A.; Isticato, R.; De Felice, M.; Moracci, M.; Ricca, E. Adsorption of β-galactosidase of Alicyclobacillus acidocaldarius on wild type and mutants spores of Bacillus subtilis. Microb. Cell Factories 2012, 11, 100. [Google Scholar] [CrossRef]
- Thyparambil, A.A.; Wei, Y.; Latour, R.A. Experimental characterization of adsorbed protein orientation, conformation, and bioactivity. Biointerphases 2015, 10, 019002. [Google Scholar] [CrossRef]
- Fears, K.P.; Sivaraman, B.; Powell, G.L.; Wu, Y.; Latour, R.A. Probing the conformation and orientation of adsorbed enzymes using side-chain modification. Langmuir 2009, 25, 9319–9327. [Google Scholar] [CrossRef]
- Ghaedmohammadi, S.; Rigi, G.; Zadmard, R.; Ricca, E.; Ahmadian, G. Immobilization of bioactive protein A from Staphylococcus aureus (SpA) on the surface of Bacillus subtilis spores. Mol. Biotechnol. 2015, 57, 756–766. [Google Scholar] [CrossRef]
- Gashtasbi, F.; Ahmadian, G.; Noghabi, K.A. New insights into the effectiveness of alpha-amylase enzyme presentation on the Bacillus subtilis spore surface by adsorption and covalent immobilization. Enzym. Microb. Technol. 2014, 64, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Falahati-Pour, S.; Lotfi, A.; Ahmadian, G.; Baghizadeh, A. Covalent immobilization of recombinant organophosphorus hydrolase on spores of Bacillus subtilis. J. Appl. Microbiol. 2015, 118, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Moshnikova, A.; Afanasyev, V.; Proussakova, O.; Chernyshov, S.; Gogvadze, V.; Beletsky, I. Cytotoxic activity of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide is underlain by DNA interchain cross-linking. Cell. Mol. Life Sci. CMLS 2006, 63, 229–234. [Google Scholar] [CrossRef]
- Becker, M.; Lütz, S.; Rosenthal, K. Environmental assessment of enzyme production and purification. Molecules 2021, 26, 573. [Google Scholar] [CrossRef] [PubMed]
- Bartels, J.; López Castellanos, S.N.; Radeck, J.; Mascher, T. Sporobeads: The utilization of the Bacillus subtilis endospore crust as a protein display platform. ACS Synth. Biol. 2018, 7, 452–461. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Yang, R. Recent progress in Bacillus subtilis spore-surface display: Concept, progress, and future. Appl. Microbiol. Biotechnol. 2017, 101, 933–949. [Google Scholar] [CrossRef]
- Yang, H.; Qu, J.; Zou, W.; Shen, W.; Chen, X. An overview and future prospects of recombinant protein production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2021, 105, 6607–6626. [Google Scholar] [CrossRef]
- Iwanicki, A.; Piątek, I.; Stasiłojć, M.; Grela, A.; Łęga, T.; Obuchowski, M.; Hinc, K. A system of vectors for Bacillus subtilis spore surface display. Microb. Cell Factories 2014, 13, 30. [Google Scholar] [CrossRef]
- Nguyen, Q.A.; Schumann, W. Use of IPTG-inducible promoters for anchoring recombinant proteins on the Bacillus subtilis spore surface. Protein Expr. Purif. 2014, 95, 67–76. [Google Scholar] [CrossRef]
- Petrillo, C.; Castaldi, S.; Lanzilli, M.; Saggese, A.; Donadio, G.; Baccigalupi, L.; Ricca, E.; Isticato, R. The temperature of growth and sporulation modulates the efficiency of spore-display in Bacillus subtilis. Microb. Cell Factories 2020, 19, 185. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, X.; Qian, Y.; Yin, L. Constructing an efficient Bacillus subtilis spore display by using Cohesin−Dockerin interactions. Molecules 2021, 26, 1186. [Google Scholar] [CrossRef]
