Abstract
The efficient production of γ-aminobutyric acid (GABA) at a neutral pH remains a challenge due to the pH sensitivity of glutamate decarboxylase (GAD) enzymes. Our study addressed this limitation by identifying and engineering GAD enzymes with high activity under neutral conditions. Through gene mining, we discovered a wild-type GAD from Enterococcus faecalis (EfGAD) with high activity at pH 7.0 and, using zero-shot (ZS) predictor-guided mutagenesis and C-terminal truncation, we developed an EfGAD variant with a significantly enhanced catalytic efficiency. This variant demonstrated a 1.3-fold increase in GABA production (~300 g/L) from monosodium glutamate (MSG) compared to the wild-type EfGAD in 5 L bioreactor experiments. The ability to operate at a neutral pH without the need for acidic conditions reduces production costs and facilitates scalability. Our findings underscore the potential of integrating machine learning tools for enzyme optimization and provide a sustainable approach to GABA biosynthesis using MSG as a substrate.
1. Introduction
γ-Aminobutyric acid (GABA) is a non-proteinogenic amino acid with essential physiological roles in microorganisms, plants, and animals. It serves as the main inhibitory neurotransmitter in the mammalian central nervous system and offers various health benefits, including lowering blood pressure, treating epilepsy, and promoting sleep [1,2,3,4]. Beyond its medical applications, GABA is a key monomer in the synthesis of biodegradable polymers such as nylon-4, making it valuable in both pharmaceutical and industrial sectors. Given its diverse applications, there is a substantial demand for GABA, necessitating advancements in its efficient and sustainable production [5].
GABA is biosynthesized through enzymatic or whole-cell catalysis, as well as direct microbial fermentation. Glutamate decarboxylase (GAD; EC 4.1.1.15), a pyridoxal 5′-phosphate (PLP)-dependent enzyme, catalyzes the conversion of L-glutamate or monosodium glutamate (MSG) to GABA (Scheme 1) [1]. However, most natural GAD enzymes exhibit optimal activity under acidic conditions (pH 4.0 to 5.0) and lose activity at pH levels above 6.0 [3]. This pH sensitivity poses a challenge since the optimal pH for growth and L-glutamate synthesis in bacteria like Corynebacterium glutamicum is neutral. The following two-stage pH control strategy is commonly employed to address such pH discrepancies: initially setting the pH at 7.0 to promote cellular growth and glutamate synthesis, then adjusting it to below 5.5 to activate GAD and facilitate GABA production [6,7,8,9]. However, this approach requires a substantial acid supplementation, increasing production costs and risking equipment corrosion.
Scheme 1.
γ-Aminobutyric acid (GABA) production from L-glutamate by glutamate decarboxylase (GAD) with pyridoxal 5′-phosphate (PLP) as a cofactor.
An alternative strategy involves enzyme engineering to obtain GADs that are active in the neutral pH range. For instance, GadB from Lactobacillus plantarum (LpGAD) exhibits an optimal pH of 5.0, while its truncated variant, LpGADΔC11, retains notable activity at a near-neutral pH, with 18.22% and 7.79% of its peak activity at pH 6.5 and 7.0, respectively [4]. Similarly, an engineered strain of C. glutamicum expressing the Escherichia coli GAD (EcGAD) mutant E89Q/Δ452–466 demonstrates a significant enhancement in GABA production at pH 7.0, achieving a 17-fold increase in the GABA yield (5.89 g/L) compared to the wild-type GAD strain (0.34 g/L) [10]. L-glutamate is commonly used as the starting material in enzymatic or whole-cell catalysis. Although MSG offers advantages such as lower cost, abundance, and superior water solubility, its use in GAD-catalyzed reactions is limited by its near-neutral pH, which significantly reduces GAD enzyme activity [2]. Thus, discovering and engineering GADs active at a neutral pH would not only enable the use of MSG as a starting material in biocatalysis, but also allow for microbial fermentation processes to proceed at a neutral pH for cell growth, L-glutamate synthesis, and GABA production, thereby eliminating the need for acidic pH adjustments [2].
Traditional enzyme engineering strategies encompass directed evolution and rational design [11,12,13]. Recently, advancements in artificial intelligence have led to the integration of machine learning (ML) tools to enhance enzyme catalytic properties [14,15,16,17]. Among these tools, zero-shot (ZS) predictors are gaining attention for their ability to forecast properties (e.g., kinetic parameters, stability, or fitness) of enzyme mutants without requiring additional labeled experimental data for the target enzyme [18,19,20,21,22]. This approach allows for the virtual screening of vast mutant libraries to design variants with enhanced characteristics. For example, ZS predictors like UniKP and DLKcat can predict the turnover number (kcat) of enzyme variants, Pythia can estimate the stability change (∆∆G) caused by amino acid mutations [23,24,25], and the protein language model ESM-1v can predict the effects of mutations on protein function [20,26]. These methodologies accelerate enzyme engineering and reduce experimental costs. To date, no studies have been reported utilizing ZS predictors for engineering GAD enzymes.
