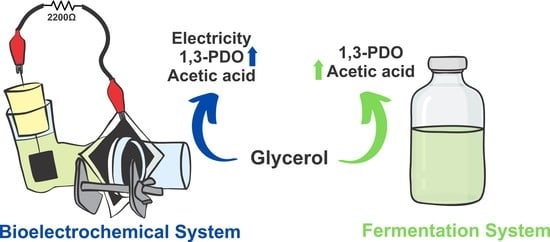
A New Pseudomonas aeruginosa Isolate Enhances Its Unusual 1,3-Propanediol Generation from Glycerol in Bioelectrochemical System
Abstract
1. Introduction
2. Results and Discussion
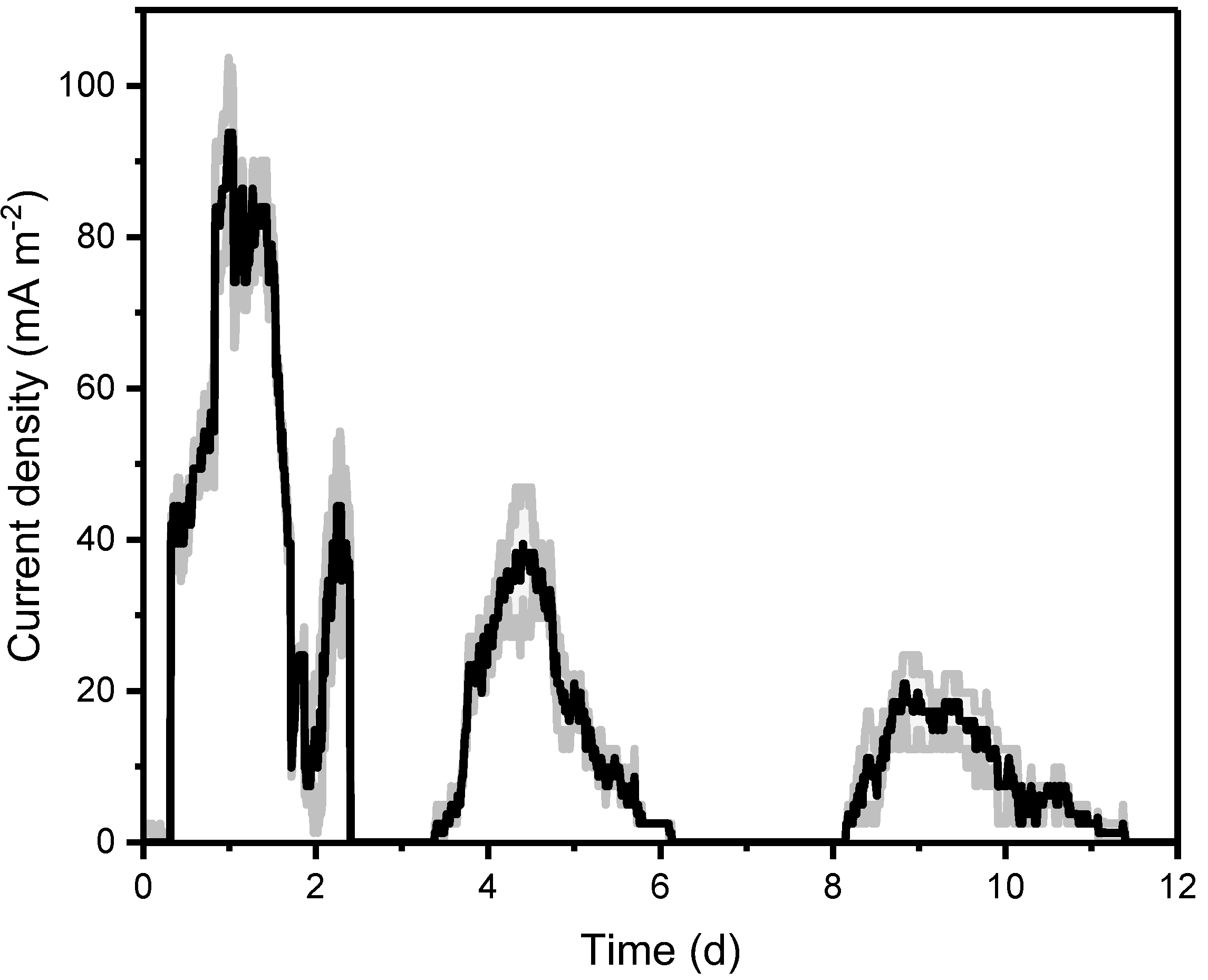
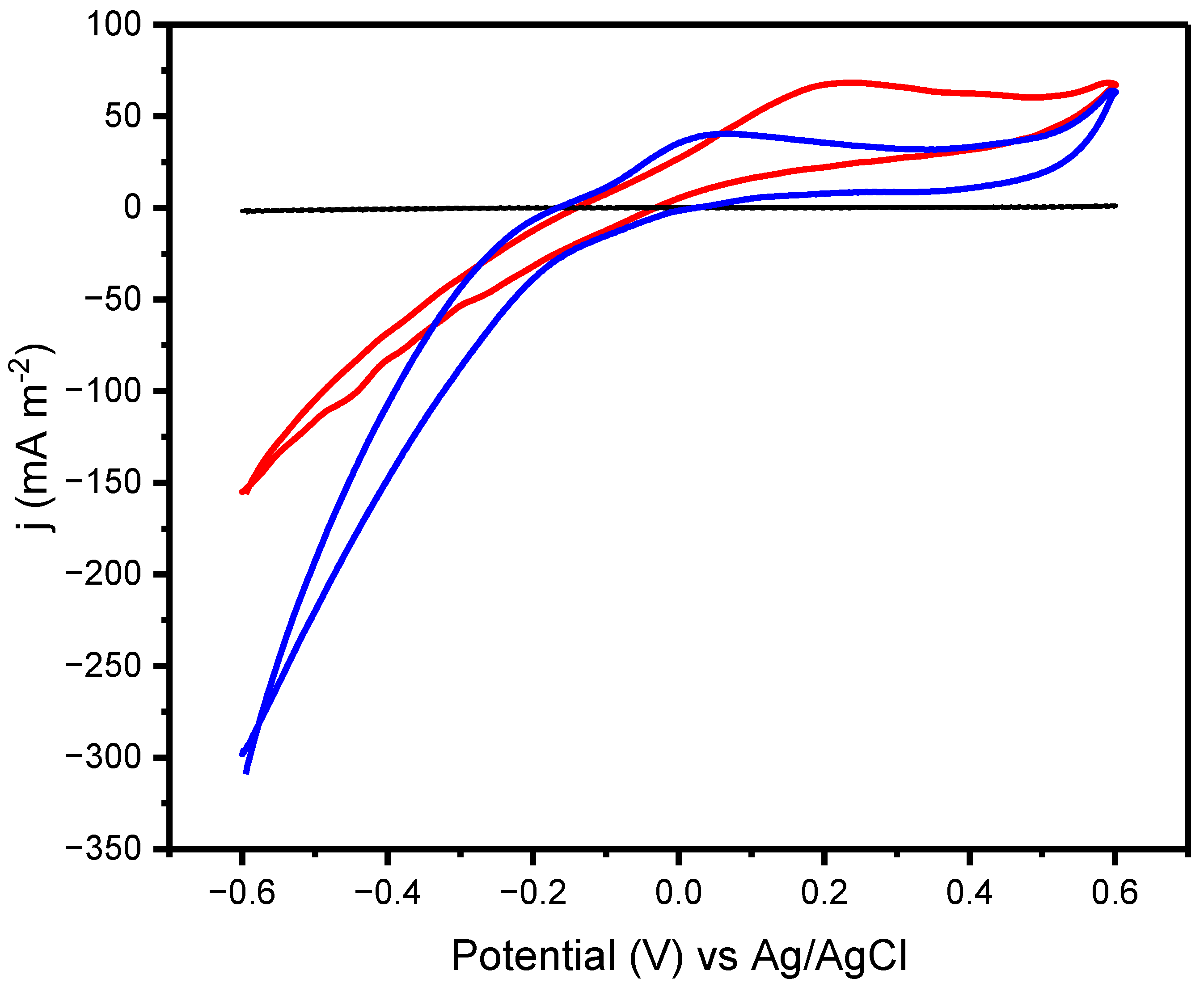
2.1. Electrochemical Activity of the Isolate
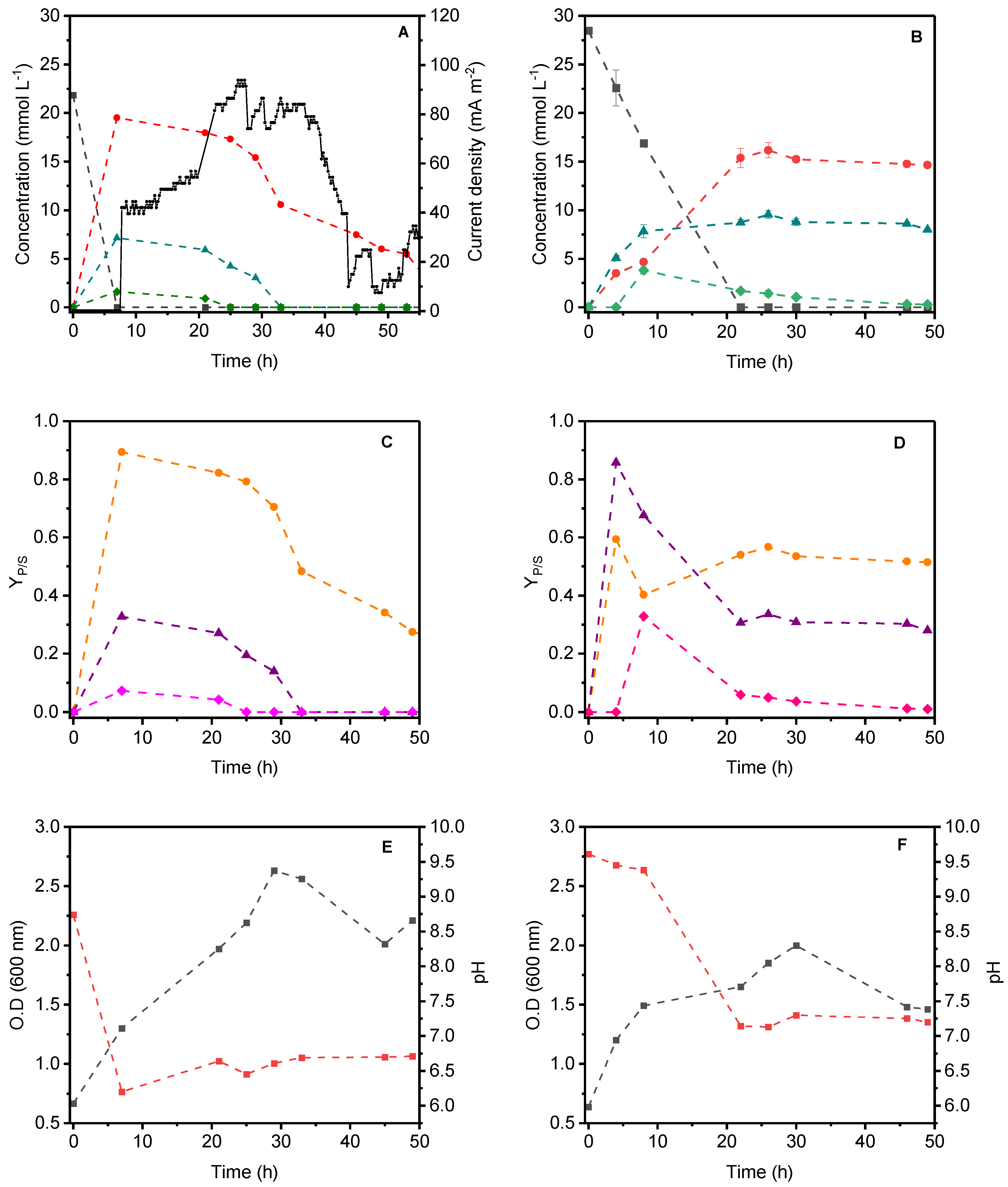
2.2. Glycerol Oxidation and by-Products Formation during MFC and Fermentative System Operation
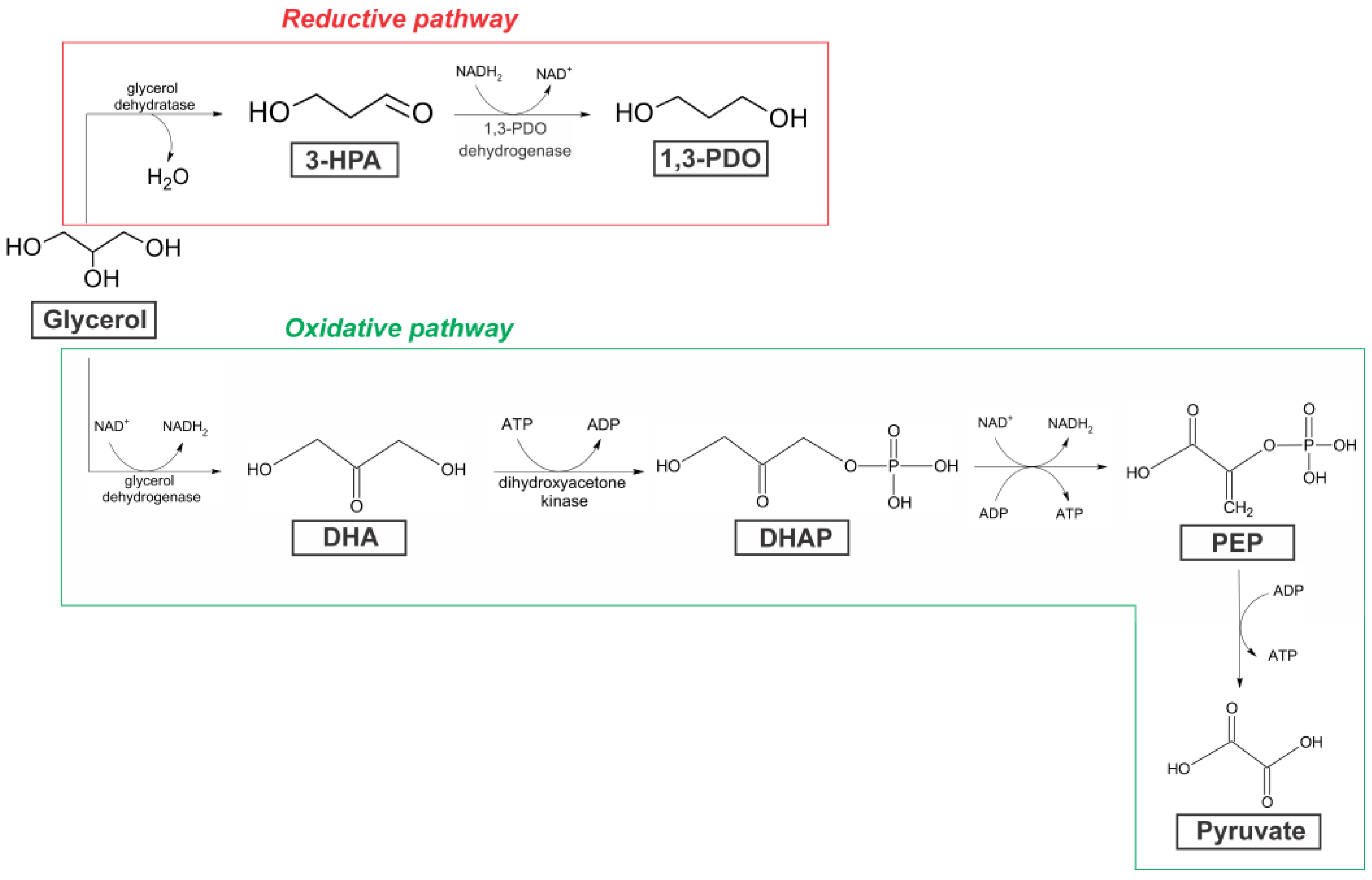
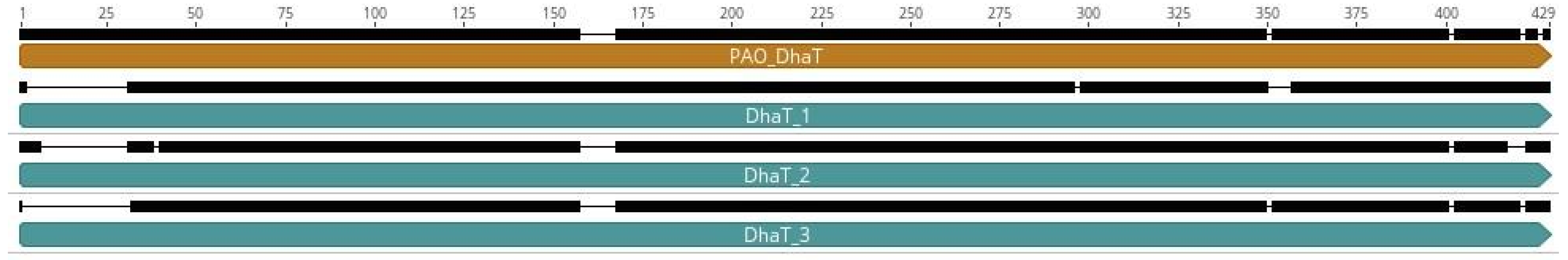
2.3. Identification of the Genes Responsible for the Production of 1,3-PDO
3. Methodology
3.1. Bacteria Isolation
3.2. Cultivation of the Isolate and Culture Medium
3.3. Microbial Fuel Cell Design and Operation
3.4. Fermentation System
3.5. Substrate and By-Product Analysis
3.6. Conversion Factor (YP/S)
3.7. Electrochemical Measurements
3.8. Coulombic Efficiency (CE)
3.9. Genome Sequencing and Identification of 1,3-PDO-Associated Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slate, A.J.; Whitehead, K.A.; Brownson, D.A.C.; Banks, C.E. Microbial Fuel Cells: An Overview of Current Technology. Renew. Sustain. Energy Rev. 2019, 101, 60–81. [Google Scholar] [CrossRef]
- Dasari, M.A.; Kiatsimkul, P.P.; Sutterlin, W.R.; Suppes, G.J. Low-Pressure Hydrogenolysis of Glycerol to Propylene Glycol. Appl. Catal. A Gen. 2005, 281, 225–231. [Google Scholar] [CrossRef]
- Attarbachi, T.; Kingsley, M.D.; Spallina, V. New Trends on Crude Glycerol Purification: A Review. Fuel 2023, 340, 127485. [Google Scholar] [CrossRef]