- Miras, I.; Schaeffer, F.; Béguin, P.; Alzari, P.M. Mapping by site-directed mutagenesis of the region responsible for cohesin−dockerin interaction on the surface of the seventh cohesin domain of Clostridium thermocellum CipA. Biochemistry 2002, 41, 2115–2119. [Google Scholar] [CrossRef]
- Chen, L.; Holmes, M.; Schaefer, E.; Mulchandani, A.; Ge, X. Highly active spore biocatalyst by self-assembly of co-expressed anchoring scaffoldin and multimeric enzyme. Biotechnol. Bioeng. 2018, 115, 557–564. [Google Scholar] [CrossRef]
- Wu, Z.; Li, P.; Chen, X.; Feng, Y.; Ma, Y.; Ni, Z.; Zhu, D.; Chen, H. Surface display system of Bacillus subtilis: A promising approach for improving the stability and applications of cellobiose dehydrogenase. Protein Expr. Purif. 2024, 218, 106448. [Google Scholar] [CrossRef]
- Gao, C.; Xu, X.; Zhang, X.; Che, B.; Ma, C.; Qiu, J.; Tao, F.; Xu, P. Chemoenzymatic synthesis of N-acetyl-D-neuraminic acid from N-acetyl-D-glucosamine by using the spore surface-displayed N-acetyl-D-neuraminic acid aldolase. Appl. Environ. Microbiol. 2011, 77, 7080–7083. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, T.; Jia, J.; Vastermark, A.; Tian, R.; Ni, Z.; Chen, Z.; Chen, K.; Yang, S. Expression and display of a novel thermostable esterase from Clostridium thermocellum on the surface of Bacillus subtilis using the CotB anchor protein. J. Ind. Microbiol. Biotechnol. 2015, 42, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tian, R.; Ni, Z.; Zhang, Q.; Zhang, T.; Chen, Z.; Chen, K.; Yang, S. Surface display of the thermophilic lipase Tm1350 on the spore of Bacillus subtilis by the CotB anchor protein. Extremophiles 2015, 19, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Mingmongkolchai, S.; Panbangred, W. Display of Escherichia coli phytase on the surface of Bacillus subtilis spore using CotG as an anchor protein. Appl. Biochem. Biotechnol. 2019, 187, 838–855. [Google Scholar] [CrossRef]
- Ullah, M.; Xia, Y.; Alshaya, D.S.; Han, J.; Attia, K.A.; Shah, T.A.; Chen, H. Display of bacterial Exochitanase on Bacillus subtilis spores improved enzyme stability and recyclability. Molecules 2024, 29, 4302. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. Surface display of lipolytic enzyme, Lipase A and Lipase B of Bacillus subtilis on the Bacillus subtilis spore. Biotechnol. Bioprocess Eng. 2017, 22, 462–468. [Google Scholar] [CrossRef]
- Song, T.; Wang, F.; Xiong, S.; Jiang, H. Surface display of organophosphorus-degrading enzymes on the recombinant spore of Bacillus subtilis. Biochem. Biophys. Res. Commun. 2019, 510, 13–19. [Google Scholar] [CrossRef]
- Yuan, Y.; Feng, F.; Chen, L.; Yao, Q.; Chen, K. Surface display of Acetobacter pasteurianus AdhA on Bacillus subtilis spores to enhance ethanol tolerance for liquor industrial potential. Eur. Food Res. Technol. 2014, 238, 285–293. [Google Scholar] [CrossRef]
- Li, S.; He, L.; Shi, N.; Ni, Z.; Bu, Q.; Zhu, D.; Chen, H. Display of Lignin Peroxidase on the Surface of Bacillus subtilis. Appl. Biochem. Biotechnol. 2024, 196, 6849–6863. [Google Scholar] [CrossRef]
- Hosseini-Abari, A.; Kim, B.G.; Lee, S.H.; Emtiazi, G.; Kim, W.; Kim, J.H. Surface display of bacterial tyrosinase on spores of Bacillus subtilis using CotE as an anchor protein. J. Basic Microbiol. 2016, 56, 1331–1337. [Google Scholar] [CrossRef]
- Liu, H.; Yang, S.; Wang, X.; Wang, T. Production of trehalose with trehalose synthase expressed and displayed on the surface of Bacillus subtilis spores. Microb. Cell Factories 2019, 18, 100. [Google Scholar] [CrossRef]
