Exploration of CYP4B1 Substrate Promiscuity Across Three Species
Abstract
1. Introduction
2. Results and Discussion
2.1. Screening of a Natural Product Library for Spectral Changes Indicating Substrate Binding
2.2. Definition of a Systematic Compound Library for In Vitro Conversion
2.3. In Vitro Conversion of Library Compounds
2.3.1. Group I and II: Saturated Fatty Acids and n-Alkanols
2.3.2. Group III: Acyclic Terpenoids
2.3.3. Groups IV and V: Monocyclic and Bicyclic Terpenes/Terpenoids
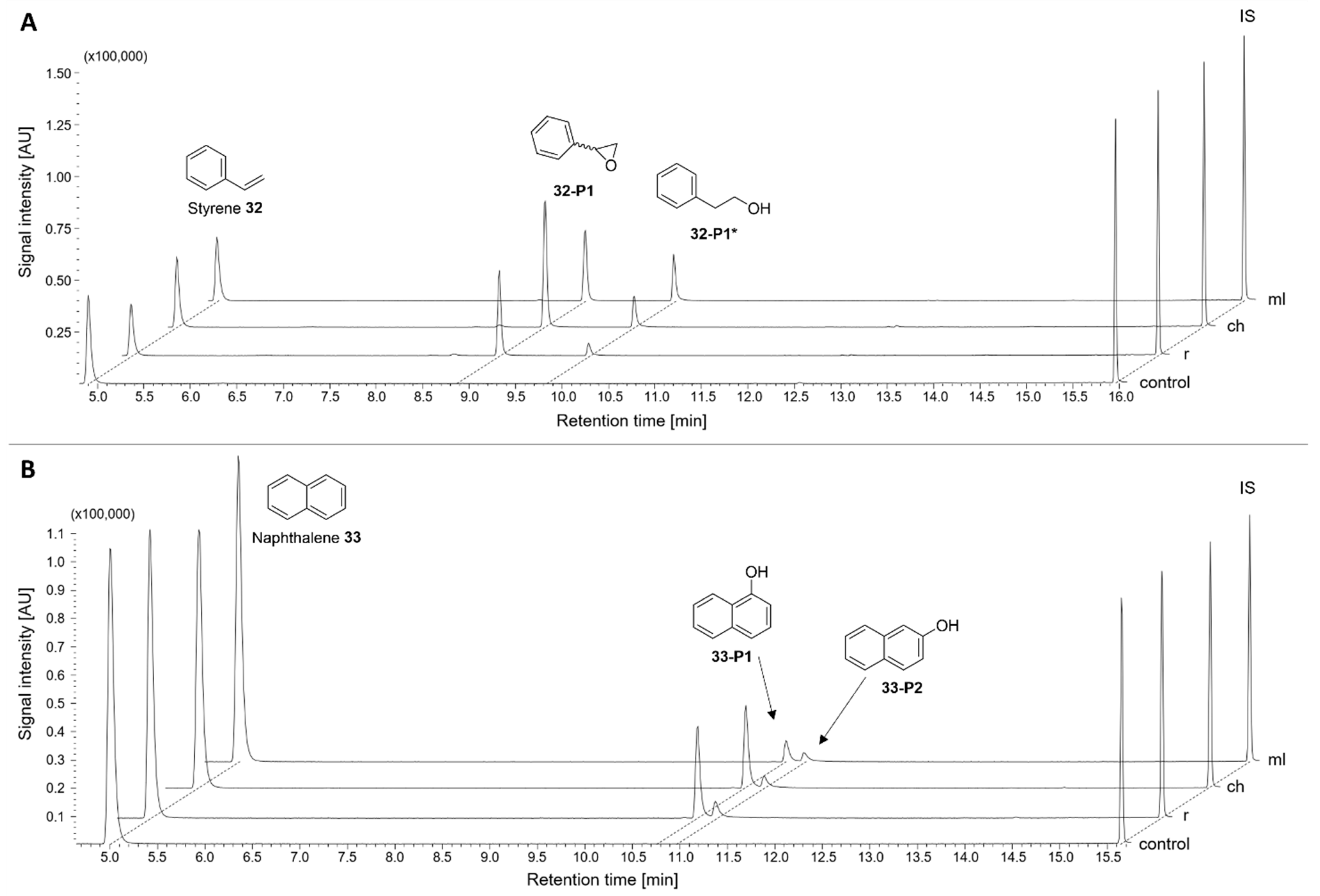
2.3.4. Group VI: Aromatic Hydrocarbons
2.3.5. Group VII: Heterocycles
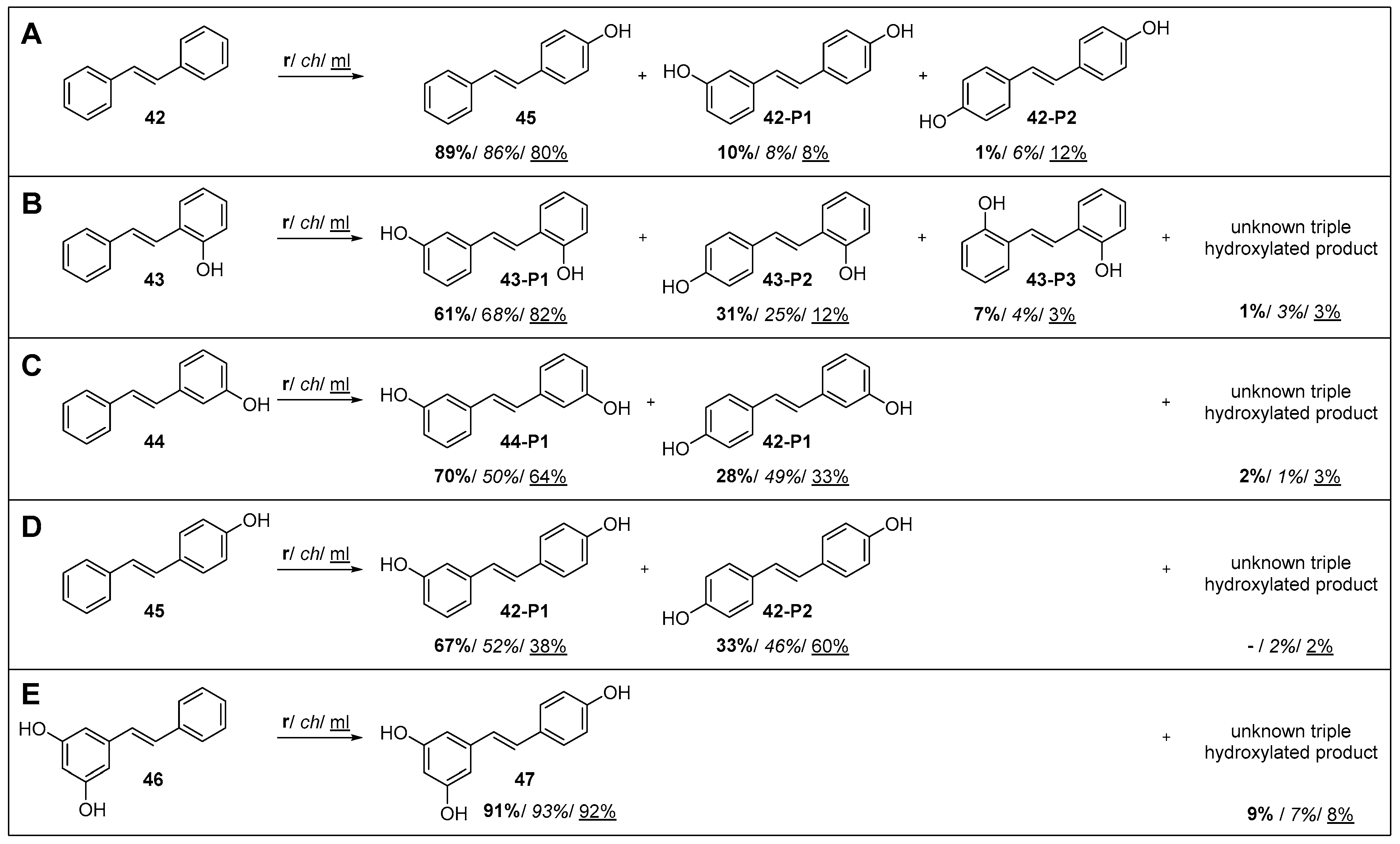
2.3.6. Group VIII: Stilbene and Stilbenoids
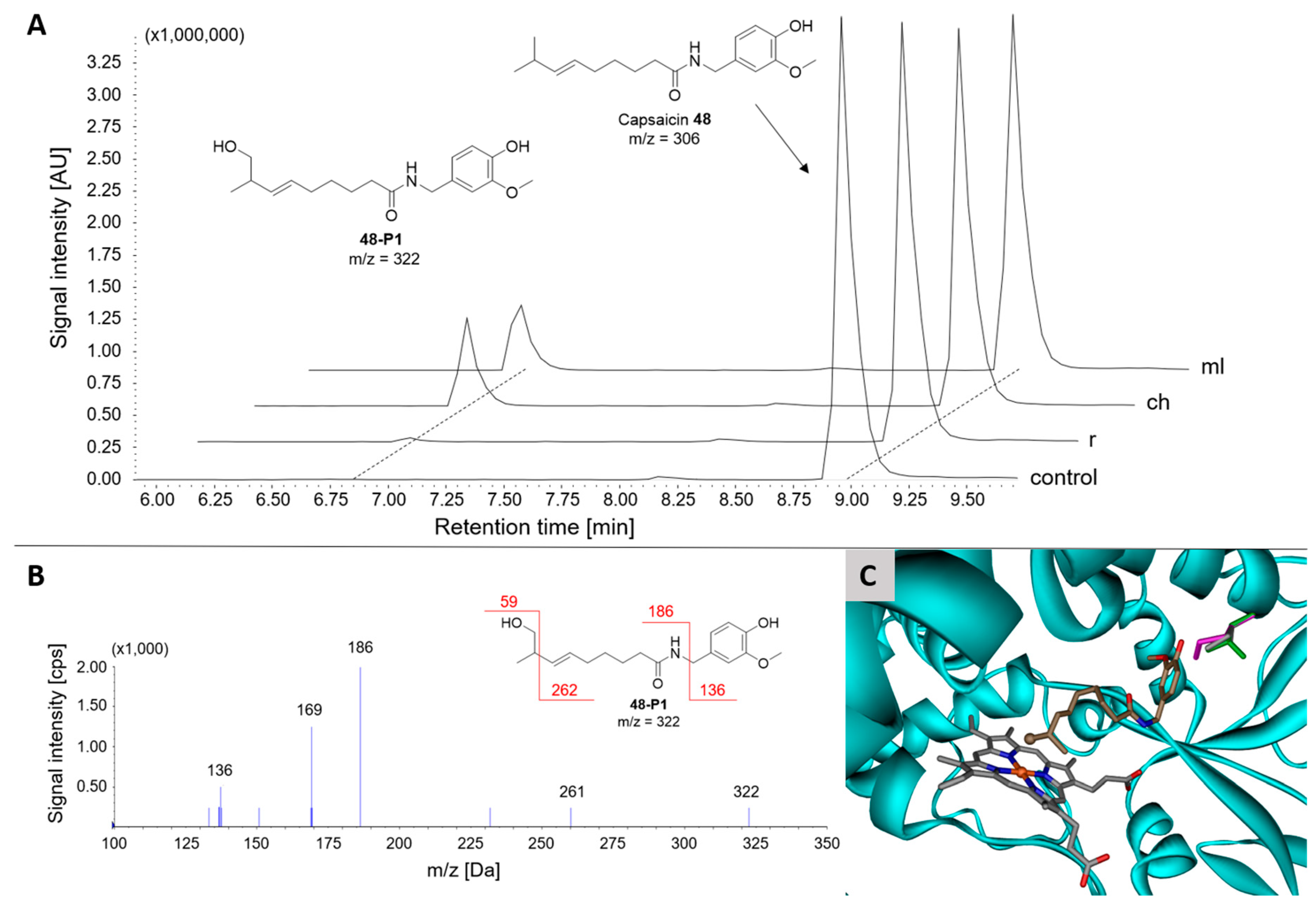
2.3.7. Group IX: Vanilloids
2.3.8. Group X: Miscellaneous
2.3.9. Summary and Overview of Substrate Conversion by CYP4B1
3. Materials and Methods
3.1. Plasmid Construction for cyp4b1 Expression in E. coli
3.2. Protein Expression in E. coli and Purification of Recombinant Enzymes
3.2.1. CYP4B1 Orthologs
3.2.2. Fdr and YkuN
3.2.3. bCYB5
3.2.4. GDH
3.3. Reconstitution of CYP4B1 Activity for Substrate Screening
3.4. Screening for Compound-Induced Spectral Changes of CYP4B1
3.5. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Akhtar, M.; Wright, J. A unified mechanistic view of oxidative reactions catalysed by P-450 and related Fe-containing enzymes. Nat. Prod. Rep. 1991, 8, 527–551. [Google Scholar] [CrossRef]
- Bernhardt, R.; Urlacher, V.B. Cytochromes P450 as promising catalysts for biotechnological application: Chances and limitations. Appl. Microbiol. Biotechnol. 2014, 98, 6185–6203. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.S.; Yadav, J.S. Microbial P450 enzymes in bioremediation and drug discovery: Emerging potentials and challenges. Curr. Protein Pept. Sci. 2018, 19, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012, 30, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 Superfamily: Update on New Sequences, Gene Mapping, Accession Numbers, Early Trivial Names of Enzymes, and Nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Hsu, M.H.; Savas, U.; Griffin, K.J.; Johnson, E.F. Human cytochrome p450 family 4 enzymes: Function, genetic variation and regulation. Drug Metab. Rev. 2007, 39, 515–538. [Google Scholar] [CrossRef]
- Simpson, A.E.C.M. The cytochrome P450 4 (CYP4) family. Gen. Pharmacol. Vasc. Syst. 1997, 28, 351–359. [Google Scholar] [CrossRef]
- Baer, B.R.; Rettie, A.E. CYP4B1: An enigmatic P450 at the interface between xenobiotic and endobiotic metabolism. Drug Metab. Rev. 2006, 38, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.P. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 2008, 75, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Röder, A.; Hüsken, S.; Hutter, M.C.; Rettie, A.E.; Hanenberg, H.; Wiek, C.; Girhard, M. Spotlight on CYP4B1. Int. J. Mol. Sci. 2023, 24, 2038. [Google Scholar] [CrossRef]
- Thesseling, F.A.; Hutter, M.C.; Wiek, C.; Kowalski, J.P.; Rettie, A.E.; Girhard, M. Novel insights into oxidation of fatty acids and fatty alcohols by cytochrome P450 monooxygenase CYP4B1. Arch. Biochem. Biophys. 2020, 679, 108216. [Google Scholar] [CrossRef]
- Scott, E.E. omega- versus (omega-1)-hydroxylation: Cytochrome P450 4B1 sterics make the call. J. Biol. Chem. 2017, 292, 5622–5623. [Google Scholar] [CrossRef]
- Boyd, M.R.; Burka, L.T.; Harris, T.M.; Willson, B.J. Lung-toxic furanoterpenoids produced by sweet potatoes (Ipomoea batatas) following microbial infection. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1974, 337, 184–195. [Google Scholar] [CrossRef]
- Wilson, B.J.; Yang, D.T.; Boyd, M.R. Toxicity of mould-damaged sweet potatoes (Ipomoea batatas). Nature 1970, 227, 521–522. [Google Scholar] [CrossRef]
- Garst, J.; Wilson, W.; Kristensen, N.; Harrison, P.; Corbin, J.; Simon, J.; Philpot, R.; Szabo, R. Species susceptibility to the pulmonary toxicity of 3-furyl isoamyl ketone (perilla ketone): In vivo support for involvement of the lung monooxygenase system. J. Anim. Sci. 1985, 60, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Devereux, T.R.; Jones, K.G.; Bend, J.R.; Fouts, J.R.; Statham, C.N.; Boyd, M.R. In vitro metabolic activation of the pulmonary toxin, 4-ipomeanol, in nonciliated bronchioal epithelial (Clara) and alveolar type ii cells isolated from rabbit lung. J. Pharmacol. Exp. Ther. 1981, 220, 223–227. [Google Scholar] [CrossRef]
