Abstract
Aminotransferases (ATs) are pyridoxal 5′-phosphate-dependent enzymes that catalyze the reversible transfer of an amino group from an amino donor to a keto substrate. ATs are promising biocatalysts that are replacing traditional chemical routes for the production of chiral amines. In this study, an in silico-screening of a metagenomic library isolated from the Curonian Lagoon identified 11 full-length fold type IV aminotransferases that were successfully expressed and used for substrate profiling. Three of them (AT-872, AT-1132, and AT-4421) were active toward (R)-methylbenzylamine. Purified proteins showed activity with L- and D-amino acids and various aromatic compounds such as (R)-1-aminotetraline. AT-872 and AT-1132 exhibited thermostability and retained about 55% and 80% of their activities, respectively, even after 24 h of incubation at 50 °C. Active site modeling revealed that AT-872 and AT-4421 have an unusual active site environment similar to the AT of Haliscomenobacter hydrossis, while AT-1132 appeared to be structurally related to the AT from thermophilic archaea Geoglobus acetivorans. Thus, we have identified and characterized PLP fold type IV ATs that were active toward both amino acids and a variety of (R)-amines.
1. Introduction
Chiral amine compounds are widely used as active pharmaceutical ingredients, agricultural chemicals, bioactive natural products, and other value-added compounds [1]. Therefore, the development of widely applicable biocatalytic strategies for their synthesis is essential. Aminotransferases (ATs), also known as transaminases (EC 2.6.1), are increasingly used as an alternative to traditional chemical catalysts. ATs are promising biocatalysts for the development of innovative routes for the production of chiral amines at an industrial scale [2,3,4,5,6]. However, the repertoire of ATs capable of catalyzing the transamination of prochiral ketones into pure chemicals is still limited. An expanded toolbox of available ATs is expected to open new routes for the synthesis of chiral amines.
ATs are pyridoxal 5′-phosphate (PLP)-dependent enzymes that catalyze the reversible transfer of an amino group from an amino donor to a keto substrate. ATs are found in all domains of life: archaea, bacteria, and eukaryotes, where they are involved in the metabolism of amino acids and other nitrogenous compounds [7,8,9]. The transamination reaction occurs in two steps. First, the amino group of the amino donor is transferred to a pyridoxal phosphate (PLP) covalently bound to a lysine in the active site via a Schiff base. This reaction yields pyridoxamine phosphate (PMP) and the keto product. The second transamination step occurs when the amino group is transferred from the PMP to the keto compound (the amine acceptor), restoring the PLP and forming a new amine compound [10].
PLP-dependent enzymes are classified into different fold types based on their structure and sequence motifs. ATs are contained in fold types I and IV. ATs of fold type IV are further subdivided into branched-chain aminotransferases (BCAATs), D-amino acid aminotransferases (DAATs), and (R)-amine-pyruvate aminotransferases (R-ATs) [11]. In higher eukaryotes, BCATs are responsible for the catabolism of leucine, isoleucine, and valine, and can also deaminate aromatic amino acids. In other organisms, BCATs are used for both the production and degradation of these amino acids [12,13]. Bacteria use DAAT to produce D-amino acids, in particular D-glutamic acid, which is essential for cell wall synthesis [14]. The most recent member of the fold type IV ATs is R-ATs. These ATs are active toward various aliphatic R-amines such as (R)-methylbenzylamine, (2R)-aminohexane, and others [15,16,17,18]. Höhne et al. [15] revealed an in silico-strategy for the detection of potential R-ATs from metagenomes. Further progress in the identification of different R-ATs was made by Steiner et al. [19] and Jiang et al. [16]. Phylogenetic analysis based on multiple sequence alignments or sequence similarity networks (SSNs) provides reliable data for subgrouping ATs; however, the sequences of potential R-ATs do not form distinct groups. In addition, it is difficult to predict the activity of ATs and the promiscuity of enzyme substrates toward donor or acceptor substrates. ATs are of great importance as potential biocatalysts for the production of pharmaceuticals and other chemical intermediates [2,20,21,22]; therefore, there is still a need to find suitable R-ATs that can use many different amine acceptors and donors.
In this study, an in silico-screening of a metagenomic library identified 11 full-length fold type IV aminotransferases that were successfully expressed and used for substrate profiling. Three of the tested ATs were active toward R-methylbenzylamine.
2. Results and Discussion
2.1. Enzymes Identification and Phylogenetic Analysis
We searched in silico for fold type IV PLP-dependent enzymes in the sequenced metagenomic DNA isolated from the Curonian Lagoon [23]. We have identified eleven genes encoding hypothetical ATs that belong to the fold type IV class. A comparison of the amino acid sequences of the selected ATs (Table S1) (between each other) showed a similarity of 31.6–100%. The homologs of the ATs were identified by a BLASTp search against the nonredundant protein sequences of the NCBI database (Table 1). Eight of the selected ATs were highly similar (91–100%) to their closest homologs and could be accurately assigned to the BCAT group. In contrast, AT-872, AT-1202, and AT-4421 did not show high homology and could not be placed with certainty in any group of fold type IV class ATs.