In this study, we identified five new GADs through gene mining, including a wild-type GAD from Enterococcus faecalis (EfGAD) with high activity at pH 7.0. By applying ZS-predictor-guided site-directed mutagenesis (SDM) and C-terminal truncation, we generated a variant, EfGAD Y39T/Δ460–469, with a significantly enhanced substrate conversion rate at pH 7.0. We evaluated the effects of reaction conditions on MSG bioconversion catalyzed by both EfGAD and its mutant Y39T/Δ460–469. Finally, we compared the performance of E. coli cells expressing EfGAD or Y39T/Δ460–469 in converting MSG to GABA in bioreactors. Our findings pave the way for an efficient GABA production system using MSG as a substrate.
2. Results
2.1. Screening Natural GADs with High Activity at a Neutral pH
A gene mining approach was employed to identify new GADs with high activity at a neutral pH, using the LpGAD protein sequence as a probe to search the non-redundant (nr) database. This initial BLAST search yielded approximately 3700 putative GADs with sequence identities over 50%. After filtering for sequence length, retaining only proteins between 450 and 470 amino acids, and removing redundant sequences using CD-HIT with a sequence identity threshold of 0.8, 205 protein sequences remained for phylogenetic analysis (Figure S1). From these, five representative GAD candidates with 50–71% sequence identity to LpGAD were selected (Table S1 and Figure S2), chemically synthesized, and cloned into the expression vector pET28a(+). Additionally, the genes for LpGADΔC11 and the EcGAD variant E89Q/Δ452–466 (Table S1), both active at pH 7.0, were synthesized as controls. These plasmids were transformed into E. coli BL21 (DE3) for overexpression (Figure S3).
The catalytic performance of E. coli whole cells expressing the different GADs was evaluated against MSG at pH 6.0 and 7.0. As shown in Figure 1A, among the five new GADs, the natural GAD from E. faecalis (EfGAD, accession no. QPF70652) exhibited the highest substrate conversion rate at both pH 6.0 and 7.0, slightly surpassing the reported LpGADΔC11, the EcGAD mutant E89Q/Δ452–466, and another highly active new enzyme, LaGAD from Ligilactobacillus aviaries. For all studied GADs, the substrate conversion was higher at pH 6.0 than at pH 7.0. Considering its highest catalytic activity at pH 7.0 and highly soluble expression, EfGAD was selected for further study.
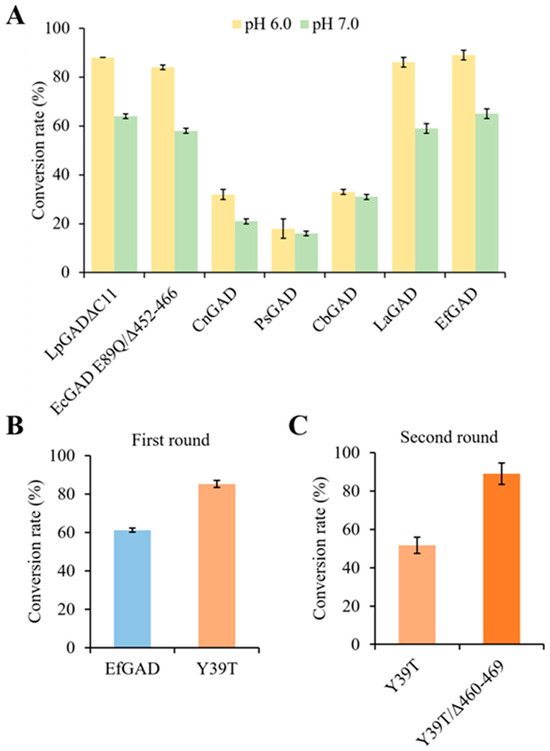
Figure 1.
Bioconversion of MSG catalyzed by GADs. (A) Catalytic performance of five new GAD candidates and two previously reported GAD mutants, LpGADΔC11 and the EcGAD variant E89Q/Δ452–466. Abbreviations: CnGAD, GAD from Companilactobacillus nuruki; PsGAD, GAD from Paucilactobacillus suebicus; CbGAD, GAD from Clostridium baratii; LaGAD, GAD from L. aviaries; and EfGAD, GAD from E. faecalis. The conversion rates of MSG (30 g/L) at pH 6.0 and 7.0 were determined after 5 h reactions. (B) Catalytic performance of EfGAD and best single variant Y39T in the first round of engineering. The conversion rates of MSG (30 g/L) at pH 7.0 were determined after 5 h reactions. (C) Catalytic performance of Y39T and truncated mutant Y39T/Δ460–469 in the second round of engineering. The conversion rates of MSG (40 g/L) at pH 7.0 were determined after 2 h reactions.
2.2. EfGAD Engineering for Improved Catalytic Performance at a Neutral pH
To enhance the catalytic performance of EfGAD towards MSG at pH 7.0, we conducted the following two rounds of enzyme engineering: (1) ZS predictor-guided design of single-point EfGAD variants and (2) C-terminal truncation. In the first round, pre-screening of EfGAD variants was performed at pH 7.0, followed by validation of the improved hits with three replicates. Subsequently, the C-terminal truncated mutant of the best single variant was experimentally constructed and evaluated.