- Slaughter, S.R.; Statham, C.N.; Philpot, R.M.; Boyd, M.R. Covalent binding of metabolites of 4-ipomeanol to rabbit pulmonary and hepatic microsomal proteins and to the enzymes of the pulmonary cytochrome P-450-dependent monooxygenase system. J. Pharmacol. Exp. Ther. 1983, 224, 252–257. [Google Scholar] [CrossRef]
- Smith, P.B.; Tiano, H.F.; Nesnow, S.; Boyd, M.R.; Philpot, R.M.; Langenbach, R. 4-Ipomeanol and 2-aminoanthracene cytotoxicity in C3H/10T12 cells expressing rabbit cytochrome P450 4B1. Biochem. Pharmacol. 1995, 50, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Garst, J.E.; Linnabary, R.D.; Channell, R.B. Perilla ketone: A potent lung toxin from the mint plant, Perilla frutescens Britton. Science 1977, 197, 573–574. [Google Scholar] [CrossRef]
- Roellecke, K.; Jager, V.D.; Gyurov, V.H.; Kowalski, J.P.; Mielke, S.; Rettie, A.E.; Hanenberg, H.; Wiek, C.; Girhard, M. Ligand characterization of CYP4B1 isoforms modified for high-level expression in Escherichia coli and HepG2 cells. Protein Eng. Des. Sel. 2017, 30, 205–216. [Google Scholar] [CrossRef]
- Kowalski, J.P.; McDonald, M.G.; Whittington, D.; Guttman, M.; Scian, M.; Girhard, M.; Hanenberg, H.; Wiek, C.; Rettie, A.E. Structure-Activity Relationships for CYP4B1 Bioactivation of 4-Ipomeanol Congeners: Direct Correlation between Cytotoxicity and Trapped Reactive Intermediates. Chem. Res. Toxicol. 2019, 32, 2488–2498. [Google Scholar] [CrossRef]
- Johannessen, C.U.; Johannessen, S.I. Valproate: Past, present, and future. CNS Drug Rev. 2003, 9, 199–216. [Google Scholar] [CrossRef]
- Rettie, A.E.; Sheffels, P.R.; Korzekwa, K.R.; Gonzalez, F.J.; Philpot, R.M.; Baillie, T.A. CYP4 isoenzyme specificity and the relationship between. Omega.-hydroxylation and terminal desaturation of valproic acid. Biochemistry 1995, 34, 7889–7895. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.R.; Rettie, A.E.; Henne, K.R. Bioactivation of 4-ipomeanol by CYP4B1: Adduct characterization and evidence for an enedial intermediate. Chem. Res. Toxicol. 2005, 18, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.; Steffens, S.; Fischer, U.; Tlolko, A.; Rainov, N.G.; Kramm, C.M. Differential cytotoxicity and bystander effect of the rabbit cytochrome P450 4B1 enzyme gene by two different prodrugs: Implications for pharmacogene therapy. Cancer Gene Ther. 2002, 9, 178–188. [Google Scholar] [CrossRef]
- Zheng, Y.-M.; Fisher, M.B.; Yokotani, N.; Fujii-Kuriyama, Y.; Rettie, A.E. Identification of a meander region proline residue critical for heme binding to cytochrome P450: Implications for the catalytic function of human CYP4B1. Biochemistry 1998, 37, 12847–12851. [Google Scholar] [CrossRef]
- Zheng, Y.i.-M.; Henne, K.R.; Charmley, P.; Kim, R.B.; McCarver, D.G.; Cabacungan, E.T.; Hines, R.N.; Rettie, A.E. Genotyping and site-directed mutagenesis of a cytochrome P450 meander Pro-X-Arg motif critical to CYP4B1 catalysis. Toxicol. Appl. Pharmacol. 2003, 186, 119–126. [Google Scholar] [CrossRef]
- Lim, S.; Alshagga, M.; Ong, C.E.; Chieng, J.Y.; Pan, Y. Cytochrome P450 4B1 (CYP4B1) as a target in cancer treatment. Hum. Exp. Toxicol. 2020, 39, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Czerwinski, M.; McLemore, T.L.; Gelboin, H.V.; Gonzalez, F.J. Quantification of CYP2B7, CYP4B1, and CYPOR messenger RNAs in normal human lung and lung tumors. Cancer Res. 1994, 54, 1085–1091. [Google Scholar] [PubMed]
- Barlin, J.N.; Jelinic, P.; Olvera, N.; Bogomolniy, F.; Bisogna, M.; Dao, F.; Barakat, R.R.; Chi, D.S.; Levine, D.A. Validated gene targets associated with curatively treated advanced serous ovarian carcinoma. Gynecol. Oncol. 2013, 128, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, S.; Yoneda, Y.; Sugimoto, T.; Hiroi, T.; Yamamoto, K.; Nakatani, T.; Funae, Y. CYP4B1 is a possible risk factor for bladder cancer in humans. Biochem. Biophys. Res. Commun. 2000, 277, 776–780. [Google Scholar] [CrossRef]
- Lin, J.T.; Chan, T.C.; Li, C.F.; Huan, S.K.H.; Tian, Y.F.; Liang, P.I.; Pan, C.T.; Shiue, Y.L. Downregulation of the cytochrome P450 4B1 protein confers a poor prognostic factor in patients with urothelial carcinomas of upper urinary tracts and urinary bladder. APMIS 2019, 127, 170–180. [Google Scholar] [CrossRef]