- Forage, R.G.; Lin, E.C.C. DHA System Mediating Aerobic and Anaerobic Dissimilation of Glycerol in Klebsiella pneumoniae NCIB 418. J. Bacteriol. 1982, 151, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Przystałowska, H.; Lipiński, D.; Słomski, R. Biotechnological Conversion of Glycerol from Biofuels to 1,3-Propanediol Using Escherichia coli. Acta Biochim. Pol. 2015, 62, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Liu, D.; Chen, Z. Recent Advances in Biological Production of 1,3-Propanediol: New Routes and Engineering Strategies. Green Chem. 2022, 24, 1390–1403. [Google Scholar] [CrossRef]
- da Silva Ruy, A.D.; de Brito Alves, R.M.; Reis Hewer, T.L.; de Aguiar Pontes, D.; Gomes Teixeira, L.S.; Magalhães Pontes, L.A. Catalysts for Glycerol Hydrogenolysis to 1,3-Propanediol: A Review of Chemical Routes and Market. Catal. Today 2021, 381, 243–253. [Google Scholar] [CrossRef]
- Marone, A.; Ayala-Campos, O.R.; Trably, E.; Carmona-Martínez, A.A.; Moscoviz, R.; Latrille, E.; Steyer, J.P.; Alcaraz-Gonzalez, V.; Bernet, N. Coupling Dark Fermentation and Microbial Electrolysis to Enhance Bio-Hydrogen Production from Agro-Industrial Wastewaters and by-Products in a Bio-Refinery Framework. Int. J. Hydrogen Energy 2017, 42, 1609–1621. [Google Scholar] [CrossRef]
- Li, Z.; Dong, Y.; Liu, Y.; Cen, X.; Liu, D.; Chen, Z. Systems Metabolic Engineering of Corynebacterium glutamicum for High-Level Production of 1,3-Propanediol from Glucose and Xylose. Metab. Eng. 2022, 70, 79–88. [Google Scholar] [CrossRef]
- Zhou, S.; Lama, S.; Sankaranarayanan, M.; Park, S. Metabolic Engineering of Pseudomonas denitrificans for the 1,3-Propanediol Production from Glycerol. Bioresour. Technol. 2019, 292, 121933. [Google Scholar] [CrossRef]
- Halfeld, G.G.; de Almeida, E.J.R.; Reginatto, V.; de Andrade, A.R. Acclimatization of a Microbial Consortium into a Stable Biofilm to Produce Energy and 1,3-Propanediol from Glycerol in a Microbial Fuel Cell. Int. J. Hydrogen Energy 2022, 47, 21241–21252. [Google Scholar] [CrossRef]