Recent reports have highlighted the potential of ML tools for enabling zero-shot prediction of mutation effects, particularly in enhancing enzyme properties [18,19]. We first utilized four ZS predictors to computationally predict the mutation effects on EfGAD’s kinetic parameters (UniKP and DLKcat), stability (Pythia), and fitness (ESM-1v). Enzyme fitness refers to an enzyme’s ability to perform its biological function efficiently, with higher fitness generally indicating enhanced catalytic efficiency, stability, and overall performance, which are critical for its functionality in practical applications [27,28]. After virtually screening all of the 8911 theoretically possible single variants of EfGAD, the potentially beneficial variants identified by each ZS predictor were cross-referenced using Venny 2.1 to compile a list of variants shared by at least three ZS predictors (Figure S4). Consequently, 107 single EfGAD variants were selected and successfully cloned for experimental testing.
The conversion of MSG (30 g/L) catalyzed by wild-type EfGAD and its variants at pH 7.0 was determined after 5 h reactions. Out of 107 single variants (I47L, V462K, I362K, M185Y, M158F, Y13S, Y13E, Y13I, D16E, Y39T, W182F, W182H, S260T, F445L, and N41K), 15 with higher substrate conversion rates were selected for further validation with three replicates. As shown in Table S2, in contrast to the wild-type of 61.2 ± 1.1%, all 15 single variants showed higher substrate conversion rates. Remarkably, the best single variant, Y39T, exhibited a significantly higher conversion rate of 85.2 ± 1.8% (Figure 1B).
C-terminal truncation has been successfully used to extend the activities of several GADs towards a near-neutral pH [10,29,30]. For instance, truncating 15 residues at the C-terminus (Δ452–466) of EcGAD extended its active pH range by 0.5 units towards alkalinity [10]. Structural analysis revealed that residues 452–466 occupy the substrate-binding site at a neutral pH, obstructing substrate access [31]. By reducing steric hindrance from the C-terminal tail, the truncation likely enhances substrate accessibility and binding, thereby improving the EcGAD activity at a near-neutral pH. In addition, given the structural similarity between EfGAD and EcGAD (Figure S5), we hypothesized that truncating the C-terminal region of EfGAD could similarly enhance its activity under neutral pH conditions. Thus, in the second round of engineering, we constructed a C-terminal truncated mutant of the best single variant Y39T. Sequence alignment analysis of EfGAD and EcGAD (Figure S6) revealed that residues 460–469 of EfGAD correspond to residues 452–466 of EcGAD. Consequently, we constructed the EfGAD mutant Y39T/Δ460–469, which was anticipated to further enhance activity at a neutral pH. After a 2 h reaction at pH 7.0, the performance of the Y39T and Y39T/Δ460–469 mutants in converting 40 g/L of MSG was evaluated. The Y39T/Δ460–469 mutant demonstrated a significantly higher conversion rate of 89.0 ± 5.6%, compared to 51.7 ± 4.2% for the single variant Y39T (Figure 1C).
2.3. Effect of Reaction Conditions on MSG Bioconversion Catalyzed by EfGAD and Its Mutant
To investigate the effect of reaction conditions on the MSG bioconversion catalyzed by EfGAD and the Y39T/Δ460–469 mutant, we determined the MSG conversion rates across varying pH, temperatures, PLP concentrations, and cell amounts. As shown in Figure 2A, the EfGAD wild-type exhibited the highest conversion rate (98.0 ± 0.4%) at pH 6.0 and only converted 10.4 ± 1.8% of substrates at pH 7.5. In contrast, the Y39T/Δ460–469 mutant displayed high conversion rates between pH 6.0 and 7.0, achieving complete MSG conversion even at pH 7.0. Notably, a conversion rate of 61.0 ± 6.5% was observed at pH 7.5 for Y39T/Δ460–469. These results indicate that the Y39T/Δ460–469 mutant significantly improved the catalytic activities of EfGAD at a neutral pH. Figure 2B illustrates the MSG bioconversion at different temperatures, revealing that both EfGAD and the Y39T/Δ460–469 mutant maintained similar conversion rates between 45 °C and 55 °C. Regarding PLP concentrations, 0.2 mM proved sufficient for the MSG conversion catalyzed by both EfGAD and Y39T/Δ460–469 (Figure 2C). The amount of whole-cell catalyst significantly influenced the conversion rates, with a general trend indicating that increased catalyst amounts enhanced substrate conversion under the tested conditions until complete conversion was achieved (Figure 2D).
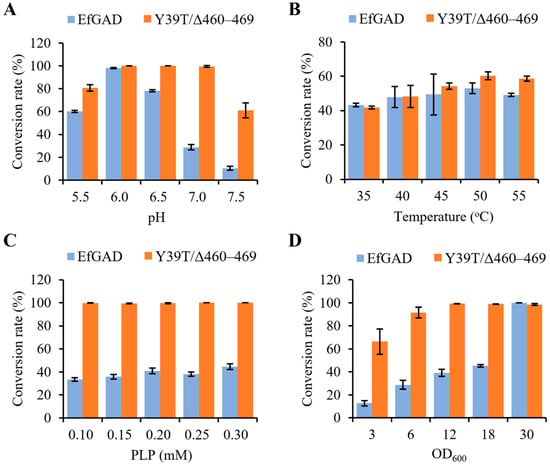
Figure 2.