- Schenkman, J.B. Nature of the type I and II spectral changes in liver microsomes. Biochemistry 1970, 9, 2081–2091. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and Chemistry of Cytochrome P450. Chem. Rev. 2005, 105, 2253–2278. [Google Scholar] [CrossRef]
- Adams, T.B.; Gavin, C.L.; McGowen, M.M.; Waddell, W.J.; Cohen, S.M.; Feron, V.J.; Marnett, L.J.; Munro, I.C.; Portoghese, P.S.; Rietjens, I.M.; et al. The FEMA GRAS assessment of aliphatic and aromatic terpene hydrocarbons used as flavor ingredients. Food Chem. Toxicol. 2011, 49, 2471–2494. [Google Scholar] [CrossRef]
- Bogazkaya, A.M.; von Bühler, C.J.; Kriening, S.; Busch, A.; Seifert, A.; Pleiss, J.; Laschat, S.; Urlacher, V.B. Selective allylic hydroxylation of acyclic terpenoids by CYP154E1 from Thermobifida fusca YX. Beilstein J. Org. Chem. 2014, 10, 1347–1353. [Google Scholar] [CrossRef]
- von Bühler, C.; Le-Huu, P.; Urlacher, V.B. Cluster screening: An effective approach for probing the substrate space of uncharacterized cytochrome P450s. Chembiochem 2013, 14, 2189–2198. [Google Scholar] [CrossRef]
- Hagvall, L.; Baron, J.M.; Borje, A.; Weidolf, L.; Merk, H.; Karlberg, A.T. Cytochrome P450-mediated activation of the fragrance compound geraniol forms potent contact allergens. Toxicol. Appl. Pharmacol. 2008, 233, 308–313. [Google Scholar] [CrossRef]
- Shimada, T.; Shindo, M.; Miyazawa, M. Species differences in the metabolism of (+)- and (−)-limonenes and their metabolites, carveols and carvones, by cytochrome P450 enzymes in liver microsomes of mice, rats, guinea pigs, rabbits, dogs, monkeys, and humans. Drug Metab. Pharmacokinet. 2002, 17, 507–515. [Google Scholar] [CrossRef]
- Yoshida, E.; Kojima, M.; Suzuki, M.; Matsuda, F.; Shimbo, K.; Onuki, A.; Nishio, Y.; Usuda, Y.; Kondo, A.; Ishii, J. Increased carvone production in Escherichia coli by balancing limonene conversion enzyme expression via targeted quantification concatamer proteome analysis. Sci. Rep. 2021, 11, 22126. [Google Scholar] [CrossRef]
- Hedenstierna, G.; Alexandersson, R.; Wimander, K.; Rosén, G. Exposure to terpenes: Effects on pulmonary function. Int. Arch. Occup. Environ. Health 1983, 51, 191–198. [Google Scholar] [CrossRef]
- Eriksson, K.; Levin, J.-O. Identification of cis- and trans-verbenol in human urine after occupational exposure to terpenes. Int. Arch. Occup. Environ. Health 1990, 62, 379–383. [Google Scholar] [CrossRef]
- Ishida, T.; Asakawa, Y.; Takemoto, T.; Aratani, T. Terpenoids biotransformation in mammals III: Biotransformation of alpha-pinene, beta-pinene, pinane, 3-carene, carane, myrcene, and p-cymene in rabbits. J. Pharm. Sci. 1981, 70, 406–415. [Google Scholar] [CrossRef]
- Schmidt, L.; Goen, T. Human metabolism of alpha-pinene and metabolite kinetics after oral administration. Arch. Toxicol. 2017, 91, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, B.; Chu, C.; Carlin, D.J. Polycyclic aromatic hydrocarbons: From metabolism to lung cancer. Toxicol. Sci. 2015, 145, 5–15. [Google Scholar] [CrossRef]
- Reid, B.C.; Ghazarian, A.A.; DeMarini, D.M.; Sapkota, A.; Jack, D.; Lan, Q.; Winn, D.M.; Birnbaum, L.S. Research opportunities for cancer associated with indoor air pollution from solid-fuel combustion. Environ. Health Perspect. 2012, 120, 1495–1498. [Google Scholar] [CrossRef]
- Piccardo, M.T.; Stella, A.; Valerio, F. Is the smokers exposure to environmental tobacco smoke negligible? Environ. Health 2010, 9, 5. [Google Scholar] [CrossRef]
- Boström, C.E.; Gerde, P.; Hanberg, A.; Jernström, B.; Johansson, C.; Kyrklund, T.; Rannug, A.; Törnqvist, M.; Victorin, K.; Westerholm, R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 2002, 110 (Suppl. 3), 451–488. [Google Scholar] [CrossRef]
- Heibati, B.; Renz, H.; Lacy, P. Wildfire and wood smoke effects on human airway epithelial cells: A scoping review. Environ. Res. 2025, 272, 121153. [Google Scholar] [CrossRef]
- Stading, R.; Gastelum, G.; Chu, C.; Jiang, W.; Moorthy, B. Molecular mechanisms of pulmonary carcinogenesis by polycyclic aromatic hydrocarbons (PAHs): Implications for human lung cancer. Semin. Cancer Biol. 2021, 76, 3–16. [Google Scholar] [CrossRef]
- Chen, L.J.; Wegerski, C.J.; Kramer, D.J.; Thomas, L.A.; McDonald, J.D.; Dix, K.J.; Sanders, J.M. Disposition and metabolism of cumene in F344 rats and B6C3F1 mice. Drug Metab. Dispos. 2011, 39, 498–509. [Google Scholar] [CrossRef]