- Allam, F.; Elnouby, M.; Sabry, S.A.; El-Khatib, K.M.; El-Badan, D.E. Optimization of Factors Affecting Current Generation, Biofilm Formation and Rhamnolipid Production by Electroactive Pseudomonas aeruginosa FA17. Int. J. Hydrogen Energy 2021, 46, 11419–11432. [Google Scholar] [CrossRef]
- Yong, X.Y.; Yan, Z.Y.; Shen, H.B.; Zhou, J.; Wu, X.Y.; Zhang, L.J.; Zheng, T.; Jiang, M.; Wei, P.; Jia, H.H.; et al. An Integrated Aerobic-Anaerobic Strategy for Performance Enhancement of Pseudomonas aeruginosa-Inoculated Microbial Fuel Cell. Bioresour. Technol. 2017, 241, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Dantas, P.V.; Peres, S.; Campos-Takaki, G.M.; La Rotta, C.E. Utilization of Raw Glycerol for Pyocyanin Production from Pseudomonas aeruginosa in Half-Microbial Fuel Cells: Evaluation of Two Electrochemical Approaches. J. Electrochem. Soc. 2013, 160, G142–G148. [Google Scholar] [CrossRef]
- Gomes, A.S.; La Rotta, C.E.; Nitschke, M.; González, E.R. Evaluation of Current Output in Pseudomonas aeruginosa Microbial Fuel Cells Using Glycerol as Susbtrate and Nafion 117 as Proton Exchange Membrane. ECS Trans. 2011, 41, 2011–2017. [Google Scholar] [CrossRef]
- Zani, A.C.B.; de Almeida, É.J.R.; Furlan, J.P.R.; Pedrino, M.; Guazzaroni, M.E.; Stehling, E.G.; de Andrade, A.R.; Reginatto, V. Electrobiochemical Skills of Pseudomonas aeruginosa Species That Produce Pyocyanin or Pyoverdine for Glycerol Oxidation in a Microbial Fuel Cell. Chemosphere 2023, 335, 139073. [Google Scholar] [CrossRef]
- dos Passos, V.F.; Marcilio, R.; Aquino-Neto, S.; Santana, F.B.; Dias, A.C.F.; Andreote, F.D.; de Andrade, A.R.; Reginatto, V. Hydrogen and Electrical Energy Co-Generation by a Cooperative Fermentation System Comprising Clostridium and Microbial Fuel Cell Inoculated with Port Drainage Sediment. Bioresour. Technol. 2019, 277, 94–103. [Google Scholar] [CrossRef]