Table 1.
The list of selected ATs.
We attempted to clarify the phylogenetic position of the selected ATs among the characterized BCATs and DAATs capable of deaminating various (R)-amines [24,25,26,27,28]. Therefore, we assembled the reviewed sequences of known BCATs and DAATs from UniProtKB, and also added ATs of interest from previous publications [15,19,24,27,29,30]. The constructed phylogenetic tree (Figure S1) revealed that most of the identified BCATs clustered together with known canonical BCATs. AT-1132 and AT-14307 attracted considerable interest, as both of these proteins were grouped with ATs of archaeal origin, which have been shown to deaminate the (R)-methylbenzylamine [29] (Figure 1a).

Figure 1.
A representative view of the phylogenetic analysis of the selected ATs (a) AT-1132 and AT-14307, (b) AT-872 and AT-4421 compared to the reviewed sequences of fold type IV aminotransferases. See Figure S1 for a full phylogenetic analysis.
AT-872 and AT-4421 clustered with Arabidopsis thaliana chloroplast DAAT and Haliscomenobacter hydrossis AT, which, as has been shown previously [30], have an unusual DAAT active site (Figure 1b). According to the phylogenetic analysis, the identified set of ATs was diverse, with a low similarity between its characterized homologs, and, potentially, contained (R)-selective counterparts. Therefore, we decided to test all identified AT using (S)- and (R)-methylbenzylamine (MBA) as substrates.
2.2. Initial Activity Screening
To determine the enzymatic activity of the selected ATs, their genes were cloned into the expression vector pLATE31 and overexpressed E. coli strain BL21 (DE3) (Figure S2 and Table S2), as described in Materials and Methods. The measurement of AT activity using cell-free extracts and a universal amino acceptor (pyruvate) showed that none of the ATs tested were active toward S-MBA. However, AT-872, AT-1132, and AT-4421 presented low activity in the presence of R-MBA. In addition, ATs showed no activity with o-xylenediamine, a common chromogenic amino donor used for AT screening and activity measurements [31]. However, some of the tested enzymes showed low but measurable activity in the presence of another chromogenic amino donor, 4-nitrophenylethylamine [32] (Figure S3). AT-14307, although very similar to AT-1132, did not display any appreciable activity toward R-MBA. For further investigation, we chose three enzymes (AT-872, AT-1132, and AT-4421) capable of deaminating R-MBA. The target proteins were expressed with 6xHis tags and purified by immobilized metal affinity chromatography (Figure S4).
2.3. Temperature Stability and Effect of pH on Activity of the Selected Aminotransferases
We began our characterization by optimizing the pH of the reaction mixture (Figure 2a). The majority of the previously characterized (R)-selective ATs were most active at pH 8 or 9 [18,19,33]. AT-1132 exhibited the maximum activity in sodium bicarbonate buffer at pH 9, while AT-4421 showed the highest activity in potassium phosphate buffer at pH 9. Although AT-1132 had a low pH optimum range, AT-4421 remained active (80%) at pH 10 (in sodium carbonate buffer). In contrast, AT-872 preferred higher pH and showed similar activity at pH 10–11.
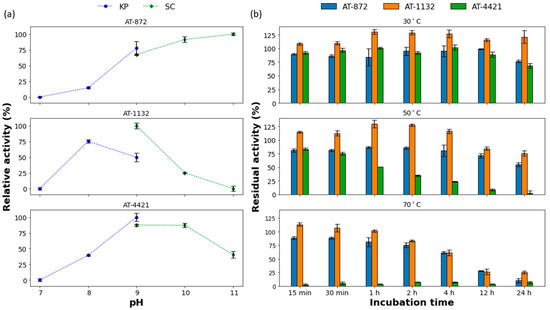
Figure 2.
(a) The effect of pH on enzyme activity. The highest activity for each enzyme was taken as 100%, KP—potassium phosphate buffer, SC—sodium carbonate buffer; (b) Thermostability of ATs. Activity before incubation was taken as 100%. For a detailed description of reaction conditions, see the Methods section. All values are mean ± SD.
The thermostability of the selected ATs was investigated at three temperatures (Figure 2b). At 30 °C, AT-4421 was moderately stable, the activity decreases by 30% after incubation for 24 h. At higher temperatures, a significant decrease with time in AT-4421 activity was observed: 25% of activity was retained after 4 h at 50 °C, while at 70 °C, complete inactivation occurred after 15 min. The other two enzymes were quite thermostable. AT-872 retained about 55% and AT-1132 around 80% of their activities, even after 24 h of incubation at 50 °C. Moreover, these two ATs retained more than 60% of their activity after incubation at 70 °C for 4 h. AT-1132 showed an increase in activity after incubation at all tested temperatures. Such phenomena were observed with certain other thermostable ATs and might be explained by the refolding of a protein that was kept at −20 °C to its natural conformation [34,35]. This increase in activity (above 100%) lasted longer at lower temperatures due to slower protein denaturation rates.