- Henne, K.R.; Fisher, M.B.; Iyer, K.R.; Lang, D.H.; Trager, W.F.; Rettie, A.E. Active site characteristics of CYP4B1 probed with aromatic ligands. Biochemistry 2001, 40, 8597–8605. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Li, P.; Zhang, X.; Ribitsch, D.; Alcalde, M.; Hollmann, F.; Wang, Y. Enantioselective Sulfoxidation of Thioanisole by Cascading a Choline Oxidase and a Peroxygenase in the Presence of Natural Deep Eutectic Solvents. Chempluschem 2020, 85, 254–257. [Google Scholar] [CrossRef]
- Yin, Y.C.; Yu, H.L.; Luan, Z.J.; Li, R.J.; Ouyang, P.F.; Liu, J.; Xu, J.H. Unusually broad substrate profile of self-sufficient cytochrome P450 monooxygenase CYP116B4 from Labrenzia aggregata. Chembiochem 2014, 15, 2443–2449. [Google Scholar] [CrossRef]
- Fruetel, J.A.; Mackman, R.L.; Peterson, J.A.; Ortiz de Montellano, P.R. Relationship of active site topology to substrate specificity for cytochrome P450terp (CYP108). J. Biol. Chem. 1994, 269, 28815–28821. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Pan, B.; Cheng, X.; Yang, J.; Chen, Y. Recent Advances in Biocatalytic Preparation of Chiral Sulfoxides. Asian J. Org. Chem. 2025, 14, e202400708. [Google Scholar] [CrossRef]
- Fruetel, J.; Chang, Y.-T.; Collins, J.; Loew, G.; Ortiz de Montellano, P.R. Thioanisole Sulfoxidation by Cytochrome P450cam (CYP101): Experimental and Calculated Absolute Stereochemistries. J. Am. Chem. Soc. 1994, 116, 11643–11648. [Google Scholar] [CrossRef]
- West, J.A.A.; Pakehham, G.; Morin, D.; Fleschner, C.A.; Buckpitt, A.R.; Plopper, C.G. Inhaled Naphthalene Causes Dose Dependent Clara Cell Cytotoxicity in Mice but Not in Rats. Toxicol. Appl. Pharmacol. 2001, 173, 114–119. [Google Scholar] [CrossRef]
- Plopper, C.G.; Suverkropp, C.; Morin, D.; Nishio, S.; Buckpitt, A. Relationship of cytochrome P-450 activity to Clara cell cytotoxicity. I. Histopathologic comparison of the respiratory tract of mice, rats and hamsters after parenteral administration of naphthalene. J. Pharmacol. Exp. Ther. 1992, 261, 353–363. [Google Scholar] [CrossRef]
- Cohen, J.T.; Carlson, G.; Charnley, G.; Coggon, D.; Delzell, E.; Graham, J.D.; Greim, H.; Krewski, D.; Medinsky, M.; Monson, R.; et al. A comprehensive evaluation of the potential health risks associated with occupational and environmental exposure to styrene. J. Toxicol. Environ. Health-Part B Crit. Rev. 2002, 5, 1–263. [Google Scholar] [CrossRef]
- Gadberry, M.G.; DeNicola, D.B.; Carlson, G.P. Pneumotoxicity and hepatotoxicity of styrene and styrene oxide. J. Toxicol. Environ. Health 1996, 48, 273–294. [Google Scholar] [CrossRef]
- Bhurta, D.; Bharate, S.B. Styryl Group, a Friend or Foe in Medicinal Chemistry. ChemMedChem 2022, 17, e202100706. [Google Scholar] [CrossRef]
- Elbarbry, F.; Espiritu, M.J.; Soo, K.; Yee, B.; Taylor, J. Inhibition of soluble epoxide hydrolase by natural isothiocyanates. Biochem. Biophys. Res. Commun. 2024, 725, 150261. [Google Scholar] [CrossRef]
- Fretland, A.J.; Omiecinski, C.J. Epoxide hydrolases: Biochemistry and molecular biology. Chem.-Biol. Interact. 2000, 129, 41–59. [Google Scholar] [CrossRef]
- Shen, S.; Li, L.; Ding, X.; Zheng, J. Metabolism of styrene to styrene oxide and vinylphenols in cytochrome P450 2F2- and P450 2E1-knockout mouse liver and lung microsomes. Chem. Res. Toxicol. 2014, 27, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Carlson, G.P. Comparison of the susceptibility of wild-type and CYP2E1 knockout mice to the hepatotoxic and pneumotoxic effects of styrene and styrene oxide. Toxicol. Lett. 2004, 150, 335–339. [Google Scholar] [CrossRef]
- Li, Q.S.; Ogawa, J.; Schmid, R.D.; Shimizu, S. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2001, 67, 5735–5739. [Google Scholar] [CrossRef]
- Baldwin, R.M.; Shultz, M.A.; Buckpitt, A.R. Bioactivation of the pulmonary toxicants naphthalene and 1-nitronaphthalene by rat CYP2F4. J. Pharmacol. Exp. Ther. 2005, 312, 857–865. [Google Scholar] [CrossRef]
- Boyd, M.R.; Wilson, B.J. Isolation and characterization of 4-ipomeanol, a lung-toxic furanoterpenoid produced by sweet potatoes (Ipomoea batatas). J. Agric. Food Chem. 1972, 20, 428–430. [Google Scholar] [CrossRef]