- Dwivedi, K.A.; Huang, S.J.; Wang, C.T. Integration of Various Technology-Based Approaches for Enhancing the Performance of Microbial Fuel Cell Technology: A Review. Chemosphere 2022, 287, 132248. [Google Scholar] [CrossRef]
- Nastro, R.A.; Flagiello, F.; Silvestri, N.; Gambino, E.; Falcucci, G.; Chandrasekhar, K. Use of Biochar-Based Cathodes and Increase in the Electron Flow by Pseudomonas aeruginosa to Improve Waste Treatment in Microbial Fuel Cells. Processes 2021, 9, 1941. [Google Scholar] [CrossRef]
- Bosire, E.M.; Rosenbaum, M.A. Electrochemical Potential Influences Phenazine Production, Electron Transfer and Consequently Electric Current Generation by Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 260405. [Google Scholar] [CrossRef]
- Gandouzi, I.; Tertis, M.; Cernat, A.; Saidane-Mosbahi, D.; Ilea, A.; Cristea, C. A Nanocomposite Based on Reduced Graphene and Gold Nanoparticles for Highly Sensitive Electrochemical Detection of Pseudomonas aeruginosa through Its Virulence Factors. Materials 2019, 12, 1180. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Yang, D.; Xu, D.; Gu, T. Anaerobic Corrosion of 304 Stainless Steel Caused by the Pseudomonas aeruginosa Biofilm. Front. Microbiol. 2017, 8, 298487. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Electromicrobiology. Annu. Rev. Microbiol. 2012, 66, 391–409. [Google Scholar] [CrossRef]
- Murray, D.B.; Haynes, K.; Tomita, M. Redox Regulation in Respiring Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA—General. Subj. 2011, 1810, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, G.N.; Altman, E.; Sangurdekar, D.P.; Khodursky, A.B.; Eiteman, M.A. Overflow Metabolism in Escherichia coli during Steady-State Growth: Transcriptional Regulation and Effect of the Redox Ratio. Appl. Environ. Microbiol. 2006, 72, 3653–3661. [Google Scholar] [CrossRef]