2.4. Substrate Scope of Aminotransferases
We measured ATs’ preferences for different amino acceptors (Table 2). AT-872 and AT-4421 were the most active in the presence of pyruvate, while AT-1132 had the highest activity toward α-ketoglutarate.

Table 2.
AT activity with different amino acceptors. The highest activity for each enzyme was taken as 100%. All values are mean ± SD.
The AT-1132 is closely related to BCATs according to protein sequence and its specificity for amino acceptor is similar to BCATs, which are most active with α-ketoglutarate. AT-872 and AT-4421 are homologous to the DAATs, which prefer pyruvate as an amino acceptor [13,36,37]. Thus, the amino acceptor preference of selected ATs correlates with protein sequences.
The conversion rates of the deamination reactions for the different amino donors were also investigated (Figure 3).
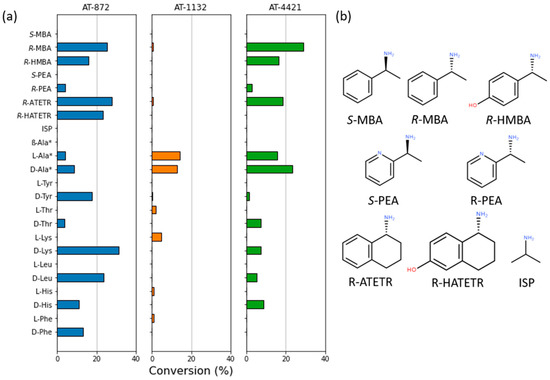
Figure 3.
(a) The substrate specificity of ATs; (b) Structures of the used substrates. Pyruvate was used as an amino acceptor for all reactions except those marked with the “*” symbol where α-ketoglutarate was used. A detailed description of reaction conditions is outlined in the Methods Section. The reaction (100 μL) conditions: 5 mM amino donor and 5 mM pyruvate or α-ketoglutarate, 10 μM PLP, 5 μg of the purified enzyme, and 50 mM sodium carbonate buffer (pH 9). The reactions were performed at 30 °C for 16 h.
AT-1132 was able to deaminate L- and D-alanine and some L-amino acids, although with very low efficiency. AT-872 and AT-4421 were active with D-amino acids and L-alanine. None of the ATs tested were able to use β-alanine or isopropylamine (ISP) as an amino donor. All three ATs were active with amino donor R-MBA (AT-872—1.0 U/mg, AT-1132—0.2 U/mg, and AT-4421—0.9 U/mg). These activity values toward R-MBA are similar to that of the other described PLP fold type IV ATs [25,29]. AT-872 and AT-4421 were also able to deaminate (R)-pyridylethylamine (R-PEA). Although a small amount of keto product was detected, the activity of AT-1132 with R-PEA was too low to give an accurate estimate of the conversion. Interestingly, AT-4421 was able to deaminate the bulky substrate (R)-aminotetralin (R-ATETR), while AT-872 was also active with the R-ATETR analog (R)-hydroxyaminotetralin (R-HATETR) and showed more than 20% conversion using equal amounts of amino donor and amino acceptor.
2.5. D-Models of the ATs Active Sites
To better understand the substrate preferences of identified ATs, we used ColabFold [38] to create 3D-structure models of the enzymes. All ATs were modeled as dimers. We also used CB-Dock2 [39,40] for template-based docking to dock the ketimine intermediate of MBA bound to PLP into the AT-872 active site.
Although AT-872 and AT-4421 have 40% similarity, their active site environments were almost identical (Figure 4a). The only notable differences were the substitutions at positions 91 and 239 (according to the AT-872 sequence), AT-4421 had Arg instead of Lys91, and Thr was replaced with Ser239. The active sites of AT-872 and AT-4421 corresponded well to the AT of Haliscomenobacter hydrossis [30] (35% overall similarity) (Figure 4b). Like all type IV AT [41], the active sites of AT-872 and AT-4421 were divided into two pockets, O and P. The O pocket of AT-872 consisted of the side chains of Phe30, Arg28 (provided by another monomer), Lys90, and Arg179, thus forming a positively charged pocket. The difference between the O pocket of AT-4421 and AT-872 was Lys91 replaced by Arg, as mentioned above. Neither ATs had the canonical DAAT “carboxylate trap” and most likely used Arg28 and Lys91 (or Arg) side chains to bind the α-carboxyl group of the substrates in a manner similar to the H. hydrossis AT. The P-pocket of AT-872 was confined by side chains of Phe36, Ser238, Ser239, and Thr240. Ser239 was replaced by Thr in the AT-4421 P-pocket. Unlike H. hydrossis AT, both AT-872 and AT-4421 were capable of deaminating R-MBA, R-PEA, or R-ATETR. This might be explained by the larger and more hydrophobic P-pocket formed when Tyr35 (in H. hydrossis AT) was replaced by Phe, and Ile240 (in H. hydrossis AT) by Ser or Thr.