- Czerwinski, M.; McLemore, T.L.; Philpot, R.M.; Nhamburo, P.T.; Korzekwa, K.; Gelboin, H.V.; Gonzalez, F.J. Metabolic activation of 4-ipomeanol by complementary DNA-expressed human cytochromes P-450: Evidence for species-specific metabolism. Cancer Res. 1991, 51, 4636–4638. [Google Scholar] [PubMed]
- Wu, Z.L.; Podust, L.M.; Guengerich, F.P. Expansion of substrate specificity of cytochrome P450 2A6 by random and site-directed mutagenesis. J. Biol. Chem. 2005, 280, 41090–41100. [Google Scholar] [CrossRef] [PubMed]
- Kunevicius, A.; Sadauskas, M.; Raudyte, J.; Meskys, R.; Burokas, A. Unraveling the Dynamics of Host-Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one. Molecules 2024, 29, 993. [Google Scholar] [CrossRef] [PubMed]
- Carpenedo, R.; Mannaioni, G.; Moroni, F. Oxindole, a sedative tryptophan metabolite, accumulates in blood and brain of rats with acute hepatic failure. J. Neurochem. 1998, 70, 1998–2003. [Google Scholar] [CrossRef]
- Jaglin, M.; Rhimi, M.; Philippe, C.; Pons, N.; Bruneau, A.; Goustard, B.; Dauge, V.; Maguin, E.; Naudon, L.; Rabot, S. Indole, a Signaling Molecule Produced by the Gut Microbiota, Negatively Impacts Emotional Behaviors in Rats. Front. Neurosci. 2018, 12, 216. [Google Scholar] [CrossRef]
- D’Agostino, J.; Zhuo, X.; Shadid, M.; Morgan, D.G.; Zhang, X.; Humphreys, W.G.; Shu, Y.Z.; Yost, G.S.; Ding, X. The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab. Dispos. 2009, 37, 2018–2027. [Google Scholar] [CrossRef]
- Yost, G.S. Bioactivation of Toxicants by Cytochrome P450-Mediated Dehydrogenation Mechanisms. In Biological Reactive Intermediates VI: Chemical and Biological Mechanisms in Susceptibility to and Prevention of Environmental Diseases; Dansette, P.M., Snyder, R., Delaforge, M., Gibson, G.G., Greim, H., Jollow, D.J., Monks, T.J., Sipes, I.G., Eds.; Springer: Boston, MA, USA, 2001; pp. 53–62. [Google Scholar] [CrossRef]
- Skiles, G.L.; Yost, G.S. Mechanistic studies on the cytochrome P450-catalyzed dehydrogenation of 3-methylindole. Chem. Res. Toxicol. 1996, 9, 291–297. [Google Scholar] [CrossRef]
- Haydock, L.A.J.; Fenton, R.K.; Sergejewich, L.; Squires, E.J.; Caswell, J.L. Acute interstitial pneumonia and the biology of 3-methylindole in feedlot cattle. Anim. Health Res. Rev. 2022, 23, 72–81. [Google Scholar] [CrossRef]
- Yost, G.S. Mechanisms of 3-methylindole pneumotoxicity. Chem. Res. Toxicol. 1989, 2, 273–279. [Google Scholar] [CrossRef]
- Lichtenstein, E.P.; Casida, J.E. Naturally Occurring Insecticides, Myristicin, an Insecticide and Synergist Occurring Naturally in the Edible Parts of Parsnips. J. Agric. Food Chem. 1963, 11, 410–415. [Google Scholar] [CrossRef]
- Götz, M.E.; Sachse, B.; Schäfer, B.; Eisenreich, A. Myristicin and Elemicin: Potentially Toxic Alkenylbenzenes in Food. Foods 2022, 11, 1988. [Google Scholar] [CrossRef] [PubMed]
- Kaur, V.; Kaushal, S.; Utreja, D. Occurrence, Isolation, Pharmacological Potential, Metabolism, and Toxicity of Myristicin: A Naturally Occurring Alkoxy-Substituted Allylbenzene. Mini-Rev. Org. Chem. 2024, 21, 477–493. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Y.K.; Yang, X.N.; Xiao, X.R.; Zhang, T.; Yang, X.W.; Qin, H.B.; Li, F. Metabolic Activation of Myristicin and Its Role in Cellular Toxicity. J. Agric. Food Chem. 2019, 67, 4328–4336. [Google Scholar] [CrossRef]
- Yun, C.-H.; Lee, H.S.; Lee, H.-Y.; Yim, S.-K.; Kim, K.-H.; Kim, E.; Yea, S.-S.; Guengerich, F.P. Roles of human liver cytochrome P450 3A4 and 1A2 enzymes in the oxidation of myristicin. Toxicol. Lett. 2003, 137, 143–150. [Google Scholar] [CrossRef]
- Rietjens, I.M.; Cohen, S.M.; Fukushima, S.; Gooderham, N.J.; Hecht, S.; Marnett, L.J.; Smith, R.L.; Adams, T.B.; Bastaki, M.; Harman, C.G.; et al. Impact of structural and metabolic variations on the toxicity and carcinogenicity of hydroxy- and alkoxy-substituted allyl- and propenylbenzenes. Chem. Res. Toxicol. 2014, 27, 1092–1103. [Google Scholar] [CrossRef]
- Akinwumi, B.C.; Bordun, K.M.; Anderson, H.D. Biological Activities of Stilbenoids. Int. J. Mol. Sci. 2018, 19, 792. [Google Scholar] [CrossRef]
- Mattio, L.M.; Catinella, G.; Dallavalle, S.; Pinto, A. Stilbenoids: A Natural Arsenal against Bacterial Pathogens. Antibiotics 2020, 9, 336. [Google Scholar] [CrossRef]