- Mason, J.T.; Kim, S.K.; Knaff, D.B.; Wood, M.J. Thermodynamic Basis for Redox Regulation of the Yap1 Signal Transduction Pathway. Biochemistry 2006, 45, 13409–13417. [Google Scholar] [CrossRef]
- Riondet, C.; Cachon, R.; Waché, Y.; Alcaraz, G.; Diviès, C. Extracellular Oxidoreduction Potential Modifies Carbon and Electron Flow in Escherichia coli. J. Bacteriol. 2000, 182, 620–626. [Google Scholar] [CrossRef]
- Kong, D.S.; Park, E.J.; Mutyala, S.; Kim, M.; Cho, Y.; Oh, S.E.; Kim, C.; Kim, J.R. Bioconversion of Crude Glycerol into 1,3-Propanediol(1,3-PDO) with Bioelectrochemical System and Zero-Valent Iron Using Klebsiella pneumoniae L17. Energies 2021, 14, 6806. [Google Scholar] [CrossRef]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Pyocyanin Alters Redox Homeostasis and Carbon Flux through Central Metabolic Pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 6372–6381. [Google Scholar] [CrossRef]
- Williams, H.D.; Zlosnik, J.E.A.; Ryall, B. Oxygen, Cyanide and Energy Generation in the Cystic Fibrosis Pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 2006, 52, 1–71. [Google Scholar] [CrossRef]
- Eschbach, M.; Schreiber, K.; Trunk, K.; Buer, J.; Jahn, D.; Schobert, M. Long-Term Anaerobic Survival of the Opportunistic Pathogen Pseudomonas aeruginosa via Pyruvate Fermentation. J. Bacteriol. 2004, 186, 4596–4604. [Google Scholar] [CrossRef]
- Head, N.E.; Yu, H. Cross-Sectional Analysis of Clinical and Environmental Isolates of Pseudomonas aeruginosa: Biofilm Formation, Virulence, and Genome Diversity. Infect. Immun. 2004, 72, 133–144. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Wittmann, C.; Nikel, P.I. Biochemistry, Genetics and Biotechnology of Glycerol Utilization in Pseudomonas Species. Microb. Biotechnol. 2020, 13, 32–53. [Google Scholar] [CrossRef]