- Wang, H.; Yang, R.; Hua, X.; Zhang, W.; Zhao, W. An approach for lactulose production using the CotX-mediated spore-displayed β-galactosidase as a biocatalyst. J. Microbiol. Biotechnol. 2016, 26, 1267–1277. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, J.; Zhang, Z.; Shi, S.; Li, D.; Shen, W.; Shen, E.; Zhou, J. Catalytic transformation of HODAs using an efficient meta-cleavage product hydrolase-spore surface display system. J. Mol. Catal. B Enzym. 2014, 102, 204–210. [Google Scholar] [CrossRef]
- Guo, Q.; An, Y.; Yun, J.; Yang, M.; Magocha, T.A.; Zhu, J.; Xue, Y.; Qi, Y.; Hossain, Z.; Sun, W. Enhanced D-tagatose production by spore surface-displayed L-arabinose isomerase from isolated Lactobacillus brevis PC16 and biotransformation. Bioresour. Technol. 2018, 247, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Song, T.; Jiang, H.; Pei, C.; Huang, Q.; Xi, H. Bacillus subtilis spore surface display of haloalkane dehalogenase DhaA. Curr. Microbiol. 2019, 76, 1161–1167. [Google Scholar] [CrossRef]
- Kwon, S.J.; Jung, H.-C.; Pan, J.-G. Transgalactosylation in a water-solvent biphasic reaction system with β-galactosidase displayed on the surfaces of Bacillus subtilis spores. Appl. Environ. Microbiol. 2007, 73, 2251–2256. [Google Scholar] [CrossRef]
- Hwang, B.-Y.; Kim, B.-G.; Kim, J.-H. Bacterial surface display of a co-factor containing enzyme, ω-transaminase from Vibrio fluvialis using the Bacillus subtilis spore display system. Biosci. Biotechnol. Biochem. 2011, 75, 1862–1865. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chang, C.; Yao, Q.; Li, G.; Qin, L.; Chen, L.; Chen, K. Display of Bombyx mori alcohol dehydrogenases on the Bacillus subtilis spore surface to enhance enzymatic activity under adverse conditions. PLoS ONE 2011, 6, e21454. [Google Scholar] [CrossRef]
- Kim, W.; Jeong, Y.; Back, S.; Kim, S.; Kim, J. Decolorization of textile dye by spore surface displayed small laccase for the enhanced thermal stability and robust repeated reaction. Biotechnol. Bioprocess Eng. 2022, 27, 930–937. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ullah, M.; Naeem, M.; Ni, Z.; Feng, Y.; Shah, T.A.; Assefa, M.; Almaary, K.S.; Chen, H. Bacillus subtilis spore surface display enhances manganese peroxidase stability and stress resistance. Bioresour. Bioprocess. 2025, 12, 57. [Google Scholar] [CrossRef]
- Shi, N.; Li, S.; He, L.; Feng, Y.; Saeed, M.; Ma, Y.; Ni, Z.; Zhu, D.; Chen, H. High-throughput screening and identification of lignin peroxidase based on spore surface display of Bacillus subtilis. J. Sci. Food Agric. 2025, 105, 2179–2189. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, H.; Kim, B.G.; Kim, J. Regioselective hydroxylation of phloretin by tyrosinase using Bacillus subtilis spore display system. Biotechnol. Bioprocess Eng. 2025, 30, 377–385. [Google Scholar] [CrossRef]
- Potot, S.; Serra, C.R.; Henriques, A.O.; Schyns, G. Display of recombinant proteins on Bacillus subtilis spores, using a coat-associated enzyme as the carrier. Appl. Environ. Microbiol. 2010, 76, 5926–5933. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Jun, J.S.; Moon, J.A.; Hong, K.W. Surface display of p75, a Lactobacillus rhamnosus GG derived protein, on Bacillus subtilis spores and its antibacterial activity against Listeria monocytogenes. AMB Express 2020, 10, 139. [Google Scholar] [CrossRef]
- Hwang, B.-Y.; Pan, J.-G.; Kim, B.-G.; Kim, J.-H. Functional display of active tetrameric β-galactosidase using Bacillus subtilis spore display system. J. Nanosci. Nanotechnol. 2013, 13, 2313–2319. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, R.; Hua, X.; Zhao, W.; Zhang, W. Functional display of active β-galactosidase on Bacillus subtilis spores using crust proteins as carriers. Food Sci. Biotechnol. 2015, 24, 1755–1759. [Google Scholar] [CrossRef]
