Enhanced Production, Purification, and Characterization of α-Glucosidase from NTG-Mutagenized Aspergillus niger for Industrial Applications
Abstract
1. Introduction
2. Results and Discussion
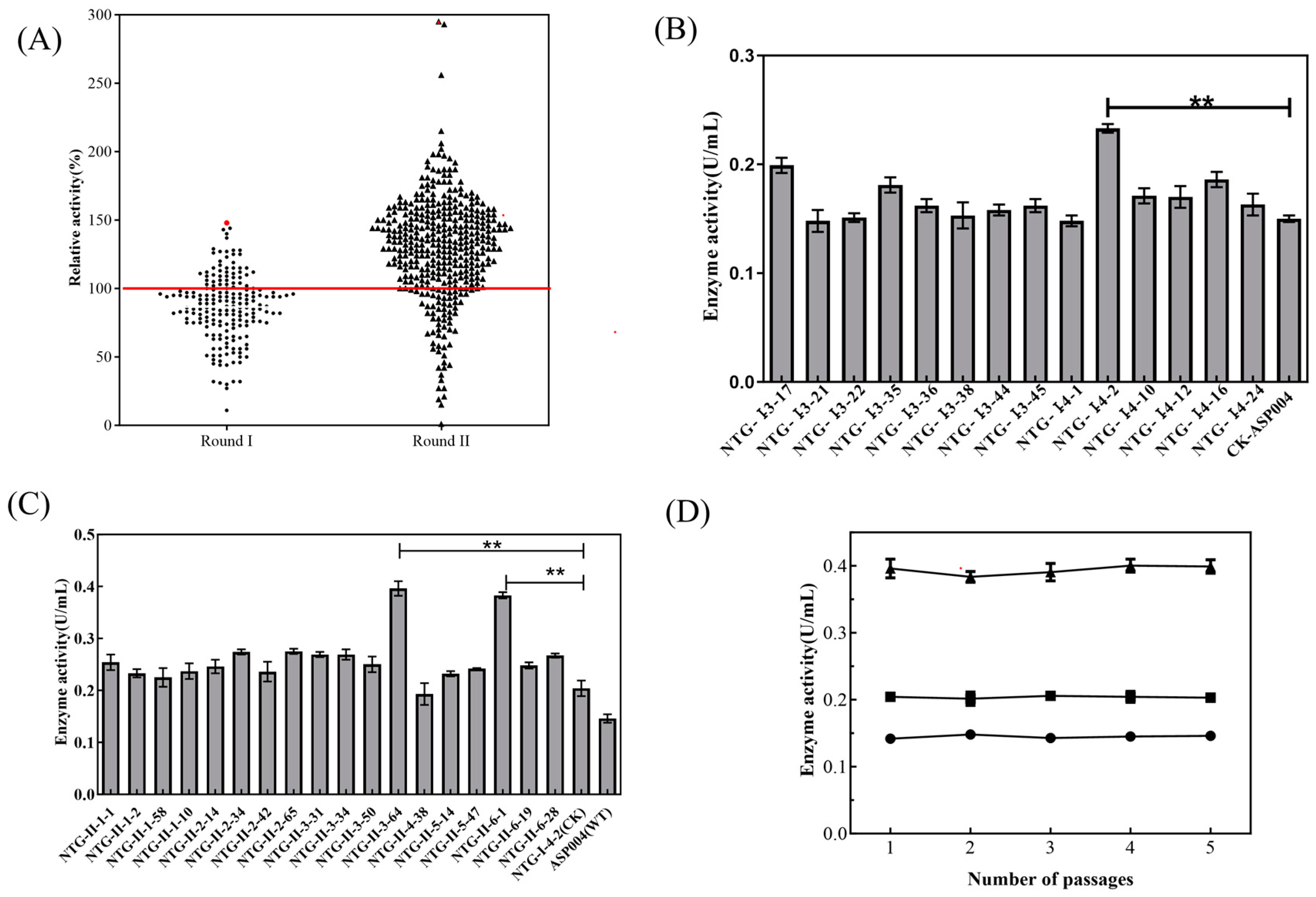
2.1. Mutagenesis and Screening of High-Yielding Strain
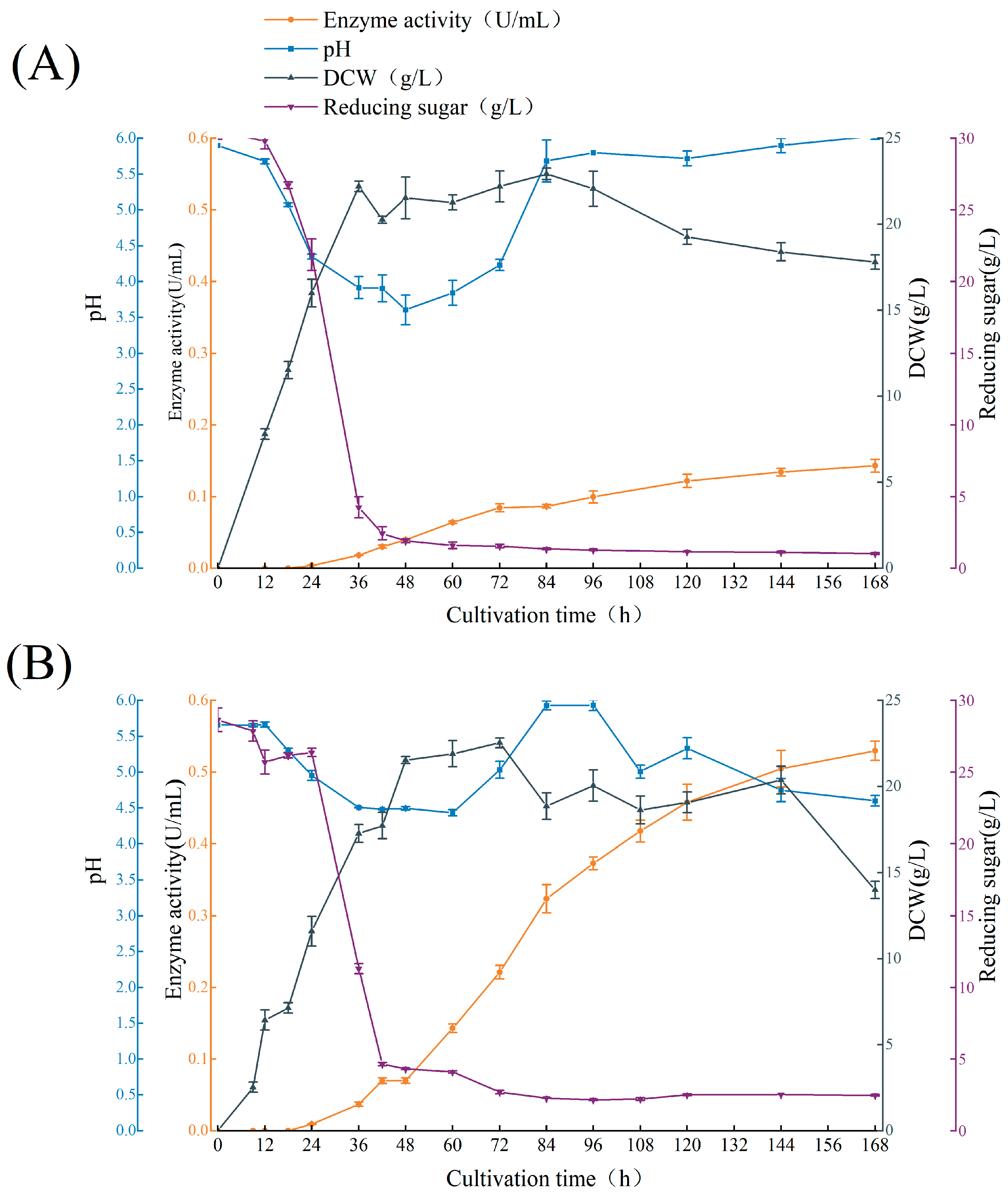
2.2. Batch Culture Profile of A. niger ASP004 and NTG-II-3-64
2.3. Isolation and Purification of α-Glucosidase
2.4. Enzymatic Properties of α-Glucosidase
2.4.1. Effect of Temperature and pH on α-Glucosidase Activity
2.4.2. Effect of Metal Ions and EDTA on α-Glucosidase Activity
2.4.3. Effect of Surfactants on α-Glucosidase Activity
2.4.4. Substrate Specificity of α-Glucosidase
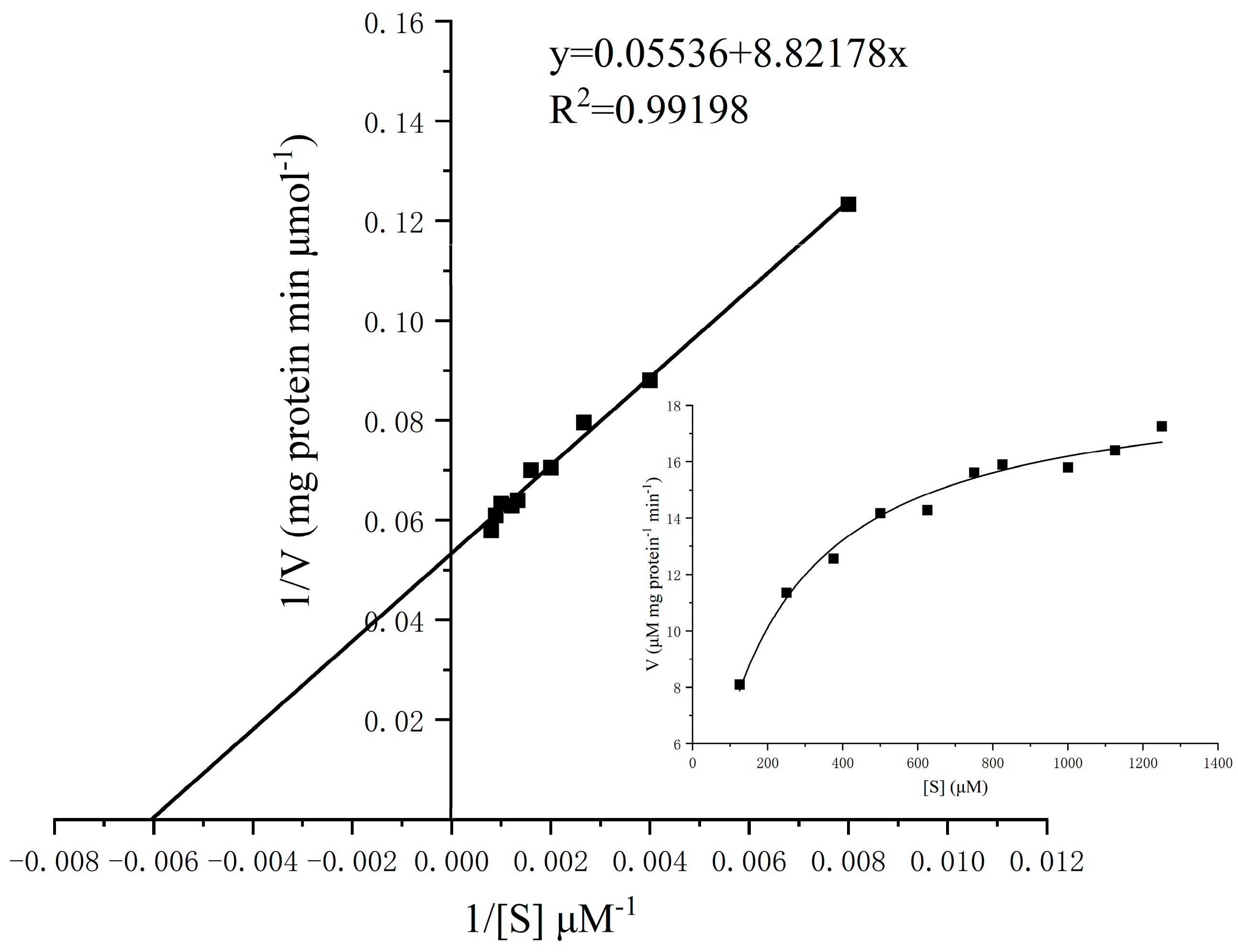
2.4.5. Kinetics Analysis of α-Glucosidase
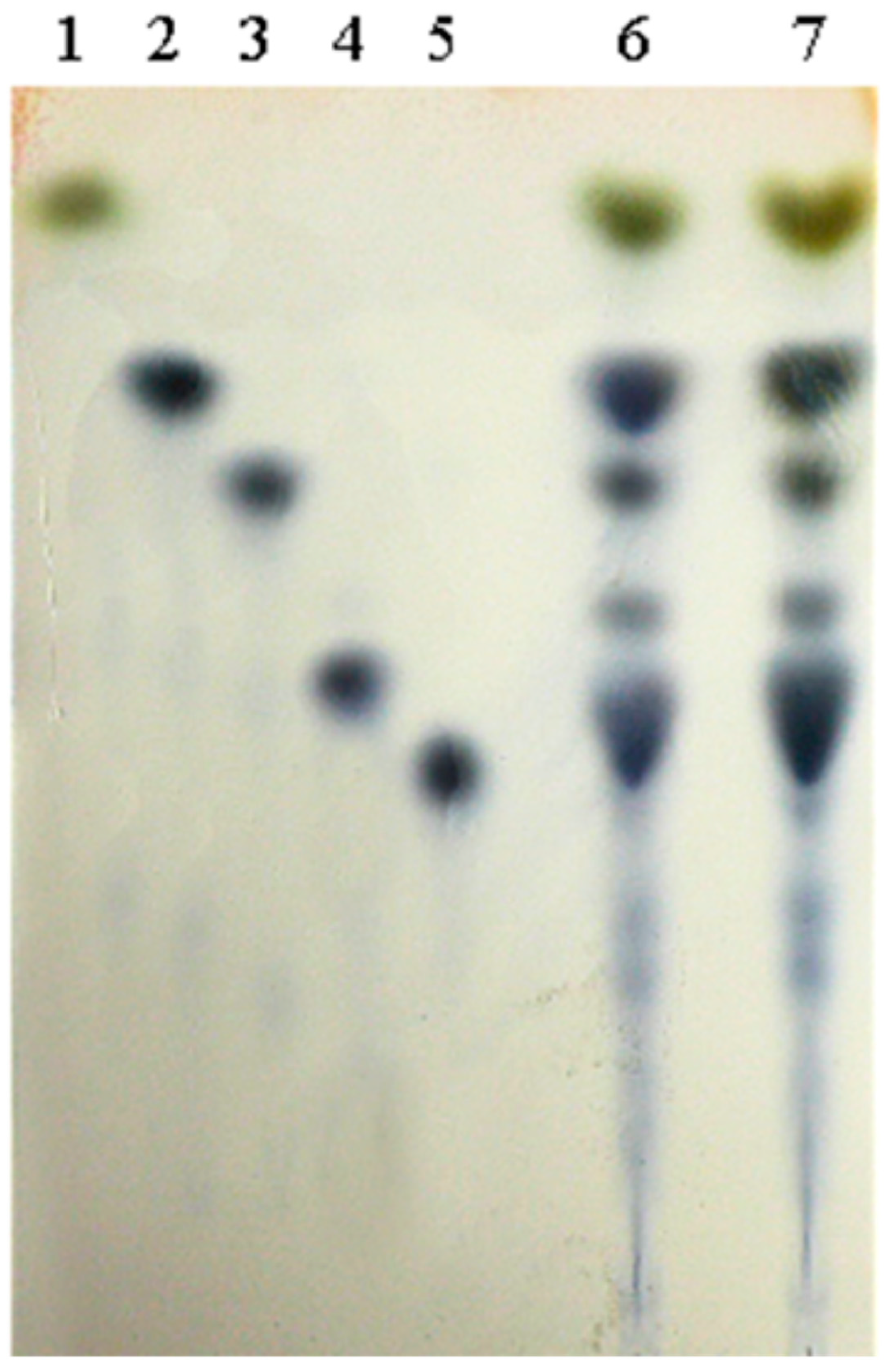
2.4.6. Detection of α-Glucosidase Transglycosidase Products
3. Materials and Methods
3.1. Strains, Chemicals and Medium
3.2. α-Glucosidase Activity Determination
3.3. Mutagenesis and Screening of High-Yield Strain of α-Glucosidase
3.4. Batch Culture Profile of High-Yield Strain of α-Glucosidase
3.5. Method of Isolation and Purification of α-Glucosidase
3.6. Enzyme Property Determination
3.6.1. Effect of Temperature and pH on Enzyme Activity
3.6.2. Effect of Metal Ions and EDTA on Enzyme Activity
3.6.3. Effect of Surfactants on Enzyme Activity
3.6.4. Substrate Specificity
3.6.5. Enzymatic Kinetic Analysis
3.6.6. Detection of α-Glucosidase Transglycosidase Products by Thin-Layer Chromatography (TLC)
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, B.; Huang, N.; Zeng, W.; Zhang, H.; Chen, G.; Liang, Z. Development of a strategy for the screening of α-glucosidase-producing microorganisms. J. Microbiol. 2020, 58, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Nimpiboon, P.; Nakapong, S.; Pichyangkura, R.; Ito, K.; Pongsawasdi, P. Synthesis of a novel prebiotic trisaccharide by a type I α-glucosidase from B. licheniformis strain TH4-2. Process Biochem. 2011, 46, 448–457. [Google Scholar] [CrossRef]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae. J. Biosci. Bioeng. 2011, 112, 545–550. [Google Scholar] [CrossRef]