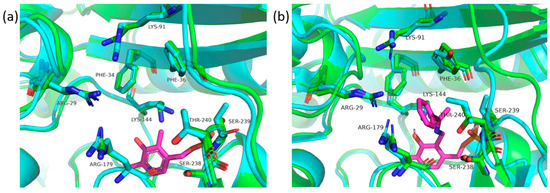
Figure 4.
(a) Comparison of AT-872 (green) and AT-4421 (cyan) active sites, where AT of Haliscomenobacter hydrossis (7p7x) was used for PLP (magenta) placement; and (b) active site of the AT-872 (green) superimposed on the AT of Haliscomenobacter hydrossis (7p7x) (cyan), with PLP bound to MBA (magenta). In both cases, the amino acid numbering was based on the AT-872.
ColabFold was also used to create a model of the AT-1132 structure. This AT is 50% similar to BCAT from the thermophilic archaea Geoglobus acetivorans [29]. Their active sites are almost identical (Figure 5) and, although less so, are similar to the AT from Thermobaculum terrenum [42].
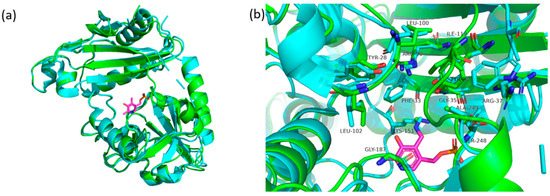
Figure 5.
(a) The monomer of the AT-1132 (green) superimposed on the monomer of the AT from Geoglobus acetivorans (5 cm0) (cyan), PLP (magenta); (b) a comparison of AT-1132 (green) and AT from Geoglobus acetivorans (cyan) active sites, PLP (magenta). Numbering based on the amino acid of the AT-1132.
The active site of AT-1132 is also divided into O and P pockets. Pocket O consists of Phe33, Arg90, IIe119, and Tyr28, while Leu100 and Leu102 are from the other monomer. The P-pocket of AT-1132 is composed of residues Gly35, Arg38, Tyr89, Gly187, Ser247, and Ala248. The major difference between AT-1132 and AT from G. acetivorans is in the loop region connecting the small subunit to the large domain, where AT-1132 contains Ile119 instead of Trp. It is worth noting that AT-14307, despite an almost identical active site composition, was unable to deaminate R-MBA or the activity of this enzyme was too low to be detectable. This might be attributed to subtle differences in the interdomain loop and other flexible regions surrounding the active site.
3. Materials and Methods
3.1. Selection of AT Genes from Metagenome Library and Phylogenetic Analysis
The sequenced metagenome of the Curonian Lagoon [23] was used as a database for the search for potential aminotransferases encoding genes. The experimentally characterized sequences of R-specific (fold type IV) ATs [15,16] were used as a query in BLAST analysis. The hits with the highest scores (>1 × 10−10) were selected for further analysis.
The phylogenetic analysis was performed with the MegaX program [43] version 10.1.8. Multiple sequence alignment was performed using the MUSCLE algorithm. The alignment was manually refined by removing variable regions at the start and the end of the protein. The evolutionary history was inferred by using the maximum likelihood method and JTT matrix-based model, and the bootstrap consensus tree was inferred from 500 replicates.
3.2. Bacterial Strains, Plasmids, Primers, and Standard Cloning Techniques
E. coli strain DH5α was used for cloning experiments. The recombinant proteins were overexpressed in E. coli strain BL21 (DE3). The bacterial strains, plasmids, and primers used in this study are listed in Table S3 in the Supplemental Material (SM). Standard molecular biology techniques were performed, as previously described [44]. The genes coding selected ATs were amplified from metagenomic DNA from the Curonian Lagoon using a Phusion Plus PCR Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania) and designed primers (Table S3). The PCR products were extracted from the gel using a GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Vilnius, Lithuania) and were cloned into pLATE31 (C-terminal His-tag) expression vector using aLICator LIC Cloning and Expression Kit 3 (Thermo Fisher Scientific, Vilnius, Lithuania). E. coli strain DH5α (Novagen, Darmstadt, Germany) cells were transformed with reaction mixtures and spread on LB agar plates supplemented with 75 µg/mL ampicillin. pDNA (plasmid DNA) from selected bacterial clones was extracted and purified using the ZR Plasmid Miniprep kit (Zymo Research, Irvine, CA, USA). Later, it was sequenced using LIC sequencing primers (Table S3) by the Sanger method (Macrogen Europe, Amsterdam, The Netherlands).