- Maccario, C.; Savio, M.; Ferraro, D.; Bianchi, L.; Pizzala, R.; Pretali, L.; Forti, L.; Stivala, L.A. The resveratrol analog 4,4′-dihydroxy-trans-stilbene suppresses transformation in normal mouse fibroblasts and inhibits proliferation and invasion of human breast cancer cells. Carcinogenesis 2012, 33, 2172–2180. [Google Scholar] [CrossRef]
- Smoliga, J.M.; Baur, J.A.; Hausenblas, H.A. Resveratrol and health—A comprehensive review of human clinical trials. Mol. Nutr. Food Res. 2011, 55, 1129–1141. [Google Scholar] [CrossRef]
- Liu, J.; Sridhar, J.; Foroozesh, M. Cytochrome P450 family 1 inhibitors and structure-activity relationships. Molecules 2013, 18, 14470–14495. [Google Scholar] [CrossRef] [PubMed]
- Sanoh, S.; Kitamura, S.; Sugihara, K.; Ohta, S. Cytochrome P450 1A1/2 mediated metabolism of trans-stilbene in rats and humans. Biol. Pharm. Bull. 2002, 25, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Savio, M.; Ferraro, D.; Maccario, C.; Vaccarone, R.; Jensen, L.D.; Corana, F.; Mannucci, B.; Bianchi, L.; Cao, Y.; Stivala, L.A. Resveratrol analogue 4,4′-dihydroxy-trans-stilbene potently inhibits cancer invasion and metastasis. Sci. Rep. 2016, 6, 19973. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Sumiyoshi, M.; Kiyoi, T.; Baba, K. Dihydroxystilbenes prevent azoxymethane/dextran sulfate sodium-induced colon cancer by inhibiting colon cytokines, a chemokine, and programmed cell death-1 in C57BL/6J mice. Eur. J. Pharmacol. 2020, 886, 173445. [Google Scholar] [CrossRef]
- Torres, P.; Avila, J.G.; Romo de Vivar, A.; Garcia, A.M.; Marin, J.C.; Aranda, E.; Cespedes, C.L. Antioxidant and insect growth regulatory activities of stilbenes and extracts from Yucca periculosa. Phytochemistry 2003, 64, 463–473. [Google Scholar] [CrossRef]
- Beltrán, L.R.; Dawid, C.; Beltrán, M.; Gisselmann, G.; Degenhardt, K.; Mathie, K.; Hofmann, T.; Hatt, H. The pungent substances piperine, capsaicin, 6-gingerol and polygodial inhibit the human two-pore domain potassium channels TASK-1, TASK-3 and TRESK. Front. Pharmacol. 2013, 4, 141. [Google Scholar] [CrossRef]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef]
- Dedov, V.N.; Tran, V.H.; Duke, C.C.; Connor, M.; Christie, M.J.; Mandadi, S.; Roufogalis, B.D. Gingerols: A novel class of vanilloid receptor (VR1) agonists. Br. J. Pharmacol. 2002, 137, 793–798. [Google Scholar] [CrossRef]
- Correa, E.A.; Hogestatt, E.D.; Sterner, O.; Echeverri, F.; Zygmunt, P.M. In vitro TRPV1 activity of piperine derived amides. Bioorg Med. Chem. 2010, 18, 3299–3306. [Google Scholar] [CrossRef]
- Dawid, C.; Henze, A.; Frank, O.; Glabasnia, A.; Rupp, M.; Buning, K.; Orlikowski, D.; Bader, M.; Hofmann, T. Structural and sensory characterization of key pungent and tingling compounds from black pepper (Piper nigrum L.). J. Agric. Food Chem. 2012, 60, 2884–2895. [Google Scholar] [CrossRef]
- Reilly, C.A.; Henion, F.; Bugni, T.S.; Ethirajan, M.; Stockmann, C.; Pramanik, K.C.; Srivastava, S.K.; Yost, G.S. Reactive intermediates produced from the metabolism of the vanilloid ring of capsaicinoids by p450 enzymes. Chem. Res. Toxicol. 2013, 26, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Reilly, C.A.; Yost, G.S. Metabolism of capsaicinoids by P450 enzymes: A review of recent findings on reaction mechanisms, bio-activation, and detoxification processes. Drug Metab. Rev. 2006, 38, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.-J.; Sup Lee, S. Capsaicin, a double-edged sword: Toxicity, metabolism, and chemopreventive potential. Life Sci. 1995, 56, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, X.N.; Liu, X.; Zhang, S.P. Capsaicin for allergic rhinitis in adults. Cochrane Database Syst. Rev. 2006, 2006, CD004460. [Google Scholar] [CrossRef]
- Derebery, M.J.; Dicpinigaitis, P.V. New horizons: Current and potential future self-treatments for acute upper respiratory tract conditions. Postgrad. Med. 2013, 125, 82–96. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Mo, Q.; Liu, J.; Yao, X.; Di, X.; Qin, Z.; He, L.; Yao, Z. Cytochrome P450 metabolism studies of [6]-gingerol, [8]-gingerol, and [10]-gingerol by liver microsomes of humans and different species combined with expressed CYP enzymes. RSC Adv. 2023, 13, 5804–5812. [Google Scholar] [CrossRef]