- Okuyama, M.; Saburi, W.; Mori, H.; Kimura, A. α-Glucosidases and α-1, 4-glucan lyases: Structures, functions, and physiological actions. Cell. Mol. Life Sci. 2016, 73, 2727–2751. [Google Scholar] [CrossRef]
- Meissner, H.; Liebl, W. Thermotoga maritima maltosyltransferase, a novel type of maltodextrin glycosyltransferase acting on starch and malto-oligosaccharides. Eur. J. Biochem. 1998, 258, 1050–1058. [Google Scholar] [CrossRef]
- Basu, A.; Mutturi, S.; Prapulla, S.G. Modeling of enzymatic production of isomaltooligosaccharides: A mechanistic approach. Catal. Sci. Technol. 2015, 5, 2945–2958. [Google Scholar] [CrossRef]
- Um, H.E.; Park, B.R.; Kim, Y.M.; Lee, B.H. Slow digestion properties of long-sized isomaltooligosaccharides synthesized by a transglucosidase from Thermoanaerobacter thermocopriae. Food Chem. 2023, 417, 135892. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Basu, A.; Anu-Appaiah, K.A.; Gnanesh Kumar, B.S.; Mutturi, S. Identification and characterization of novel transglycosylating α-glucosidase from Aspergillus neoniger. J. Appl. Microbiol. 2020, 129, 1644–1656. [Google Scholar] [CrossRef]
- Song, Y.B.; Lamothe, L.M.; Rodriguez, N.E.; Rose, D.R.; Lee, B.H. New insights suggest isomaltooligosaccharides are slowly digestible carbohydrates, rather than dietary fibers, at constitutive mammalian α-glucosidase levels. Food Chem. 2022, 383, 132456. [Google Scholar] [CrossRef]
- Song, K.M.; Okuyama, M.; Kobayashi, K.; Mori, H.; Kimura, A. Characterization of a glycoside hydrolase family 31 α-glucosidase involved in starch utilization in Podospora anserine. Biosci. Biotechnol. Biochem. 2013, 77, 2117–2124. [Google Scholar] [CrossRef]