- Malinowski, J.J. Evaluation of Liquid Extraction Potentials for Downstream Separation of 1,3-Propanediol. Biotechnol. Tech. 1999, 13, 127–130. [Google Scholar] [CrossRef]
- Biebl, H.; Zeng, A.P.; Menzel, K.; Deckwer, W.D. Fermentation of Glycerol to 1,3-Propanediol and 2,3-Butanediol by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 1998, 50, 24–29. [Google Scholar] [CrossRef]
- Cheng, K.K.; Zhang, J.A.; Liu, D.H.; Sun, Y.; Liu, H.J.; De Yang, M.; Xu, J.M. Pilot-Scale Production of 1,3-Propanediol Using Klebsiella pneumoniae. Process. Biochem. 2007, 42, 740–744. [Google Scholar] [CrossRef]
- Homann, T.; Tag, C.; Biebl, H.; Deckwer, W.D.; Schink, B. Fermentation of Glycerol to 1,3-Propanediol by Klebsiella and Citrobacter Strains. Appl. Microbiol. Biotechnol. 1990, 33, 121–126. [Google Scholar] [CrossRef]
- Biebl, H. Fermentation of Glycerol by Clostridium pasteurianum—Batch and Continuous Culture Studies. J. Ind. Microbiol. Biotechnol. 2001, 27, 18–26. [Google Scholar] [CrossRef]
- Nakas, J.P.; Schaedle, M.; Parkinson, C.M.; Coonley, C.E.; Tanenbaum, S.W.; Tertiolecta, D.; Primolecta, D.; Parva, D.; Bardawil, D.; Salina, D. System Development for Linked-Fermentation Production of Solvents from Algal Biomass. Appl. Environ. Microbiol. 1983, 46, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, S.; Fick, M.; Aggelis, G. The Effect of Raw Glycerol Concentration on the Production of 1,3-Propanediol by Clostridium butyricum. J. Chem. Technol. Biotechnol. 2004, 79, 1189–1196. [Google Scholar] [CrossRef]
- Altafini, R.D.M.; Martins, T.M.; Bruni, A.T.; Reginatto, V. Upgraded medium composition highlights the relevance of iron sulfate for 1, 3-propanediol production by a Clostridium beijerinckii strain. Biocatal. Agric. Biotechnol. 2022, 43, 102388. [Google Scholar] [CrossRef]