- Tavassoli, S.; Hinc, K.; Iwanicki, A.; Obuchowski, M.; Ahmadian, G. Investigation of spore coat display of Bacillus subtilis β-galactosidase for developing of whole cell biocatalyst. Arch. Microbiol. 2013, 195, 197–202. [Google Scholar] [CrossRef]
- Yan, M.; Wang, Z.; Zhou, H.; Chen, Y.; Saeed, M.; Xu, Y.; Chen, Y.; Ni, Z.; Fang, Z.; Chen, H. Exploring the potential of spore surface-displayed keratinase for feather waste degradation using high-throughput screening. J. Sci. Food Agric. 2025, 105, 5714–5727. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, M.; Saeed, M.; Li, K.; Chen, Y.; Okoye, C.O.; Fang, Z.; Ni, Z.; Chen, H. The flexible linker and CotG were more effective for the spore surface display of keratinase KERQ7. World J. Microbiol. Biotechnol. 2024, 40, 35. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, T.; Sun, T.; Ni, Z.; Le, Y.; Tian, R.; Chen, Z.; Zhang, C. Clostridium thermocellum nitrilase expression and surface display on Bacillus subtilis spores. J. Mol. Microbiol. Biotechnol. 2015, 25, 381–387. [Google Scholar]
- Chen, H.; Chen, Z.; Ni, Z.; Tian, R.; Zhang, T.; Jia, J.; Chen, K.; Yang, S. Display of Thermotoga maritima MSB8 nitrilase on the spore surface of Bacillus subtilis using out coat protein CotG as the fusion partner. J. Mol. Catal. B Enzym. 2016, 123, 73–80. [Google Scholar] [CrossRef]
- Chen, H.; Chen, Z.; Wu, B.; Ullah, J.; Zhang, T.; Jia, J.; Wang, H.; Tan, T. Influences of various peptide linkers on the Thermotoga maritima MSB8 nitrilase displayed on the spore surface of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2017, 27, 64–71. [Google Scholar] [CrossRef]
- Karava, M.; Gockel, P.; Kabisch, J. Bacillus subtilis spore surface display of photodecarboxylase for the transformation of lipids to hydrocarbons. Sustain. Energy Fuels 2021, 5, 1727–1733. [Google Scholar] [CrossRef]
- Gu, J.; Yang, R.; Hua, X.; Zhang, W.; Zhao, W. Adsorption-based immobilization of C. aldicellulosiruptor saccharolyticus cellobiose 2-epimerase on B acillus subtilis spores. Biotechnol. Appl. Biochem. 2015, 62, 237–244. [Google Scholar] [CrossRef]
- He, W.; Jiang, B.; Mu, W.; Zhang, T. Production of D-allulose with D-psicose 3-epimerase expressed and displayed on the surface of Bacillus subtilis spores. J. Agric. Food Chem. 2016, 64, 7201–7207. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Hwang, E.T.; Lee, S. Multienzymatic cascade reactions via enzyme complex by immobilization. ACS Catal. 2019, 9, 4402–4425. [Google Scholar] [CrossRef]
- Chauhan, K.; Zárate-Romero, A.; Sengar, P.; Medrano, C.; Vazquez-Duhalt, R. Catalytic kinetics considerations and molecular tools for the design of multienzymatic cascade nanoreactors. ChemCatChem 2021, 13, 3732–3748. [Google Scholar] [CrossRef]
- Cai, X.; Huang, Y.; Zhu, C. Immobilized Multi-Enzyme/Nanozyme Biomimetic Cascade Catalysis for Biosensing Applications. Adv. Healthc. Mater. 2025, 14, 2401834. [Google Scholar] [CrossRef]
- Chen, L.; Mulchandani, A.; Ge, X. Spore-displayed enzyme cascade with tunable stoichiometry. Biotechnol. Prog. 2017, 33, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-Y.; Hsieh, H.-Y.; Stewart, G.C.; Lin, C.-H. Bioremediation of Atrazine and its Metabolite Using Multiple Enzymes Delivered by a Bacillus thuringiensis Spore Display System. bioRxiv 2023. [Google Scholar] [CrossRef]