- Krohn, B.M.; Lindsay, J.A. Purification and characterization of a thermostable α-glucosidase from a Bacillus subtilis high-temperature growth transformant. Current Microbiol. 1991, 22, 273–278. [Google Scholar] [CrossRef]
- Marín, D.; Linde, D.; Fernández-Lobato, M. Purification and biochemical characterization of an α-glucosidase from Xanthophyllomyces dendrorhous. Yeast 2006, 23, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Larsbrink, J.; Izumi, A.; Hemsworth, G.R.; Davies, G.J.; Brumer, H. Structural enzymology of Cellvibrio japonicus Agd31B protein reveals α-transglucosylase activity in glycoside hydrolase family 31. J. Biol. Chem. 2012, 287, 43288–43299. [Google Scholar] [CrossRef]
- Nakamura, A.; Nishimura, I.; Yokoyama, A.; Lee, D.G.; Hidaka, M.; Masaki, H.; Kimura, A.; Chiba, S.; Uozumi, T. Cloning and sequencing of an α-glucosidase gene from Aspergillus niger and its expression in A. nidulans. J. Biotechnol. 1997, 53, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Nishio, T.; Minoura, K.; Uozumi, T.; Wada, M.; Hashimoto, N.; Kawachi, R.; Oku, T. Recombinant α-Glucosidase from Aspergillus niger. Overexpression by Emericella nidulans, Purification and Characterization. J. Appl. Glycosci. 2006, 53, 13–16. [Google Scholar] [CrossRef]
- Zhao, N.; Xu, Y.; Wang, K.; Zheng, S. Synthesis of isomalto-oligosaccharides by Pichia pastoris displaying the Aspergillus niger α-glucosidase. J. Agric. Food Chem. 2017, 65, 9468–9474. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, W.; Bah, F.B.; Song, C.; Zhou, Y.; Ji, L.; Yuan, Y. Heterologous expression of a thermostable α-glucosidase from Geobacillus sp. strain HTA-462 by Escherichia coli and its potential application for isomaltose–oligosaccharide synthesis. Molecules 2019, 24, 1413. [Google Scholar]