- Ahrens, K.; Menzel, K.; Zeng, A.-P.; Deckwer, W.-D. Kinetic, Dynamic, and Pathway Studies of Glycerol Metabolism by Klebsiella pneumoniae in Anaerobic Continuous Culture: III. Enzymes and Fluxes of Glycerol Dissimilation and 1,3-Propanediol Formation. Biotechnol. Bioeng. 1998, 59, 544–545. [Google Scholar] [CrossRef]
- Skraly, F.A.; Lytle, B.L.; Cameron, D.C. Construction and Characterization of a 1,3-Propanediol Operon. Appl. Environ. Microbiol. 1998, 64, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.; Zheng, Z.; Liu, D. 1,3-Propanediol and Its Copolymers: Research, Development and Industrialization. Biotechnol. J. 2010, 5, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Phillips, E.J.P. Novel Mode of Microbial Energy Metabolism: Organic Carbon Oxidation Coupled to Dissimilatory Reduction of Iron or Manganese. Appl. Environ. Microbiol. 1988, 54, 1472–1480. [Google Scholar] [CrossRef]
- Egoburo, D.E.; Diaz Peña, R.; Kolender, A.; Pettinari, M.J. Optimization and Validation of a GC–FID Method for Quantitative Determination of 1,3-Propanediol in Bacterial Culture Aqueous Supernatants Containing Glycerol. Chromatographia 2017, 80, 1121–1127. [Google Scholar] [CrossRef]
- Schmidell, W.; de Almeida Lima, U.; Walter Borzani, E.A. Biotecnologia Industrial—Vol. 2: Engenharia Bioquímica; Editora Blucher: São Paulo, Brazil, 2001; Volume 2, pp. 1689–1699. [Google Scholar]
- Liu, H.; Logan, B.E. Electricity Generation Using an Air-Cathode Single Chamber Microbial Fuel Cell in the Presence and Absence of a Proton Exchange Membrane. Environ. Sci. Technol. 2004, 38, 4040–4046. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, 102. [Google Scholar] [CrossRef]
| Strain | iMAX (mA m−2) | CE (%) | Glycerol (mM) * | REXT (ohms) | Anode | Salt Bridge + Cathode | Reference |
|---|---|---|---|---|---|---|---|
| EW819 | 91.3 | 1.49 | 11 | 1000 | Carbon cloth | PEM + Pt-Carbon cloth (air) | [16] |
| ATCC27853 | 110 | --- | 101 | 500 | Carbon cloth | PEM + Pt-Carbon black (air) | [15] |