3.3. Expression and Purification of Recombinant Proteins
The E. coli BL21 (DE3) (Novagen, Darmstadt, Germany) bacteria harboring the appropriate plasmid were grown with shaking (180 rpm) at 37 °C in 200 mL of LB medium supplemented with 100 μg/mL ampicillin. When OD600 reached 0.6–0.8, protein expression was induced by adding IPTG (final concentration 1.0 mM). After the induction for 12 h at 20 °C temperature, the cells were collected by centrifugation for 20 min, 4000× g at 4 °C. The cell pellet was resuspended in 25 mL of 50 mM PBS buffer (pH 7.2) and disrupted using ultrasound. The cell debris was removed by centrifugation for 20 min at 15,000× g and 4 °C. The supernatant was loaded onto a 1 mL of HiTrap Nickel Chelating HP column (GE Healthcare, Uppsala, Sweden) that had been pre-equilibrated with PBS pH 7.2. The column was washed with five column volumes of PBS pH 7.2 containing 30 mM imidazole. The recombinant protein was eluted in PBS pH 7.2 containing 400 mM imidazole. The collected recombinant protein fraction was desalted on a 5 mL Sephadex G25 column (GE Healthcare, Uppsala, Sweden) using PBS pH 7.2. The protein solution was diluted with glycerol (50% final concentration) and supplemented with 50 μM PLP and stored at –20 °C for further use. The purity of the proteins was determined by the SDS-PAGE [45] standard procedure using 12% gels. The protein bands were visualized using Coomassie blue G-250 (Fluka, Buchs, Switzerland) staining. The concentration of purified proteins was determined using a Pierce Coomassie (Bradford) Protein Assay Kit (Thermo Fisher, Rockford, IL, USA) [46].
3.4. Enzyme Assays
The ATs capable of deaminating methylbenzylamine were screened by measuring the amount of formed acetophenone. The reaction (volume 100 μL) conditions were as follows: 10 mM (S)- or (R)-methylbenzylamine, 10 mM pyruvate as amino acceptor, 10 μM PLP, 50 μL of cell-free lysate (in PBS pH 7.2 buffer), and 50 mM potassium phosphate buffer (pH 9). The reactions were incubated at 30 °C for 16 h. Then, 100 μL of acetonitrile was added to stop the reactions. The reaction mixture was centrifugated for 10 min at 15,000× g and transferred to the 96-well plates. The formation of acetophenone was determined by the change in UV absorbance at 240 nm using a PowerWave XS plate reader (BioTek, Santa Clara, CA, USA).
To study the effect of pH on enzyme activity, reactions were carried out in buffers of different pH values (50 mM potassium phosphate pH 7–8; 50 mM sodium bicarbonate pH 9–11). The relative enzyme activity was determined by the amount of formed acetophenone. The reaction mixture (100 μL) contained 5 mM (R)-methylbenzylamine, 5 mM pyruvate, 10 μM PLP, and 5 μg of the purified enzyme. The reaction was performed at 30 °C for 20 min. The measurement of the acetophenone formed was carried out, as mentioned above. The experiments were conducted in triplicate.
The thermostability of the enzyme was determined by incubating the enzyme at set temperatures (30, 50, or 70 °C) for a specific amount of time (up to 24 h). The remaining enzyme activity after incubation was determined by the amount of formed acetophenone. The reaction mixture (100 μL) contained 5 mM (R)-methylbenzylamine, 5 mM pyruvate, 10 μM PLP, 5 μg of the incubated enzyme, and 50 mM sodium carbonate buffer (pH 9). Reactions were performed at 30 °C for 20 min. The measurement of the acetophenone formed was carried out as mentioned above. The experiments were conducted in triplicate.
3.5. Scope of Amino Acceptors and Donors
The optimal amino acceptor was determined by measuring the amount of formed acetophenone. The reactions were carried out using different amino acceptors under the following conditions: 10 mM (R)-methylbenzylamine, 5 mM amino acceptor, 10 μM PLP, 5 μg of the purified enzyme, and 50 mM sodium carbonate buffer (pH 9), with a total reaction volume of 100 μL. The reactions were performed at 30 °C for 1 h. The measurement of the acetophenone formed was carried out as mentioned above. The experiments were conducted in triplicate.
Amino donor specificity was determined by measuring the amount of formed alanine or glutamic acid. The reaction (100 μL) conditions: 5 mM amino donor, 5 mM pyruvate or α-ketoglutarate, 10 μM PLP, 5 μg of the purified enzyme, and 50 mM sodium carbonate buffer (pH 9). The reactions were performed at 30 °C for 16 h. The reaction was stopped by adding 500 μL of acetonitrile. The reaction mixture was centrifugated for 20 min at 15,000× g. The amount of formed alanine or glutamic acid was determined by HPLC–MS.
3.6. Analytical Methods
The HPLC–MS analysis was performed using a high-performance liquid chromatography system (Shimadzu, Kyoto, Japan) equipped with a photodiode array (PDA; Shimadzu, Kyoto, Japan) detector and a mass spectrometer (LCMS 2020; Shimadzu, Kyoto, Japan). The samples were mixed with an equal volume of acetonitrile, vortexed for 1 min, clarified by centrifugation at 10,000× g for 10 min, and subjected to HPLC–MS. The chromatographic separation was conducted using a 150 × 3 mm YMC-Pack Pro C18 column (YMC, Kyoto, Japan) at 40 °C and a mobile phase that consisted of water with 0.1% formic acid (solvent A) and acetonitrile (solvent B) delivered in gradient elution mode at a flow rate of 0.45 mL/min. The elution program was used as follows: isocratic 5% B for 1 min, from 5 to 95% B over 5 min, isocratic 95% B for 2 min, from 95 to 5% B over 1 min, isocratic 5% B for 4 min. The data were analyzed using LabSolutions LCMS software version 5.42.