- Kumar, S.; Mutturi, S. Expression of a novel α-glucosidase from Aspergillus neoniger in Pichia pastoris and its efficient recovery for synthesis of isomaltooligosaccharides. Enzym. Microb. Technol. 2020, 141, 109653. [Google Scholar] [CrossRef] [PubMed]
- Tissopi, T.; Kumar, S.; Sadhu, A.; Mutturi, S. Surface display of novel transglycosylating α-glucosidase from Aspergillus neoniger on Pichia pastoris for synthesis of isomaltooligosaccharides. Biochem. Eng. J. 2022, 181, 108400. [Google Scholar] [CrossRef]
- Kimura, A.; Takenishi, S.; Takata, M.; Tsujisaka, Y.; Chiba, S. Properties and Substrate Specificity of a-Glucosidase from Aspergillus niger GRM 3. J. Appl. Glycosci. 1997, 44, 303–312. [Google Scholar]
- EFSA Panel on Food Contact Materials, Enzymes and Processing Aids (CEP); Silano, V.; Barat Baviera, J.M.; Bolognesi, C.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lambré, C.; Lampi, E.; et al. Safety evaluation of the food enzyme α-glucosidase from the Aspergillus niger strain AE-TGU. EFSA J. 2022, 20, e07240. [Google Scholar] [PubMed]
- Li, W.; Chen, G.; Gu, L.; Zeng, W.; Liang, Z. Genome Shuffling of Aspergillus niger for Improving Transglycosylation Activity. Appl. Biochem. Biotechnol. 2014, 172, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.G.; Li, W.; Zhang, Y.K.; Qin, Y.L.; Wu, K.Y.; Liang, Z.Q. A high-throughput method for screening of Aspergillus niger mutants with high transglycosylation activity by detecting non-fermentable reducing sugar. World J. Microbiol. Biotechnol. 2011, 27, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Ko, E.H.; Lee, J.S.; Kim, S.W. Over-production of β-glucosidase by Aspergillus niger mutant from lignocellulosic biomass. Biotechnol. Lett. 1999, 21, 647–650. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Q.; Zhou, Y.; Lu, Z.; Zhang, G.; Ma, Y. A new GH13 α-glucosidase from alkaliphilic Bacillus pseudofirmus 703 with both exo-α-l, 4-glucosidase and oligo-l, 6-glucosidase activities toward amylopectin. Int. J. Biol. Macromol. 2017, 101, 973–982. [Google Scholar] [CrossRef]