| EW603 | 141.2 | 5.16 | 11 | 1000 | Carbon cloth | PEM + Pt-Carbon cloth (air) | [16] |
| ATCC27853 | 153 | --- | 101 | 500 | Carbon cloth | PEM + Pt-Carbon black (air) | [15] |
| ATCC27853 | 399.0 | 46.70 | 271.5 | 1000 | Carbon felt | SB + Carbon felt (ferrocyanide) | [14] |
| ATCC27853 | 418.3 | 48.63 | 271.5 | 1000 | Carbon felt | PEM + Pt-Black on a carbon felt with carbon vulcan powder and Teflon (air) | [14] |
| EL14 | 82.4 | 4.54 | 22 | 2200 | Carbon cloth | PEM + Pt-Carbon cloth (air) | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narcizo, J.P.; Mancilio, L.B.K.; Pedrino, M.; Guazzaroni, M.-E.; de Andrade, A.R.; Reginatto, V. A New Pseudomonas aeruginosa Isolate Enhances Its Unusual 1,3-Propanediol Generation from Glycerol in Bioelectrochemical System. Catalysts 2023, 13, 1133. https://doi.org/10.3390/catal13071133
Narcizo JP, Mancilio LBK, Pedrino M, Guazzaroni M-E, de Andrade AR, Reginatto V. A New Pseudomonas aeruginosa Isolate Enhances Its Unusual 1,3-Propanediol Generation from Glycerol in Bioelectrochemical System. Catalysts. 2023; 13(7):1133. https://doi.org/10.3390/catal13071133
Chicago/Turabian StyleNarcizo, Julia Pereira, Lucca Bonjy Kikuti Mancilio, Matheus Pedrino, María-Eugenia Guazzaroni, Adalgisa Rodrigues de Andrade, and Valeria Reginatto. 2023. "A New Pseudomonas aeruginosa Isolate Enhances Its Unusual 1,3-Propanediol Generation from Glycerol in Bioelectrochemical System" Catalysts 13, no. 7: 1133. https://doi.org/10.3390/catal13071133
APA StyleNarcizo, J. P., Mancilio, L. B. K., Pedrino, M., Guazzaroni, M.-E., de Andrade, A. R., & Reginatto, V. (2023). A New Pseudomonas aeruginosa Isolate Enhances Its Unusual 1,3-Propanediol Generation from Glycerol in Bioelectrochemical System. Catalysts, 13(7), 1133. https://doi.org/10.3390/catal13071133