4. Conclusions
Several PLP fold type IV ATs were identified and characterized in this study. These enzymes are not only active toward D-amino acids but can also transaminate various (R)-amines, which makes them potential biocatalysts. Based on the high thermostability of AT-872 and AT-1132, and the broad substrate spectrum of AT-872 and AT-4421, the engineering of these ATs for application in the production of high-value amino compounds seems feasible. Our results suggest that BCAT and DAAT have biocatalytic potential and that these groups of fold type IV aminotransferases should not be overlooked in the search for R-selective ATs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal13030587/s1, Figure S1: Evolutionary analysis by Maximum Likelihood method; Figure S2: Protein expression of ATs in E. coli BL-21 (DE3). Analysis of cleared lysates using SDS-PAGE; Figure S3: Activities with: (S)-methylbenzylamine (S-MBA), (R)-methylbenzylamine (R-MBA) and 4-nitrophenylethylamine (4NPEA); Figure S4: Protein purification SDS-PAGE; Table S1: Percent similarity matrix of the chosen ATs; Table S2: AT protein theoretical mass and observed activities and Table S3: Bacterial strains, plasmids, and primers used in this study.
Author Contributions
Conceptualization, R.M., J.V. and R.S. (Rokas Statkevičius); methodology, R.S. (Rokas Statkevičius), R.S. (Rūta Stanislauskienė) and J.V.; software, J.V. and R.S. (Rokas Statkevičius); validation, J.V. and R.M.; formal analysis, R.S. (Rokas Statkevičius); investigation, R.S. (Rokas Statkevičius); resources, J.V. and R.S. (Rūta Stanislauskienė); data curation, R.S. (Rokas Statkevičius) and J.V.; writing—original draft preparation, R.S. (Rokas Statkevičius) and J.V.; writing—review and editing, R.M. and R.S. (Rūta Stanislauskienė); visualization, R.S. (Rokas Statkevičius); supervision, R.M.; project administration, R.M. and J.V.; funding acquisition, R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This project received funding from the European Social Fund (project No. 09.3.3.-LMT-K-712-22-0229) under a grant agreement with the Research Council of Lithuania (LMTLT).
Data Availability Statement
Data is contained within the article or the Supplementary Materials.
Acknowledgments
We thank J. Stankevičiūtė for her technical assistance and help with draft preparation and review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rossino, G.; Robescu, M.S.; Licastro, E.; Tedesco, C.; Martello, I.; Maffei, L.; Vincenti, G.; Bavaro, T.; Collina, S. Biocatalysis: A Smart and Green Tool for the Preparation of Chiral Drugs. Chirality 2022, 34, 1403–1418. [Google Scholar] [CrossRef]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Koszelewski, D.; Tauber, K.; Faber, K.; Kroutil, W. ω-Transaminases for the Synthesis of Non-Racemic α-Chiral Primary Amines. Trends Biotechnol. 2010, 28, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.S.; Park, E.S.; Shin, J.S. Features and Technical Applications of ω-Transaminases. Appl. Microbiol. Biotechnol. 2012, 94, 1163–1171. [Google Scholar] [CrossRef]
- Slabu, I.; Galman, J.L.; Lloyd, R.C.; Turner, N.J. Discovery, Engineering, and Synthetic Application of Transaminase Biocatalysts. ACS Catal. 2017, 7, 8263–8284. [Google Scholar] [CrossRef]
- Truppo, M.D.; David Rozzell, J.; Turner, N.J. Efficient Production of Enantiomerically Pure Chiral Amines at Concentrations of 50 g/L Using Transaminases. Org. Process Res. Dev. 2010, 14, 234–237. [Google Scholar] [CrossRef]
- Cooper, A.J.L.; Meister, A. An Appreciation of Professor Alexander E. Braunstein. The Discovery and Scope of Enzymatic Transamination. Biochimie 1989, 71, 387–404. [Google Scholar] [CrossRef]
- Braunstein, A.E.; Kritzmann, M.G. Formation and Breakdown of Amino-Acids by Inter-Molecular Transfer of the Amino Group. Nature 1937, 140, 503–504. [Google Scholar] [CrossRef]
- Hayashi, H. Pyridoxal Enzymes: Mechanistic Diversity and Uniformity. J. Biochem. 1995, 118, 463–473. [Google Scholar] [CrossRef]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal Phosphate Enzymes: Mechanistic, Structural, and Evolutionary Considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef]