- Krohn, B.M.; Lindsay, J.A. Purification, characterization, and comparison of an α-glucosidase (endo-oligo-1, 4-glucosidase) from the mesophile Bacillus subtilis and the obligate thermophile Bacillus caldolyticus. Curr. Microbiol. 1991, 22, 133–140. [Google Scholar] [CrossRef]
- Mccleary, B.V.; Gibson, T.S.; Sheehan, H.; Casey, A.; Horgan, L.; O’Flaherty, J. Purification, properties, and industrial significance of transglucosidase from Aspergillus niger. Carbohydr. Res. 1989, 185, 147–162. [Google Scholar] [CrossRef]
- Kato, N.; Suyama, S.; Shirokane, M.; Kato, M.; Kobayashi, T.; Tsukagoshi, N. Novel α-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl. Environ. Microbiol. 2002, 68, 1250–1256. [Google Scholar] [CrossRef]
- Sheu, D.C.; Duan, K.J.; Lin, C.T. Purification and characterization of α-glucosidase from Aspergillus carbonarious. Biotechnol. Tech. 1994, 8, 515–520. [Google Scholar] [CrossRef]
- Botstein, D.; Jones, E.W. Nonrandom Mutagenesis of the Escherichia coli Genome by Nitrosoguanidine. J. Bacteriol. 1969, 98, 847–848. [Google Scholar] [CrossRef]
- Oeschger, M.P.; Berlyn, M.K.B. A simple procedure for localized mutagenesis using nitrosoguanidine. Mol. Gen. Genet. MGG 1974, 134, 77–83. [Google Scholar] [CrossRef]
- Anindyawati, T.; Ann, Y.G.; Ito, K.; Iizuka, M.; Minamiura, N. Two kinds of novel a-glucosidases from Aspergillus awamori KT-11: Their purifications, properties and specificities. J. Ferment. Bioeng. 1998, 85, 465–469. [Google Scholar] [CrossRef]
- Ma, M.; Okuyama, M.; Sato, M.; Tagami, T.; Klahan, P.; Kumagai, Y.; Mori, H.; Kimura, A. Effects of mutation of Asn694 in Aspergillus niger α-glucosidase on hydrolysis and transglucosylation. Appl. Microbiol. Biotechnol. 2017, 101, 6399–6408. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Lee, S.L.; Tung, P.Y. Effects of initial maltose concentration, agitation and aeration rates on the production of glucosyltransferase by Aspergillus niger CCRC 31494. Biotechnol. Lett. 1996, 18, 489–494. [Google Scholar] [CrossRef]