Effect of reaction conditions on MSG bioconversion catalyzed by EfGAD and the mutant Y39T/Δ460–469. (A) The conversion rates of MSG (40 g/L) at 37 °C and different pH buffers (5.5, 6.0, 6.5, 7.0, and 7.5) were determined after 2 h reactions. (B) The conversion rates of MSG (40 g/L) at pH 6.0 and different temperatures (35, 40, 45, 50, and 55 °C) were determined after 20 min reactions. The OD600 of 6.0 and 3.0 was used for E. coli whole cells expressing EfGAD wild-type and Y39T/Δ460–469, respectively. (C) The conversion rates of MSG (40 g/L) at pH 6.0 and different PLP concentrations (0.10, 0.15, 0.20, 0.25, and 0.30 mM) were determined after 1 h reactions. (D) The conversion rates of MSG (40 g/L) at pH 6.0 and different amounts of recombinant E. coli whole cells (OD600 of 3.0, 6.0, 12.0, 18.0, and 30.0) were determined after 1 h reactions. More details are shown in Section 4.6.
2.4. Evaluation of EfGAD and Y39T/Δ460–469 for GABA Production in 5 L Fermenters
To evaluate and compare the catalytic performance of EfGAD and Y39T/Δ460–469 in GABA production, the biotransformation was performed in 5 L bioreactors using recombinant E. coli whole-cell catalysts (7 g DCW L−1) at a near-neutral pH (pH 6.0 for EfGAD and pH 6.5 for Y39T/Δ460–469) (Figure 3). During the first 2.5 h of the reaction, the Y39T/Δ460–469 mutant nearly completely consumed 1125× g of MSG, while 83.1 g/L of MSG remained for EfGAD. After multiple substrate additions, nearly 300 g/L of GABA was produced after 9 h using E. coli whole cells expressing the Y39T/Δ460–469 mutant, representing a 1.3-fold increase compared to the 226 g/L produced with the EfGAD wild-type. These results demonstrated that the Y39T/Δ460–469 mutant exhibits an outstanding catalytic performance at a near-neutral pH and has significant potential for GABA production in industrial applications, whether through biocatalysis or microbial fermentation, without the need for acidic pH adjustment.
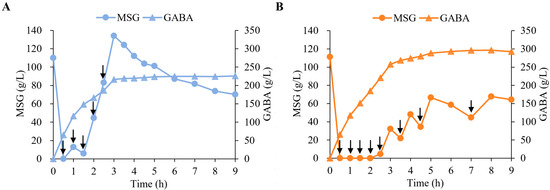
Figure 3.
Biotransformation of MSG to GABA using E. coli whole cells expressing EfGAD and the mutant Y39T/Δ460–469. The reaction mixture initially contained 1.5 L, with 7 g DCW (dry cell weight) L−1 of either EfGAD (A) or Y39T/Δ460–469 (B), 225 g of MSG, and 0.2 mM PLP. The reaction was carried out at 45 °C under near-neutral conditions (pH 6.0 for EfGAD and pH 6.5 for Y39T/Δ460–469). During the first 2.5 h, 225 g of MSG was added every 30 min for both EfGAD and Y39T/Δ460–469. Due to the rapid substrate consumption in the Y39T/Δ460–469-catalyzed reaction, additional MSG (150 g, 150 g, and 75 g) was supplied at 3.5, 4.5, and 7 h, respectively. Arrows indicate MSG addition time points.
3. Discussion
In this study, we successfully addressed the challenge of producing GABA at a neutral pH by identifying and engineering GAD enzymes with high catalytic activity under such conditions. Our discovery of EfGAD and LaGAD, both of which demonstrated comparable or superior performance to two existing engineered variants (LpGADΔC11 and the EcGAD mutant E89Q/Δ452–466) at pH 6.0 and 7.0, highlights the potential of naturally occurring GADs that are active in neutral environments. Notably, the engineered EfGAD variant Y39T/Δ460–469 exhibited a significant enhancement in MSG conversion efficiency, achieving a nearly 1.3-fold increase in GABA production compared to the wild-type EfGAD in 5 L bioreactor experiments. This improvement underscores the value of integrating ML tools, such as ZS predictors, to guide site-directed mutagenesis and enzyme optimization.
The C-terminal truncation (Δ460–469) likely enhances EfGAD’s catalytic performance by reducing steric hindrance from its flexible C-terminal tail, thereby improving substrate accessibility and binding. Structural studies of EcGAD [31], a homolog of EfGAD, support this hypothesis. As shown in Figure S7A, the crystal structure of EcGAD at neutral pH reveals that its C-terminal residues (452–466) extend into the active site funnel, potentially obstructing substrate access [31]. In contrast, at low pH, the electron density for these residues is absent, indicating increased flexibility. Experimental data further corroborate this, showing that truncating 15 C-terminal residues in EcGAD shifts its active pH range 0.5 units toward alkalinity [10]. Given the structural similarity between EfGAD and EcGAD (Figure S7B), the flexible C-terminus of EfGAD likely influences substrate access in a similar manner at neutral pH. Thus, the enhanced catalytic performance of the truncated EfGAD mutant at neutral pH is likely due to improved substrate accessibility. Additionally, the Y39T mutation may contribute to enhanced catalytic performance by influencing the enzyme’s oligomerization state. Previous research indicates that the N-terminal domain (residues 1–57) of EcGAD is crucial for hexamer formation [31]. It is plausible that the Y39T mutation in EfGAD similarly affects oligomerization, altering catalytic behavior at neutral pH. However, further structural analyses of the mutants are required to elucidate the underlying mechanisms.