- Percudani, R.; Peracchi, A. The B6 Database: A Tool for the Description and Classification of Vitamin B6-Dependent Enzymatic Activities and of the Corresponding Protein Families. BMC Bioinform. 2009, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Goto, M.; Miyahara, I.; Hayashi, H.; Kagamiyama, H.; Hirotsu, K. Crystal Structures of Branched-Chain Amino Acid Aminotransferase Complexed with Glutamate and Glutarate: True Reaction Intermediate and Double Substrate Recognition of the Enzyme. Biochemistry 2003, 42, 3725–3733. [Google Scholar] [CrossRef] [PubMed]
- Hutson, S. Structure and Function of Branched Chain Aminotransferases. Prog. Nucleic Acid Res. Mol. Biol. 2001, 70, 175–206. [Google Scholar] [CrossRef] [PubMed]
- Radkov, A.D.; Moe, L.A. Bacterial Synthesis of D-Amino Acids. Appl. Microbiol. Biotechnol. 2014, 98, 5363–5374. [Google Scholar] [CrossRef] [PubMed]
- Höhne, M.; Schätzle, S.; Jochens, H.; Robins, K.; Bornscheuer, U.T. Rational Assignment of Key Motifs for Function Guides in Silico Enzyme Identification. Nat. Chem. Biol. 2010, 6, 807–813. [Google Scholar] [CrossRef]
- Jiang, J.; Chen, X.; Zhang, D.; Wu, Q.; Zhu, D. Characterization of (R)-Selective Amine Transaminases Identified by in Silico Motif Sequence Blast. Appl. Microbiol. Biotechnol. 2015, 99, 2613–2621. [Google Scholar] [CrossRef] [PubMed]
- Sayer, C.; Martinez-Torres, R.J.; Richter, N.; Isupov, M.N.; Hailes, H.C.; Littlechild, J.A.; Ward, J.M. The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine: Pyruvate Transaminase from Nectria Haematococca. FEBS J. 2014, 281, 2240–2253. [Google Scholar] [CrossRef]
- Pavkov-Keller, T.; Strohmeier, G.A.; Diepold, M.; Peeters, W.; Smeets, N.; Schürmann, M.; Gruber, K.; Schwab, H.; Steiner, K. Discovery and Structural Characterisation of New Fold Type IV-Transaminases Exemplify the Diversity of This Enzyme Fold. Sci. Rep. 2016, 6, 38183. [Google Scholar] [CrossRef]
- Telzerow, A.; Paris, J.; Håkansson, M.; González-Sabín, J.; Ríos-Lombardía, N.; Gröger, H.; Morís, F.; Schürmann, M.; Schwab, H.; Steiner, K. Expanding the Toolbox of R-Selective Amine Transaminases by Identification and Characterization of New Members. ChemBioChem 2020, 22, 1232–1242. [Google Scholar] [CrossRef]
- Fuchs, M.; Farnberger, J.E.; Kroutil, W. The Industrial Age of Biocatalytic Transamination. Eur. J. Org. Chem. 2015, 2015, 6965–6982. [Google Scholar] [CrossRef]
- Kohls, H.; Steffen-Munsberg, F.; Höhne, M. Recent Achievements in Developing the Biocatalytic Toolbox for Chiral Amine Synthesis. Curr. Opin. Chem. Biol. 2014, 19, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Ghislieri, D.; Turner, N.J. Biocatalytic Approaches to the Synthesis of Enantiomerically Pure Chiral Amines. Top. Catal. 2014, 57, 284–300. [Google Scholar] [CrossRef]
- Zilius, M.; Samuiloviene, A.; Stanislauskienė, R.; Broman, E.; Bonaglia, S.; Meškys, R.; Zaiko, A. Depicting Temporal, Functional, and Phylogenetic Patterns in Estuarine Diazotrophic Communities from Environmental DNA and RNA. Microb. Ecol. 2021, 81, 36–51. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Dibrova, D.V.; Nikolaeva, A.Y.; Rakitina, T.V.; Popov, V.O. Identification of Branched-Chain Amino Acid Aminotransferases Active towards (R)-(+)-1-Phenylethylamine among PLP Fold Type IV Transaminases. J. Biotechnol. 2018, 271, 26–28. [Google Scholar] [CrossRef]
- Zhai, L.; Xie, Z.; Tian, Q.; Guan, Z.; Cai, Y.; Liao, X. Structural and Functional Analysis of the Only Two Pyridoxal 5′-Phosphate-Dependent Fold Type IV Transaminases in Bacillus altitudinis W3. Catalysts 2020, 10, 1308. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Boyko, K.M.; Nikolaeva, A.Y.; Zeifman, Y.S.; Rakitina, T.V.; Suplatov, D.A.; Popov, V.O. Biochemical and Structural Insights into PLP Fold Type IV Transaminase from Thermobaculum Terrenum. Biochimie 2019, 158, 130–138. [Google Scholar] [CrossRef]