- Qin, Y.; Wu, G.; Guo, Y.; Ke, D.; Yin, J.; Wang, D.; Fan, X.; Liu, Z.; Ruan, L.; Hu, Y. Engineered glyphosate oxidase coupled to spore-based chemiluminescence system for glyphosate detection. Anal. Chim. Acta 2020, 1133, 39–47. [Google Scholar] [CrossRef]
- Jia, H.; Lee, F.S.; Farinas, E.T. Bacillus subtilis spore display of laccase for evolution under extreme conditions of high concentrations of organic solvent. ACS Comb. Sci. 2014, 16, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Jia, H.; Topiol, S.; Farinas, E.T. Engineering CotA laccase for acidic pH stability using Bacillus subtilis spore display. J. Microbiol. Biotechnol. 2017, 27, 507–513. [Google Scholar] [CrossRef]
- Xiong, X.; Xiong, S.; Zhu, R.; Liu, S.; Gao, F.; Chen, Y. Engineering Bacillus Spores to Display Nicotine Oxidase: In Situ Specific and Sensitive Nicotine Detection. ACS Sens. 2025, 10, 3589–3599. [Google Scholar] [CrossRef]
- Tang, C.; Wang, L.; Sun, J.; Chen, G.; Shen, J.; Wang, L.; Han, Y.; Luo, J.; Li, Z.; Zhang, P. Degradable living plastics programmed by engineered spores. Nat. Chem. Biol. 2025, 21, 1006–1011. [Google Scholar] [CrossRef]
- Kawada, M.; Jo, H.; Medina, A.M.; Sim, S. Catalytic materials enabled by a programmable assembly of synthetic polymers and engineered bacterial spores. J. Am. Chem. Soc. 2023, 145, 16210–16217. [Google Scholar] [CrossRef] [PubMed]
- Nemiwal, M.; Zhang, T.C.; Kumar, D. Enzyme immobilized nanomaterials as electrochemical biosensors for detection of biomolecules. Enzym. Microb. Technol. 2022, 156, 110006. [Google Scholar] [CrossRef]
- Kim, H.S.; White, E.M.; Crane, G.; Patel, K.; Noh, M.H.; Rahman, M.A.; Feist, A.M.; Locklin, J.J.; Pokorski, J.K. Scalable fabrication of a tough and recyclable spore-bearing biocomposite thermoplastic polyurethane. Chem. Eng. J. 2025, 505, 159863. [Google Scholar] [CrossRef]
- Kim, H.S.; Noh, M.H.; White, E.M.; Kandefer, M.V.; Wright, A.F.; Datta, D.; Lim, H.G.; Smiggs, E.; Locklin, J.J.; Rahman, M.A. Biocomposite thermoplastic polyurethanes containing evolved bacterial spores as living fillers to facilitate polymer disintegration. Nat. Commun. 2024, 15, 3338. [Google Scholar] [CrossRef]
- Cui, Z.; Kawada, M.; Hui, Y.; Sim, S. Programming aliphatic polyester degradation by engineered bacterial spores. Biomacromolecules 2025, 26, 1882–1891. [Google Scholar] [CrossRef]
- González, L.M.; Mukhitov, N.; Voigt, C.A. Resilient living materials built by printing bacterial spores. Nat. Chem. Biol. 2020, 16, 126–133. [Google Scholar] [CrossRef]
- Rheem, H.B.; Kim, N.; Nguyen, D.T.; Baskoro, G.A.; Roh, J.H.; Lee, J.K.; Kim, B.J.; Choi, I.S. Single-Cell Nanoencapsulation: Chemical Synthesis of Artificial Cell-in-Shell Spores. Chem. Rev. 2025, 125, 6366–6396. [Google Scholar] [CrossRef] [PubMed]
- Iturralde, M.; Ripoll, M.; Silvio, D.d.; Gallego, M.; Grajales-Hernández, D.A.; López, X.; Betancor, L.; López-Gallego, F. Artificial Spores as Multi-Functional Biocatalysts to Perform Biosynthetic Cascades. Adv. Funct. Mater. 2024, 34, 2406097. [Google Scholar] [CrossRef]
- Liu, S.-R.; Cai, L.-F.; Wang, L.-Y.; Yi, X.-F.; Peng, Y.-J.; He, N.; Wu, X.; Wang, Y.-P. Polydopamine coating on individual cells for enhanced extracellular electron transfer. Chem. Commun. 2019, 55, 10535–10538. [Google Scholar] [CrossRef]
- Sun, Z.; Hübner, R.; Li, J.; Wu, C. Artificially sporulated Escherichia coli cells as a robust cell factory for interfacial biocatalysis. Nat. Commun. 2022, 13, 3142. [Google Scholar] [CrossRef]