The integration of ZS predictors represents a transformative advancement in enzyme engineering [19,20,22]. These tools enable the prediction of mutation effects on critical enzyme properties, such as kinetic parameters, stability, and overall fitness, without requiring additional labeled data for the target enzyme [20,21,23,24]. Notably, UniKP and DLKcat predict kcat by analyzing protein sequences and substrate information [23,24], while Pythia estimates stability changes (∆∆G) induced by mutations [25]. Additionally, ESM-1v, a protein language model, captures the functional consequences of mutations across diverse protein families, providing valuable insights into how mutations affect protein function and fitness [20,26]. While these tools predict the effects of amino acid mutations on various enzyme properties, they all contribute to the identification of improved enzyme variants. By identifying the top variants predicted by each ZS tool and cross-referencing them, we compiled a list of promising mutations that were consistently highlighted across multiple predictors. Experimental validation revealed that 15 out of 107 single EfGAD mutants displayed improved substrate conversion rates. Analysis of the top 500 predicted mutants from each ZS tool revealed that 9 of the 15 experimentally improved mutants were identified by Pythia, while ESM-1v, UniKP, and DLKcat identified 5, 2, and 1 improved mutants, respectively. As more experimental studies evaluate the predictive power of ZS tools, and as advanced ML algorithms continually refine these predictions, their accuracy is expected to improve. This progress will unleash even greater potential for ML in enzyme engineering, driving innovations in biocatalyst development.
Additionally, the ability of the EfGAD Y39T/Δ460–469 mutant to achieve high GABA yields at a neutral pH eliminates the need for the acidic conditions typically required by natural GADs. This not only reduces production costs but also simplifies process scalability, making it more commercially viable. Our findings provide a framework for the continued optimization of GAD enzymes and pave the way for more sustainable and cost-effective GABA production using MSG as a substrate. By combining ML-guided predictions with traditional experimental methods, this approach opens up new opportunities for the discovery and optimization of industrially relevant biocatalysts.
4. Materials and Methods
4.1. Strains, Plasmids, and Chemicals
The pET-28a(+) vector was used in this study for cloning and expression purposes. Chemically competent E. coli DH5 and E. coli BL21 (DE3) were purchased from TransGen Biotech (Beijing, China). All chemicals were analytical grade or higher and purchased from Sigma-Aldrich (St. Louis, MO, USA) and AppliChem (Darmstadt, Germany) unless otherwise specified. The primer STAR Max DNA Polymerase, KOD Plus Neo DNA Polymerase, and restriction enzyme DpnI were acquired from Takara (Shanghai, China). The T5 Exonuclease was obtained from New England Biolabs (Beverley, MA, USA). All primers were purchased from Genecreate (Wuhan, China).
4.2. Cloning Glutamate Decarboxylase-Encoding Gene
The genes encoding the two previously reported GAD variants, LpGADΔC11 and EcGAD E89Q/Δ452–466, were chemically synthesized (Genewiz, Suzhou, China) and each ligated separately into the NdeI and HindIII restriction sites of the pET-28a(+) vector. Additionally, five putative GAD genes (Table S1) identified through gene mining were also chemically synthesized and ligated into the same vector at identical restriction sites. All seven gene products fused with His-tag at their N-terminal. In addition, the gene for EfGAD with an Δ460–469 mutation was prepared by mutating the codon encoding residue 460 to a stop codon. The resulting plasmids were then transformed into E. coli BL21 (DE3) cells for expression.
4.3. ZS Prediction-Guided Engineering of EfGAD
Four ML tools were utilized to enable ZS prediction of mutation effects on enzyme properties or functions. Specifically, UniKP and DLKcat were employed to predict the turnover number (kcat) of enzyme variants [23,24]. Pythia (https://pythia.wulab.xyz, accessed on 7 December 2023) was used to predict the stability (∆∆G) of enzyme variants [25], and ESM-1v (https://www.findproteinstar.com/Zeroshot, accessed on 8 December 2023) to predict enzyme function [26]. To identify potentially beneficial EfGAD variants, all 8911 theoretically possible single variants (469 residues × 19 amino acids) were virtually screened using each ZS predictor. The results were cross-referenced using Venny 2.1 (http://www.liuxiaoyuyuan.cn/, accessed on 15 December 2023) to compile a list of variants predicted to be beneficial by at least three of the four ZS predictors. These potentially beneficial variants were then selected and cloned for experimental testing. The primers used are summarized in Table S3.
4.4. Expression of GADs
The positive transformants were cultivated in TB medium supplemented with kanamycin (50 µg/mL) at 37 °C and shaken at 220 rpm. When the culture reached an OD600 of 0.6–0.8, isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, followed by incubation at 28 °C for an additional 20 h. Cells were then harvested by centrifugation (4 °C, 4000× g, 20 min) and used for biotransformation or expression analysis. The supernatants and cell pellets from E. coli cultures expressing GADs were analyzed by SDS-PAGE.