- Reilly, C.A.; Ehlhardt, W.J.; Jackson, D.A.; Kulanthaivel, P.; Mutlib, A.E.; Espina, R.J.; Moody, D.E.; Crouch, D.J.; Yost, G.S. Metabolism of capsaicin by cytochrome P450 produces novel dehydrogenated metabolites and decreases cytotoxicity to lung and liver cells. Chem. Res. Toxicol. 2003, 16, 336–349. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.; Fan, J.; Feng, Z.; Song, X. Pharmacological activity of capsaicin: Mechanisms and controversies (Review). Mol. Med. Rep. 2024, 29, 38. [Google Scholar] [CrossRef]
- Nagayoshi, H.; Murayama, N.; Kim, V.; Kim, D.; Takenaka, S.; Yamazaki, H.; Guengerich, F.P.; Shimada, T. Oxidation of Naringenin, Apigenin, and Genistein by Human Family 1 Cytochrome P450 Enzymes and Comparison of Interaction of Apigenin with Human P450 1B1.1 and Scutellaria P450 82D.1. Chem. Res. Toxicol. 2023, 36, 1778–1788. [Google Scholar] [CrossRef]
- Rendic, S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002, 34, 83–448. [Google Scholar] [CrossRef] [PubMed]
- Basheer, L.; Kerem, Z. Interactions between CYP3A4 and Dietary Polyphenols. Oxid. Med. Cell Longev. 2015, 2015, 854015. [Google Scholar] [CrossRef] [PubMed]
- Ronis, M.J. Effects of soy containing diet and isoflavones on cytochrome P450 enzyme expression and activity. Drug Metab. Rev. 2016, 48, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.Y.; Leung, L.K. A potential protective mechanism of soya isoflavones against 7,12-dimethylbenz[a]anthracene tumour initiation. Br. J. Nutr. 2003, 90, 457–465. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol Chem. 1964, 239, 2370–2378. [Google Scholar] [CrossRef]
- Girhard, M.; Klaus, T.; Khatri, Y.; Bernhardt, R.; Urlacher, V.B. Characterization of the versatile monooxygenase CYP109B1 from Bacillus subtilis. Appl. Microbiol. Biotechnol. 2010, 87, 595–607. [Google Scholar] [CrossRef]
- Neunzig, J.; Sanchez-Guijo, A.; Mosa, A.; Hartmann, M.F.; Geyer, J.; Wudy, S.A.; Bernhardt, R. A steroidogenic pathway for sulfonated steroids: The metabolism of pregnenolone sulfate. J. Steroid Biochem. Mol. Biol. 2014, 144 Pt B, 324–333. [Google Scholar] [CrossRef]
- Mulrooney, S.B.; Waskell, L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5). Protein Expr. Purif. 2000, 19, 173–178. [Google Scholar] [CrossRef]
- Zöllner, A.; Kagawa, N.; Waterman, M.R.; Nonaka, Y.; Takio, K.; Shiro, Y.; Hannemann, F.; Bernhardt, R. Purification and functional characterization of human 11beta hydroxylase expressed in Escherichia coli. FEBS J. 2008, 275, 799–810. [Google Scholar] [CrossRef]
- Kranz-Finger, S.; Mahmoud, O.; Ricklefs, E.; Ditz, N.; Bakkes, P.J.; Urlacher, V.B. Insights into the functional properties of the marneral oxidase CYP71A16 from Arabidopsis thaliana. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 2–10. [Google Scholar] [CrossRef]
- Hsu, M.H.; Baer, B.R.; Rettie, A.E.; Johnson, E.F. The Crystal Structure of Cytochrome P450 4B1 (CYP4B1) Monooxygenase Complexed with Octane Discloses Several Structural Adaptations for omega-Hydroxylation. J. Biol. Chem. 2017, 292, 5610–5621. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2010, 27, 343–350. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Khatri, Y.; Hannemann, F.; Girhard, M.; Kappl, R.; Hutter, M.; Urlacher, V.B.; Bernhardt, R. A natural heme-signature variant of CYP267A1 from Sorangium cellulosum So ce56 executes diverse omega-hydroxylation. FEBS J. 2015, 282, 74–88. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
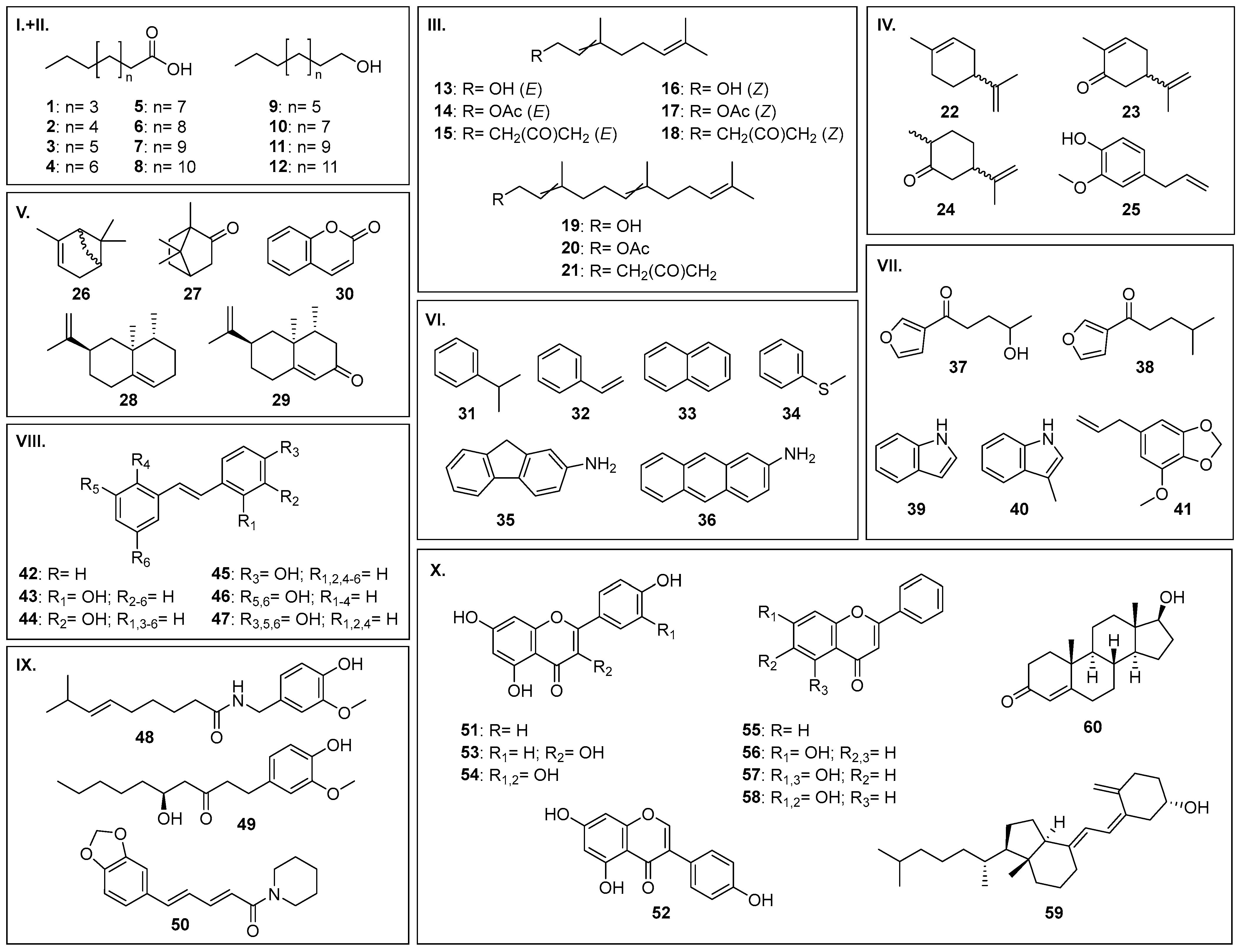
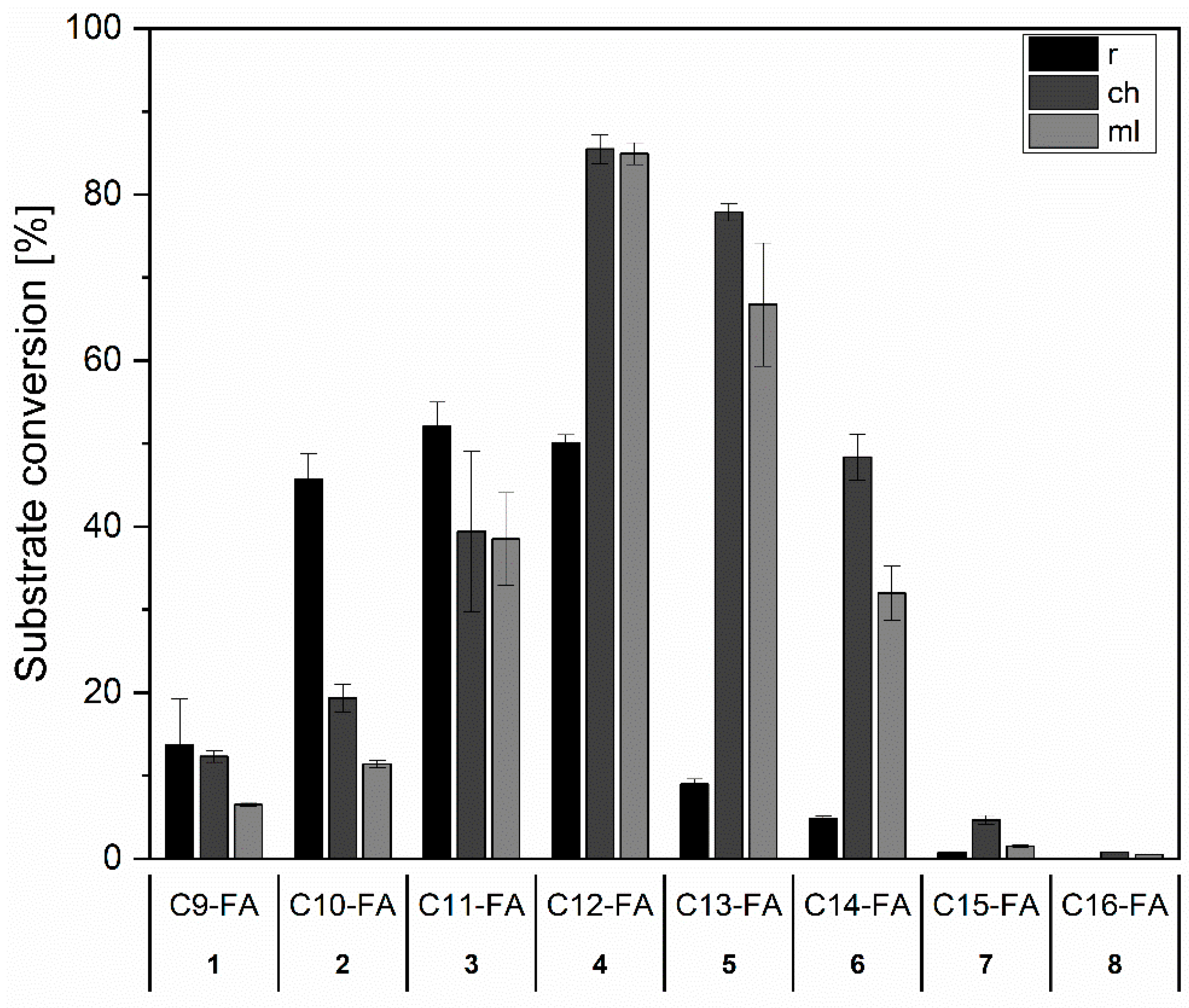
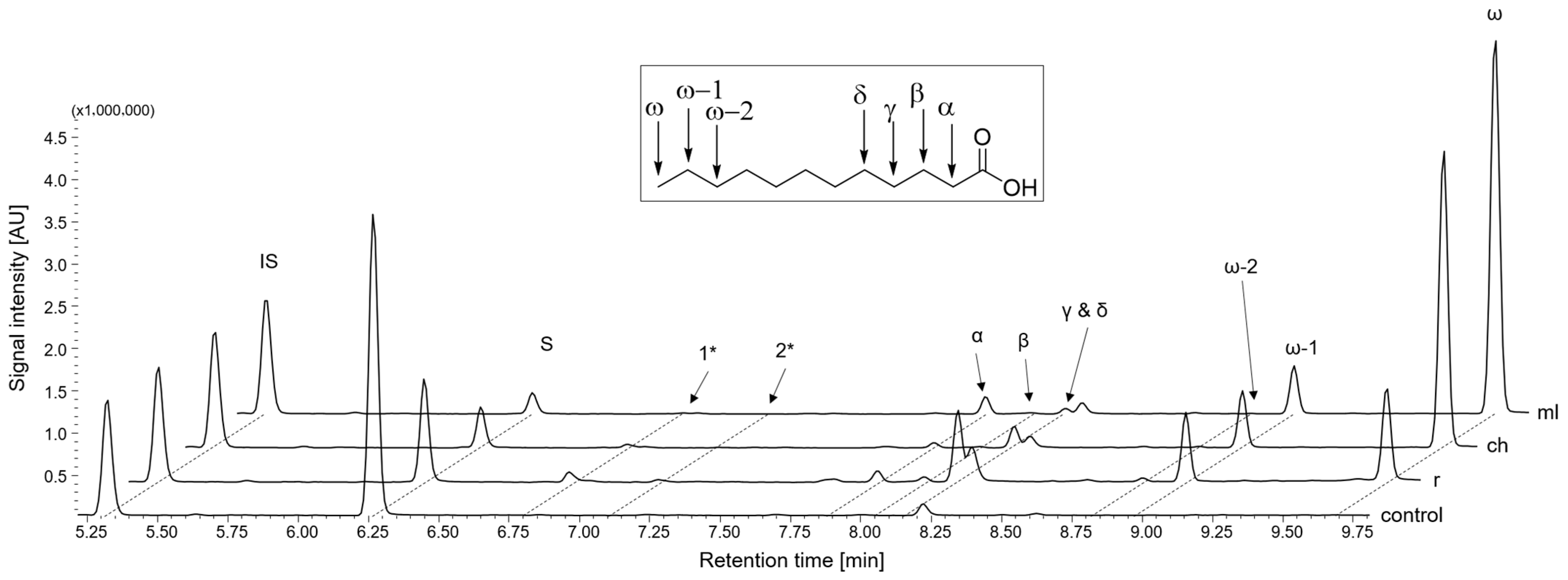

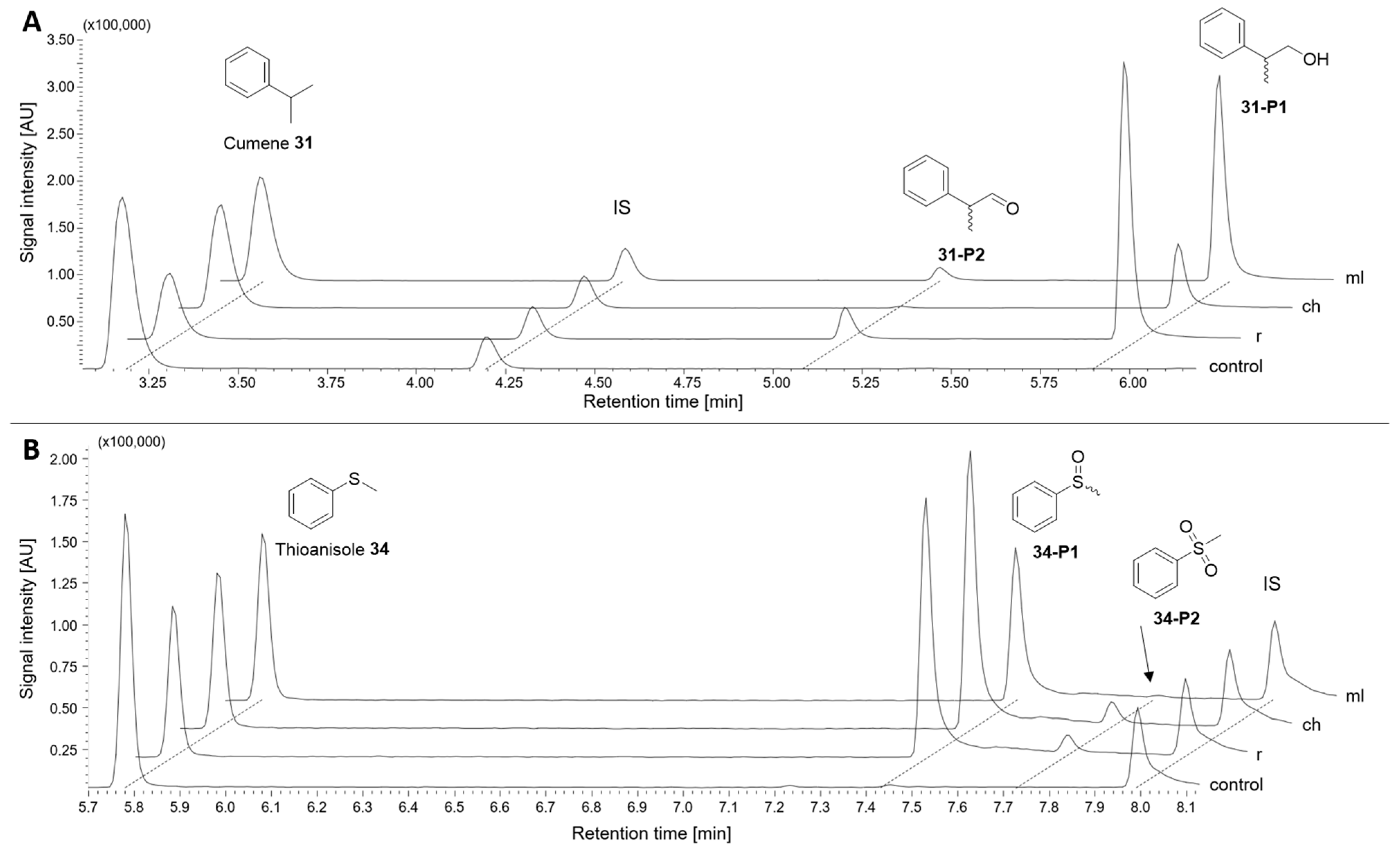
| No | Substance Group | Compound | Conversion [%] | Main Product(s) Observed | |||
|---|---|---|---|---|---|---|---|
| r | ch | mL | |||||
| 1 | I | Saturated fatty acid | Nonanoic acid | 14 ± 6 | 12 ± 1 | 6 ± <1 | ω-, ω-1-OH |
| 2 | Decanoic acid | 46 ± 3 | 19 ± 2 | 11 ± <1 | ω-, ω-1-OH | ||
| 3 | Undecanoic acid | 52 ± 3 | 39 ± 10 | 38 ± 6 | ω-, ω-1-OH | ||
| 4 | Dodecanoic acid | 50 ± 1 | 85 ± 2 | 85 ± 1 | ω-, ω-1-OH | ||
| 5 | Tridecanoic acid | 9 ± 1 | 78 ± 1 | 67 ± 7 | ω-, ω-1-OH | ||
| 6 | Tetradecanoic acid | 5 | 48 ± 3 | 32 ± 3 | ω-, ω-1-OH | ||
| 7 | Pentadecanoic acid | 1 | 5 | 2 | ω-, ω-1-OH | ||
| 8 | Hexadecanoic acid | - | 1 | 1 | ω-, ω-1-OH | ||
| 9 | II | Fatty alcohol (n-alkanol) | 1-Decanol | 10 ± 2 | 11 ± 2 | 9 ± <1 | ω-OH |
| 10 | 1-Dodecanol | 18 ± 1 | 9 ± 1 | 16 ± 1 | ω-, ω-1-OH | ||
| 11 | 1-Tetradecanol | 5 | 15 ± 4 | 18 ± 1 | ω-1-OH, ω-OH, ω-2-OH | ||