- Yan, Q.J.; Han, P.; Yang, S.Q.; Jiang, Z.Q. Purification and characterization of a novel α-glucosidase from Malbranchea cinnamomea. Biotechnol. Lett. 2015, 37, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- del Moral, S.; Barradas-Dermitz, D.M.; Aguilar-Uscanga, M.G. Production and biochemical characterization of α-glucosidase from Aspergillus niger ITV-01 isolated from sugar cane bagasse. 3 Biotech 2018, 8, 7. [Google Scholar] [CrossRef]
- Kita, A.; Matsui, H.; Somoto, A.; Takata, M.; Chiba, S. Substrate Specificity and Subsite Affinities of Crystalline α-glucosidase from Aspergillus niger. Agric. Biol. Chem. 1991, 55, 2327–2335. [Google Scholar] [CrossRef]
- Piller, K.; Daniel, R.M.; Petach, H.H. Properties and stabilization of an extracellular α-glucosidase from the extremely thermophilic archaebacteria Thermococcus strain AN 1: Enzyme activity at 130 °C. Biochim. Biophys. Acta 1996, 1292, 197–205. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Li, W.; Wu, K.Y.; Chen, G.G.; Liang, Z.Q. Purification and characterization of an intracellular α-glucosidase with high transglycosylation activity from Aspergillus niger M-1. Prep. Biochem. Biotechnol. 2011, 41, 201–217. [Google Scholar] [CrossRef]
- Chen, D.L.; Tong, X.; Chen, S.W.; Chen, S.; Wu, D.; Fang, S.G.; Wu, J.; Chen, J. Heterologous expression and biochemical characterization of α-glucosidase from Aspergillus niger by Pichia pastroris. J. Agric. Food Chem. 2010, 58, 4819–4824. [Google Scholar] [CrossRef]
- Yoon, S.H.; Robyt, J.F. Activation and stabilization of 10 starch-degrading enzymes by Triton X-100, polyethylene glycols, and polyvinyl alcohols. Enzym. Microb. Technol. 2005, 37, 556–562. [Google Scholar] [CrossRef]