4.5. Conversion of MSG to GABA in Tubes Catalyzed by GADs
The recombinant E. coli cell pellets were resuspended in phosphate buffer (pH 7.0) to an OD600 of 6.0. During the initial round of engineering or screening of new GAD variants, the biotransformation reaction was performed in 10 mL Falcon tubes with 30 g/L MSG, 0.1 mM PLP, and a total volume of 5 mL. This reaction was conducted at 37 °C with shaking at 300 rpm for 5 h, after which the samples were collected to measure the residual MSG concentration. In the second round, the MSG concentration was increased to 40 g/L, and samples were taken after 2 h. MSG concentrations were measured using an SBA biosensor (Shandong Academy of Sciences, Jinan, China).
4.6. Effect of Reaction Conditions on the Catalytic Performance of GADs
To evaluate the effect of pH on the MSG (40 g/L) bioconversion catalyzed by EfGAD or the Y39T/Δ460–469 mutant, reactions were conducted in phosphate buffer (pH 6.0–7.5) or acetate buffer (pH 5.5) with 0.1 mM PLP at 37 °C for 2 h. The effect of temperature was assessed by carrying out reactions in phosphate buffer (pH 6.0) with 0.1 mM PLP across a temperature range of 35–55 °C for 20 min. The impact of PLP concentration on the MSG (40 g/L) bioconversion was tested in phosphate buffer (pH 6.0) at 37 °C with varying PLP concentrations (0.10–0.30 mM) over 1 h. To determine the effect of biocatalyst loading on the MSG conversion, reactions were performed in phosphate buffer (pH 6.0) with 0.1 mM PLP at 37 °C for 1 h, using different amounts of recombinant E. coli cells at OD600 values of 3.0, 6.0, 12.0, 18.0, and 30.0. For reactions examining the effect of temperature on the MSG conversion by the Y39T/Δ460–469 mutant, an OD600 of 3.0 was used; all other reactions were conducted at an OD600 of 6.0. The substrate conversion rates were calculated based on the initial and remaining MSG concentrations after the reaction. All experiments were performed in triplicate.
4.7. Whole-Cell Biotransformation in 5 L Fermenters
The recombinant E. coli whole-cell biocatalysts were prepared as follows: Seed cultures were grown overnight at 37 °C and 220 rpm in 250 mL shake flasks containing 30 mL TB medium with 50 μg/mL kanamycin. A 4% inoculum was transferred into 5 L bioreactors containing 1.5 L of fermentation medium (glucose, 10 g/L; yeast extract, 2 g/L; peptone, 1 g/L; KH2PO4, 13.5 g/L; (NH4)2HPO4, 4 g/L; MgSO4·7H2O, 1.4 g/L; C6H8O7·H2O, 1.7 g/L; and a solution of trace elements, 10 mL). The temperature was maintained at 37 °C, and the pH was automatically controlled at 7.0 using NH₄OH. Dissolved oxygen (DO) levels were held at 30% by adjusting the agitation speed. A concentrated glucose solution (500 g/L) was fed at appropriate rates to keep the glucose concentration between 3 and 5 g/L. When the OD600 reached 30, IPTG was added to a final concentration of 0.1 mM, and the cultures were grown for an additional 14–16 h at 25 °C. Cells were then harvested by centrifugation (4 °C, 4000× g, 30 min) and prepared for biotransformation.
The biotransformation of MSG to GABA was carried out in a 5 L fermenter (Shanghai BaoXing Bio-engineering Equipment Co., Ltd., Shanghai, China) at 45 °C using E. coli whole cells harboring either EfGAD or the EfGAD Y39T/Δ460–469 variant. The reaction mixture, with an initial volume of 1.5 L, contained 7 g DCW L−1 recombinant E. coli cells, 225 g MSG, and 0.2 mM PLP. The pH was controlled at 6.0 for EfGAD and 6.5 for EfGAD Y39T/Δ460–469 using 85% H3PO4. During the first 2.5 h, 225 g of MSG was added every 30 min for both EfGAD and Y39T/Δ460–469. For Y39T/Δ460–469, additional MSG (150 g, 150 g, and 75 g) was supplied at 3.5, 4.5, and 7 h, respectively, to match the rapid substrate consumption. Samples were periodically withdrawn to measure the MSG and GABA concentrations, with the MSG determined by the SBA biosensor (Shandong Academy of Sciences, Jinan, China) and GABA determined by HPLC.
The GABA concentrations were determined as follows: 5 mL of the sample solution was added to a 50 mL volumetric flask, followed by 5 mL of 0.05 mol/L sodium bicarbonate buffer and 2.5 mL of 1% 2,4-dinitrofluorobenzene acetonitrile solution. The mixture was heated at 60 °C for 1 h, then cooled. After cooling, 0.01 mol/L potassium dihydrogen phosphate solution was added to reach a final volume of 50 mL. The solution was filtered through a 0.22 μm membrane, and the filtrate was used for analysis. GABA analysis was performed using a Thermo U3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) with a Hypersil GOLD column (4.6 × 250 mm, 5 μm) and detection at 360 nm. A 10 μL sample was injected with a flow rate of 0.8 mL/min. The mobile phase consisted of A: 0.05 mol/L sodium acetate with 60 μL/L acetic acid and B: acetonitrile. The gradient elution was: 0–10 min, 10–30% B; 10–15 min, 30% B; 15–16 min, 30–10% B; and 16–30 min, 10% B.