- Zeifman, Y.S.; Boyko, K.M.; Nikolaeva, A.Y.; Timofeev, V.I.; Rakitina, T.V.; Popov, V.O.; Bezsudnova, E.Y. Functional Characterization of PLP Fold Type IV Transaminase with a Mixed Type of Activity from Haliangium Ochraceum. Biochim. Biophys. Acta-Proteins Proteom. 2019, 1867, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Stekhanova, T.N.; Rakitin, A.L.; Mardanov, A.V.; Bezsudnova, E.Y.; Popov, V.O. A Novel Highly Thermostable Branched-Chain Amino Acid Aminotransferase from the Crenarchaeon Vulcanisaeta Moutnovskia. Enzym. Microb. Technol. 2017, 96, 127–134. [Google Scholar] [CrossRef]
- Isupov, M.N.; Boyko, K.M.; Sutter, J.-M.; James, P.; Sayer, C.; Schmidt, M.; Schönheit, P.; Nikolaeva, A.Y.; Stekhanova, T.N.; Mardanov, A.V.; et al. Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus Acetivorans and Archaeoglobus Fulgidus: Biochemical and Structural Characterization. Front. Bioeng. Biotechnol. 2019, 7, 7. [Google Scholar] [CrossRef]
- Bakunova, A.K.; Nikolaeva, A.Y.; Rakitina, T.V.; Isaikina, T.Y.; Khrenova, M.G.; Boyko, K.M.; Popov, V.O.; Bezsudnova, E.Y. The Uncommon Active Site of D-amino Acid Transaminase from Haliscomenobacter Hydrossis: Biochemical and Structural Insights into the New Enzyme. Molecules 2021, 26, 5053. [Google Scholar] [CrossRef]
- Green, A.P.; Turner, N.J.; O’Reilly, E. Chiral Amine Synthesis Using ω-Transaminases: An Amine Donor That Displaces Equilibria and Enables High-Throughput Screening. Angew. Chem. Int. Ed. 2014, 53, 10714–10717. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Ladkau, N.; Moody, T.S.; Ward, J.M.; Hailes, H.C. A Rapid, Sensitive Colorimetric Assay for the High-Throughput Screening of Transaminases in Liquid or Solid-Phase. Chem. Commun. 2015, 51, 17225–17228. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.L.; Zhang, N.N.; Ye, G.Y.; Zheng, Y.G. Efficient Biosynthesis of (R)-3-Amino-1-Butanol by a Novel (R)-Selective Transaminase from Actinobacteria sp. J. Biotechnol. 2019, 295, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.; Deepankumar, K.; Shin, G.; Hong, E.Y.; Kim, B.G.; Chung, T.; Yun, H. Identification of Novel Thermostable ω-Transaminase and Its Application for Enzymatic Synthesis of Chiral Amines at High Temperature. RSC Adv. 2016, 6, 69257–69260. [Google Scholar] [CrossRef]
- Wang, C.; Tang, K.; Dai, Y.; Jia, H.; Li, Y.; Gao, Z.; Wu, B. Identification, Characterization, and Site-Specific Mutagenesis of a Thermostable ω-Transaminase from Chloroflexi Bacterium. ACS Omega 2021, 6, 17058–17070. [Google Scholar] [CrossRef]
- Inoue, K.; Kuramitsu, S.; Aki, K.; Watanabe, Y.; Takagi, T.; Nishigai, M.; Ikai, A.; Kagamiyama, H. Branched-Chain Amino Acid Aminotransferase of Escherichia Coli: Overproduction and Properties. J. Biochem. 1988, 104, 777–784. [Google Scholar] [CrossRef]
- Tanizawa, K.; Masu, Y.; Asano, S.; Tanaka, H.; Soda, K. Thermostable D-Amino Acid Aminotransferase from a Thermophilic Bacillus Species: Purification, Characterization, and Active Site Sequence Determination. J. Biol. Chem. 1989, 264, 2445–2449. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein-Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, 159–164. [Google Scholar] [CrossRef]
- Yang, X.; Liu, Y.; Gan, J.; Xiao, Z.-X.; Cao, Y. Problem Solving Protocol FitDock: Protein-Ligand Docking by Template Fitting. Brief. Bioinform. 2022, 23, 1–11. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Popov, V.O.; Boyko, K.M. Structural Insight into the Substrate Specificity of PLP Fold Type IV Transaminases. Appl. Microbiol. Biotechnol. 2020, 104, 2343–2357. [Google Scholar] [CrossRef] [PubMed]
- Bezsudnova, E.Y.; Nikolaeva, A.Y.; Bakunova, A.K.; Rakitina, T.V.; Suplatov, D.A.; Popov, V.O.; Boyko, K.M. Probing the Role of the Residues in the Active Site of the Transaminase from Thermobaculum Terrenum. PLoS ONE 2021, 16, e0255098. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J. Molecular Cloning: A Laboratory Manual/Joseph Sambrook, David W. Russell; Russell 1957-; David, W., Cold Spring Harbor Laboratory, Eds.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001; ISBN 0879695773. [Google Scholar]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).