- Carvalho, A.F.A.; Boscolo, M.; da Silva, R.; Ferreira, H.; Gomes, E. Purification and characterization of the α-glucosidase produced by thermophilic fungus Thermoascus aurantiacus CBMAI 756. J. Microbiol. 2010, 48, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Wu, D.; Hu, N.; Li, N.; Dai, C.; Chen, X.; Suo, Y.; Li, G.; Wu, Y. Robust hybrid enzyme nanoreactor mediated plasmonic sensing strategy for ultrasensitive screening of anti-diabetic drug. Biosens. Bioelectron. 2018, 99, 653–659. [Google Scholar] [CrossRef] [PubMed]




| Steps | Total Protein (mg) | Total Enzyme Activity (U) | Specific Enzyme Activity (U·mg−1) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|
| Crude enzyme | 375.34 | 16.82 | 0.045 | 100 | 1 |
| Ultrafiltration | 112.01 | 10.13 | 0.090 | 60.23 | 2.00 |
| Ethanol precipitation | 49.75 | 9.24 | 0.186 | 54.93 | 4.13 |
| DEAE—fast flow | 5.58 | 4.85 | 0.869 | 28.83 | 19.31 |
| Sephacryl S-200 | 2.57 | 2.57 | 1.0 | 15.28 | 22.22 |
| Peak 1 | Peak 2 | Peak 3 | Peak 4 | |
|---|---|---|---|---|
| Enzyme activity (U·mL−1) | 0.485 | 0.058 | 0.028 | 0.052 |
| Total enzyme activity (U) | 2.18 | 0.18 | 0.09 | 0.47 |
| Total protein (mg) | 2.51 | 1.48 | 1.96 | 10.08 |
| Specific enzyme activity (U·mg−1) | 0.87 | 0.12 | 0.04 | 0.05 |
| Peak 1 | Peak 2 | Peak 3 | |
|---|---|---|---|
| Enzyme activity (U·mL−1) | 0 | 0.002 | 0.23 |
| Total enzyme activity (U) | 0.003 | 0.006 | 2.57 |
| Total protein (mg) | 0.006 | 0.045 | 2.57 |
| Specific enzyme activity (U·mg−1) | 0 | 0.13 | 1.0 |
| Compound | Concentration (mmol·L−1) | Residual Activity (%) |
|---|---|---|
| Control | 0 | 100 |
| KCl | 1 | 85.8 ± 2.3 ** |
| MgCl2 | 1 | 88.9 ± 3.2 ** |
| ZnCl2 | 1 | 85.8 ± 2.4 ** |
| Mncl2 | 1 | 90.9 ± 1.2 * |
| NaCl | 1 | 98.2 ± 1.7 |
| LiCl | 1 | 93.4 ± 4.4 |
| CaCl2 | 1 | 109 ± 2 * |
| CoCl2 | 1 | 91.6 ± 2.8 |
| FeCl3 | 1 | 76.1 ± 3.2 ** |
| CuCl2 | 1 | 80.1 ± 2.9 ** |
| EDTA | 20 | 110 ± 2 * |