5. Conclusions
In conclusion, this study successfully identified and engineered new GAD enzymes with high activity at a neutral pH, addressing a key limitation in GABA production. Through the integration of ML-guided mutagenesis and C-terminal truncation, we developed the EfGAD Y39T/Δ460–469 mutant, which significantly enhanced GABA yields in biotransformation using MSG as a substrate, achieving 1.3 times higher production (~300 g/L) than the wild-type EfGAD (226 g/L). This work not only demonstrates the potential of neutral-pH-active GADs for efficient GABA biosynthesis using MSG as the starting material but also highlights the power of ML in enzyme optimization, offering valuable insights for future biocatalytic and fermentation processes.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/catal14120905/s1, Table S1: Summary of enzymes in this study. Table S2: Substrate conversion of EfGAD wild-type and 15 single variants. Table S3: List of primers used in this study. Figure S1: Phylogenetic analysis of GAD from lactobacillus plantarum (LpGAD) and homologs. Figure S2: Pairwise identity matrices of five new enzymes and two previously reported GAD variants. Figure S3: SDS-PAGE analysis of five new enzymes and two previously reported GAD variants. Figure S4: Venny analysis of LpGAD variants predicted to be potentially beneficial by four zero-shot predictors. Figure S5: Sequence alignment of GAD from Enterococcus faecalis (EfGAD) and GAD from E. coli (EcGAD). Figure S6: Sequence alignment of GADs from E. faecalis (EfGAD) and E. coli (EcGAD). Figure S7: Superposition of EcGAD structures at different pH and predicted EfGAD structure.
Author Contributions
Conceptualization, J.Z.; methodology, L.M.; software, H.Y.; validation, L.M., Y.Z., J.L. and Z.Z.; formal analysis, Y.Z.; data curation, L.M.; writing—original draft preparation, L.M. and H.Y.; writing—review and editing, R.C. and J.Z.; visualization, J.L.; supervision, H.Y. and J.Z.; project administration, R.C.; and funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Innovation Talent Program of Hubei Province (No. 2023DJC122) and the National Key Research and Development Program of China (No. 2019YFA0906400).
Data Availability Statement
Data are contained within this article.
Conflicts of Interest
Y.Z., R.C., J.L., Z.Z. were employed by company Road Environment Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Yuan, H.; Wang, H.; Fidan, O.; Qin, Y.; Xiao, G.; Zhan, J. Identification of new glutamate decarboxylases from Streptomyces for efficient production of γ-aminobutyric acid in engineered Escherichia coli. J. Med. Biol. Eng. 2019, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Park, Y.-L.; Choi, T.-R.; Kim, H.J.; Song, H.-S.; Han, Y.-H.; Lee, S.M.; Park, S.L.; Lee, H.S.; Bhatia, S.K.; et al. Production of γ-aminobutyric acid from monosodium glutamate using Escherichia coli whole-cell biocatalysis with glutamate decarboxylase from Lactobacillus brevis KCTC 3498. Korean J. Chem. Eng. 2020, 37, 2225–2231. [Google Scholar] [CrossRef]
- Yarabbi, H.; Mortazavi, S.A.; Yavarmanesh, M.; Javadmanesh, A. Molecular cloning, gene overexpression and characterization of glutamate decarboxylase from Enterococcus faecium DO. LWT 2021, 148, 111699. [Google Scholar] [CrossRef]
- Lyu, C.; Yao, L.; Zhu, Q.; Mei, J.; Cao, Y.; Hu, S.; Zhao, W.; Huang, J.; Mei, L.; Yao, S.; et al. Reconstruction of the glutamate decarboxylase system in Lactococcus lactis for biosynthesis of food-grade γ-aminobutyric acid. Appl. Microbiol. Biotechnol. 2021, 105, 4127–4140. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-D.; Li, X.; Shi, H.-L.; Jia, Y.-Y.; Dong, Z.-X.; Jiao, Z.-J.; Wang, L.-F.; Xu, J.-H.; Yao, L.-G.; Kan, Y.-C. Efficient expression of novel glutamate decarboxylases and high level production of γ-aminobutyric acid catalyzed by engineered Escherichia coli. Int. J. Biol. Macromol. 2020, 160, 372–379. [Google Scholar] [CrossRef]
- Choi, J.W.; Yim, S.S.; Lee, S.H.; Kang, T.J.; Park, S.J.; Jeong, K.J. Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell Factories. 2015, 14, 21. [Google Scholar] [CrossRef]
- Yang, H.; Xing, R.; Hu, L.; Liu, S.; Li, P. Accumulation of γ-aminobutyric acid by Enterococcus avium 9184 in scallop solution in a two-stage fermentation strategy. Microb. Biotechnol. 2015, 9, 478–485. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, L.; Gao, Q.; Yu, S.M.; Li, L.; Gao, N.F. The two-step biotransformation of monosodium glutamate to GABA by Lactobacillus brevis growing and resting cells. Appl. Microbiol. Biotechnol. 2012, 94, 1619–1627. [Google Scholar] [CrossRef]