| 12 | 1-Hexadecanol | 5 | 15 ± 2 | 13 ± 1 | ω-OH | ||
| 13 | III | Acyclic terpenoid | Geraniol | 61 ± 3 | 69 ± 2 | 54 ± 3 | 8-Hydroxygeraniol 13-P1 |
| 14 | Geranyl acetate | 24 ± 1 | 49 ± 7 | 44 ± 6 | Epoxides c | ||
| 15 | Geranyl acetone | 24 ± 1 | 51 ± 2 | 38 ± 4 | Epoxides c | ||
| 16 | Nerol | 15 ± 2 | 13 ± 1 | 6 ± 1 | 8-Hydroxynerol 16-P1 | ||
| 17 | Neryl acetate | 10 ± 1 | 42 ± 4 | 14 ± 1 | Epoxides c | ||
| 18 | Neryl acetone | 8 ± <1 | 16 ± 1 | 8 ± 2 | Epoxides c | ||
| 19 | Farnesol * | - | - | - | |||
| 20 | Farnesyl acetate * | - | - | - | |||
| 21 | Farnesyl acetone * | - | - | - | |||
| 22a/b | IV | Monocyclic terpene/ terpenoid | Limonene (R/S) | -/- | -/- | -/- | |
| 23a/b | Carvone (R/S) | 73 ± 2/44 ± 2 | 50 ± 3/29 ± 1 | 29 ± 2/25 ± 2 | n.d. c | ||
| 24 | Dihydrocarvone * (mixture of isomers) | 62 ± 2 | 44 ± 2 | 29 ± 2 | n.d. c | ||
| 25 | Eugenol | - | - | - | |||
| 26a/b | V | Biyclic terpene/ terpenoid | α-Pinene (R/S) | 34 ± 1/- | 30 ± 1/- | 9 ± 2/- | Myrtenol 26a-P1 |
| 27 | Camphor | - | - | - | |||
| 28 | Valencene | - | - | - | |||
| 29 | Nootkatone | - | - | - | |||
| 30 | Coumarin | - | - | - | |||
| 31 | VI | Aromatic hydrocarbon | Cumene | 62 ± 5 | 30 ± <1 | 37 ± 8 | 2-Phenyl-1-propanol 31-P1 |
| 56 ± 6% ee (S) | 64 ± 2% ee (S) | 36 ± 8% ee (S) | |||||
| 32 | Styrene | 25 ± 4 | 36 ± 2 | 24 ± 1 | 7,8-Styrene epoxide 32-P1 | ||
| 11 ± 4% ee (S) | 9 ± 3% ee (S) | 15 ± 5% ee (S) | |||||
| 33 | Naphthalene | 23 ± 2 | 22 ± 3 | 12 ± 3 | 1-/2-Naphthol 33-P1/33-P2 | ||
| 34 | Thioanisole | 50 ± 5 | 57 ± 9 | 43 ± 6 | Sulfoxide 34-P1, sulfone 34-P2 | ||
| 46 ± 3% ee (R) | 49 ± 1% ee (R) | 53 ± 2% ee (R) | |||||
| 35 | 2-Aminofluorene | 45 ± 2 | 39 ± 3 | 26 ± 10 | n.d. c | ||
| 36 | 2-Aminoanthracene | - | - | - | |||
| 37 | VII | Heterocycle | 4-Ipomeanol | 51 ± 6 | 26 ± 4 | 33 ± 2 | Furan epoxide d |
| 38 | Perilla ketone b | 81 ± 3 | 92 ± 2 | 94 ± 2 | Furan epoxide d | ||
| 39 | Indole | 46 ± 2 | 65 ± 1 | 18 ± <1 | Indoline-2-one 39-P1 | ||
| 40 | 3-Methylindole | 38 ± 8 | 47 ± 4 | 18 ± 7 | 3-Methyleneindolenine d | ||
| 41 | Myristicin a | 52 ± 2 | 38 ± 2 | 28 ± 1 | 5-Allyl-2,3-dihydroxyanisole 41-P1 | ||
| 42 | VIII | Stilbene/ stilbenoid | trans-Stilbene | 27 ± 3 | 77 ± 3 | 87 ± 3 | 4-OH 45, 3,4′-OH 42-P1, 4,4′-OH 42-P2 |
| 43 | trans-2-Hydroxystilbene | 40 ± 5 | 77 ± 5 | 47 ± 6 | 2,3′-OH 43-P1, 2,4′-OH 43-P2, 2,2′-OH 43-P3 | ||
| 44 | trans-3-Hydroxystilbene | 14 ± 1 | 26 ± 1 | 36 ± 5 | 3,3’-OH 44-P1, 3,4’-OH 42-P1 | ||
| 45 | trans-4-Hydroxystilbene | 5 ± 1 | 30 ± 4 | 31 ± 5 | 3,4’-OH 42-P1, 4,4’-OH 42-P2 | ||
| 46 | Pinosylvin | 6 ± 1 | 2 | <1 | Resveratrol 47 | ||
| 47 | Resveratrol | - | - | - | |||
| 48 | IX | Vanilloid | Capsaicin | 1 | 17 ± 2 | 16 ± 1 | 17-Hydroxycapsaicin 48-P1 |
| 49 | [6]-Gingerol | 0 | 70 ± <1 | 51 ± 4 | 10-Hydroxy-[6]-gingerol 49-P1 | ||
| 50 | Piperine a | <1 | 2 | 1 | n.d. c | ||
| 51 | X | Miscellaneous | Apigenin a | <1 | <1 | <1 | n.d. c |
| 52 | Genistein a | 11 ± 5 | 5 | 2 | n.d. c | ||
| 53 | Kaempferol a | <1 | <1 | <1 | n.d. c | ||
| 54 | Quercetin a | - | - | - | |||
| 55 | Flavone a | - | - | - | |||
| 56 | 7-Hydroxyflavone a | 3 | 2 | 1 | n.d. c | ||
| 57 | 5-,7-Dihydroxyflavone a | <1 | <1 | <1 | n.d. c | ||
| 58 | 6-,7-Dihydroxyflavone a | - | - | - | |||
| 59 | Cholecalciferol a | - | - | - | |||
| 60 | Testosterone a | - | - | - | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röder, A.; Hutter, M.C.; Heitzer, E.; Franz, P.J.; Hüsken, S.; Wiek, C.; Girhard, M. Exploration of CYP4B1 Substrate Promiscuity Across Three Species. Catalysts 2025, 15, 454. https://doi.org/10.3390/catal15050454
Röder A, Hutter MC, Heitzer E, Franz PJ, Hüsken S, Wiek C, Girhard M. Exploration of CYP4B1 Substrate Promiscuity Across Three Species. Catalysts. 2025; 15(5):454. https://doi.org/10.3390/catal15050454
Chicago/Turabian StyleRöder, Annika, Michael C. Hutter, Eva Heitzer, Pia Josephine Franz, Saskia Hüsken, Constanze Wiek, and Marco Girhard. 2025. "Exploration of CYP4B1 Substrate Promiscuity Across Three Species" Catalysts 15, no. 5: 454. https://doi.org/10.3390/catal15050454
APA StyleRöder, A., Hutter, M. C., Heitzer, E., Franz, P. J., Hüsken, S., Wiek, C., & Girhard, M. (2025). Exploration of CYP4B1 Substrate Promiscuity Across Three Species. Catalysts, 15(5), 454. https://doi.org/10.3390/catal15050454