| 40 | 102 ± 5.13 | |
| 60 | 93 ± 4.35 * | |
| 80 | 98.6 ± 5.03 | |
| 100 | 99.5 ± 5.5 |
| Surfactants | Concentration (%) | Residual Activity (%) |
|---|---|---|
| Control | 0 | 100 |
| Tween 20 | 1.0 | 116 ± 2.2 ** |
| Tween 40 | 1.0 | 117 ± 4.8 * |
| Tween 80 | 1.0 | 120 ± 2.3 ** |
| Span 80 | 1.0 | 131 ± 0.7 *** |
| Triton X-100 | 1.0 | 122 ± 4.3 ** |
| SDS | 1.0 | 50 ± 0.8 *** |
| PEG 2000 | 1.0 | 127 ± 0.67 *** |
| Substrate | Relative Activity (%) |
|---|---|
| maltose | 100 |
| α-D-glucopyranoside | 16.8 ± 0.19 *** |
| sucrose | 60.5 ± 1.3 ** |
| lactose | 0 |
| soluble starch | 59.1 ± 1.2 ** |
| isomaltose | 38.2 ± 2.3 *** |
| panose | 39.4 ± 2.8 *** |
| isomaltotriose | 38.8 ± 1.2 *** |
| 4-Nitrophenyl β-D-glucuronide | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, B.; Liu, Q.; Xiong, J.; Feng, G.; Chang, Z.; Gao, H. Enhanced Production, Purification, and Characterization of α-Glucosidase from NTG-Mutagenized Aspergillus niger for Industrial Applications. Catalysts 2025, 15, 450. https://doi.org/10.3390/catal15050450
Yao B, Liu Q, Xiong J, Feng G, Chang Z, Gao H. Enhanced Production, Purification, and Characterization of α-Glucosidase from NTG-Mutagenized Aspergillus niger for Industrial Applications. Catalysts. 2025; 15(5):450. https://doi.org/10.3390/catal15050450
Chicago/Turabian StyleYao, Bowei, Qian Liu, Junjie Xiong, Guangming Feng, Zhongyi Chang, and Hongliang Gao. 2025. "Enhanced Production, Purification, and Characterization of α-Glucosidase from NTG-Mutagenized Aspergillus niger for Industrial Applications" Catalysts 15, no. 5: 450. https://doi.org/10.3390/catal15050450
APA StyleYao, B., Liu, Q., Xiong, J., Feng, G., Chang, Z., & Gao, H. (2025). Enhanced Production, Purification, and Characterization of α-Glucosidase from NTG-Mutagenized Aspergillus niger for Industrial Applications. Catalysts, 15(5), 450. https://doi.org/10.3390/catal15050450