- Wen, J.; Sun, W.; Leng, G.; Li, D.; Feng, C.; Tian, Z.; Wang, X. Enhanced fermentative γ-aminobutyric acid production by a metabolic engineered Corynebacterium glutamicum. Biotechnol. Bioprocess Eng. 2024, 29, 129–140. [Google Scholar] [CrossRef]
- Thu Ho, N.A.; Hou, C.Y.; Kim, W.H.; Kang, T.J. Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness. J. Biosci. Bioeng. 2013, 115, 154–158. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, Q.; Wu, W.; Pu, Z.; Yu, H. Rational design of enzyme activity and enantioselectivity. Front. Bioeng. Biotechnol. 2023, 11, 1129149. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Liu, B.; Shen, Y.; Jing, K.; Savage, T.R. Protein engineering design from directed evolution to de novo synthesis. Biochem. Eng. J. 2021, 174, 108096. [Google Scholar] [CrossRef]
- Qu, G.; Li, A.; Acevedo-Rocha, C.G.; Sun, Z.; Reetz, M.T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. Int. Ed. 2020, 59, 13204–13231. [Google Scholar] [CrossRef] [PubMed]
- Siedhoff, N.E.; Schwaneberg, U.; Davari, M.D. Machine learning-assisted enzyme engineering. Meth. Enzymol. 2020, 643, 281–315. [Google Scholar]
- Kouba, P.; Kohout, P.; Haddadi, F.; Bushuiev, A.; Samusevich, R.; Sedlar, J.; Damborsky, J.; Pluskal, T.; Sivic, J.; Mazurenko, S. Machine learning-guided protein engineering. ACS Catal. 2023, 13, 13863–13895. [Google Scholar] [CrossRef]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef]
- Kang, L.; Tan, P.; Hong, L. Enzyme engineering in the age of artificial intelligence. ACS Synth. Biol. 2023, 4, 524–534. [Google Scholar]
- Wittmann, B.J.; Johnston, K.E.; Wu, Z.; Arnold, F.H. Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol. 2021, 69, 11–18. [Google Scholar] [CrossRef]
- Wittmann, B.J.; Yue, Y.; Arnold, F.H. Informed training set design enables efficient machine learning-assisted directed protein evolution. Cell Syst. 2021, 12, 1026–1045. [Google Scholar] [CrossRef]
- Meier, J.; Rao, R.; Verkuil, R.; Liu, J.; Sercu, T.; Rives, A. Language models enable zero-shot prediction of the effects of mutations on protein function. Adv. Neural inf. Process. Syst. 2021, 34, 29287–29303. [Google Scholar]
- Mansoor, S.; Baek, M.; Juergens, D.; Watson, J.L.; Baker, D. Zero-shot mutation effect prediction on protein stability and function using RoseTTAFold. Protein Sci. 2023, 32, e4780. [Google Scholar] [CrossRef]
- Cheng, P.; Mao, C.; Tang, J.; Yang, S.; Cheng, Y.; Wang, W.; Gu, Q.; Han, W.; Chen, H.; Li, S.; et al. Zero-shot prediction of mutation effects with multimodal deep representation learning guides protein engineering. Cell Res. 2024, 34, 630–647. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Deng, H.; He, J.; Keasling, J.D.; Luo, X. UniKP: A unified framework for the prediction of enzyme kinetic parameters. Nat. Commun. 2023, 14, 8211. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yuan, L.; Lu, H.; Li, G.; Chen, Y.; Engqvist, M.K.M.; Kerkhoven, E.J.; Nielsen, J. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat. Catal. 2022, 5, 662–672. [Google Scholar] [CrossRef]
- Sun, J.; Zhu, T.; Cui, Y.; Wu, B. Structure-based self-supervised learning enables ultrafast prediction of stability changes upon mutation at the protein universe scale. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yang, L.; Liang, X.; Zhang, N.; Lu, L. STAR: A web server for assisting directed protein evolution with machine learning. ACS Omega 2023, 8, 44751–44756. [Google Scholar] [CrossRef]
- Firnberg, E.; Labonte, J.W.; Gray, J.J.; Ostermeier, M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 2014, 31, 1581–1592. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef]
- Yu, K.; Lin, L.; Hu, S.; Huang, J.; Mei, L. C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH. Enzyme Microb. Technol. 2012, 50, 263–269. [Google Scholar] [CrossRef]
- Shin, S.-M.; Kim, H.; Joo, Y.; Lee, S.-J.; Lee, Y.-J.; Lee, S.J.; Lee, D.-W. Characterization of glutamate decarboxylase from Lactobacillus plantarum and its C-terminal function for the pH dependence of activity. J. Agric. Food Chem. 2014, 62, 12186–12193. [Google Scholar] [CrossRef]
- Capitani, G.; Biase, D.D.; Aurizi, C.; Gut, H.; Bossa, F.; Grütter, M.G. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003, 22, 4027–4037. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).