- Wang, S.; Gong, Z.; Hübner, R.; Karring, H.; Wu, C. Pickering Emulsion Biocatalysis with Engineered Living Cells for Degrading Polycarbonate Plastics. Small 2025, 21, 2504376. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Sun, Z.; Hübner, R.; Karring, H.; Ebbesen, M.F.; Dimde, M.; Wu, C. Engineering living cells with polymers for recyclable photoenzymatic catalysis. Nat. Catal. 2024, 7, 1404–1416. [Google Scholar] [CrossRef]
- Bolmanis, E.; Grigs, O.; Didrihsone, E.; Senkovs, M.; Nikolajeva, V. Pilot-scale production of Bacillus subtilis MSCL 897 spore biomass and antifungal secondary metabolites in a low-cost medium. Biotechnol. Lett. 2024, 46, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Naveen, A.K.; Sontakke, M. A review on regulatory aspects, challenges and public perception in acceptance of genetically modified foods. Food Sci. Biotechnol. 2024, 33, 791–804. [Google Scholar] [CrossRef]
- Kauffmann, F.; Van Damme, P.; Leroux-Roels, G.; Vandermeulen, C.; Berthels, N.; Beuneu, C.; Mali, S. Clinical trials with GMO-containing vaccines in Europe: Status and regulatory framework. Vaccine 2019, 37, 6144–6153. [Google Scholar] [CrossRef] [PubMed]
- Vojnovic, S.; Aleksic, I.; Ilic-Tomic, T.; Stevanovic, M.; Nikodinovic-Runic, J. Bacillus and Streptomyces spp. as hosts for production of industrially relevant enzymes. Appl. Microbiol. Biotechnol. 2024, 108, 185. [Google Scholar] [CrossRef]


| Description of Patent | Patent | Year | Application Filed by |
|---|---|---|---|
| Methods for surface display of target proteins on bacterial spores | WO2020232316A1 | 2019 | Bayer Cropscience LP |
| WO2020102642A2 | 2018 | Bayer Cropscience LP | |
| WO2019099635A1 | 2017 | Bayer Cropscience LP | |
| WO2016140702A1 | 2015 | US Department of Health & Human Services | |
| WO2012001000A1 | 2010 | DSM IP Assets B.V. | |
| WO2011160026A2 | 2010 | Research Development Foundation | |
| WO2006012366A2 | 2004 | Phyllom LLC (Piedmont, CA, USA) | |
| WO2005028654A1 | 2006 | Korea Advanced Institute of Science and Technology | |
| WO2002046388A1 | 2000 | Genofocus Co., Ltd. (Daejeon, Republic of Korea) | |
| WO2002000232A2 | 2000 | Maxygen, Inc. (Newark, CA, USA) | |
| KR102173586B1 | 2019 | Korean Agency for Defense Development | |
| Specific applications of spore-based biocatalysts. | WO2022049442A1 | 2020 | 3M Innovative Properties Company (St. Paul, MN, USA) |
| WO2019060574A1 | 2017 | Spogen Biotech Inc. (Saint Louis, MO, USA) | |
| KR101244433B1 | 2009 | Korea Research Institute of Bioscience and Biotechnology | |
| KR100758209B1 | 2004 | HLB Genex (Daejeon, Republic of Korea) | |
| CN109593695A | 2018 | Qilu University of Technology | |
| CN103014054A | 2012 | Jiangnan University |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantelic, B.; Radivojevic, N.; Aleksic, I.; Simic, J.; Nikodinovic-Runic, J. Microbial Spore-Based Biocatalysts: Properties, Applications and New Trends. Catalysts 2025, 15, 894. https://doi.org/10.3390/catal15090894
Pantelic B, Radivojevic N, Aleksic I, Simic J, Nikodinovic-Runic J. Microbial Spore-Based Biocatalysts: Properties, Applications and New Trends. Catalysts. 2025; 15(9):894. https://doi.org/10.3390/catal15090894
Chicago/Turabian StylePantelic, Brana, Nikola Radivojevic, Ivana Aleksic, Jelena Simic, and Jasmina Nikodinovic-Runic. 2025. "Microbial Spore-Based Biocatalysts: Properties, Applications and New Trends" Catalysts 15, no. 9: 894. https://doi.org/10.3390/catal15090894
APA StylePantelic, B., Radivojevic, N., Aleksic, I., Simic, J., & Nikodinovic-Runic, J. (2025). Microbial Spore-Based Biocatalysts: Properties, Applications and New Trends. Catalysts, 15(9), 894. https://doi.org/10.3390/catal15090894









