Bayesian Optimization for an ATP-Regenerating In Vitro Enzyme Cascade
Abstract
1. Introduction
2. Results and Discussion
2.1. Specification of the Cascade and Its Optimization
2.2. Iterative Optimization for Specific Activity of MVK
3. Materials and Methods
3.1. Materials
3.2. Enzyme Production
3.3. Enzyme Assays
3.4. Analytics
3.5. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sheldon, R.A.; Brady, D.; Bode, M.L. The Hitchhiker’s guide to biocatalysis: Recent advances in the use of enzymes in organic synthesis. Chem. Sci. 2020, 11, 2587–2605. [Google Scholar] [CrossRef]
- Wang, Z.; Sekar, B.S.; Li, Z. Recent advances in artificial enzyme cascades for the production of value-added chemicals. Bioresour. Technol. 2021, 323, 124551. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, K.; Bornscheuer, U.T.; Lütz, S. Cascades of Evolved Enzymes for the Synthesis of Complex Molecules. Angew. Chem. Int. Ed. 2022, 61, e202208358. [Google Scholar] [CrossRef] [PubMed]
- Siedentop, R.; Claaßen, C.; Rother, D.; Lütz, S.; Rosenthal, K. Getting the Most Out of Enzyme Cascades: Strategies to Optimize In Vitro Multi-Enzymatic Reactions. Catalysts 2021, 11, 1183. [Google Scholar] [CrossRef]
- Zhu, F.; Zhong, X.; Hu, M.; Lu, L.; Deng, Z.; Liu, T. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol. Bioeng. 2014, 111, 1396–1405. [Google Scholar] [CrossRef]
- Mandenius, C.-F.; Brundin, A. Bioprocess optimization using design-of-experiments methodology. Biotechnol. Prog. 2008, 24, 1191–1203. [Google Scholar] [CrossRef]
- Shen, L.; Kohlhaas, M.; Enoki, J.; Meier, R.; Schönenberger, B.; Wohlgemuth, R.; Kourist, R.; Niemeyer, F.; van Niekerk, D.; Bräsen, C.; et al. A combined experimental and modelling approach for the Weimberg pathway optimisation. Nat. Commun. 2020, 11, 1098. [Google Scholar] [CrossRef]
- Korman, T.P.; Opgenorth, P.H.; Bowie, J.U. A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun. 2017, 8, 15526. [Google Scholar] [CrossRef]
- Greenhill, S.; Rana, S.; Gupta, S.; Vellanki, P.; Venkatesh, S. Bayesian Optimization for Adaptive Experimental Design: A Review. IEEE Access 2020, 8, 13937–13948. [Google Scholar] [CrossRef]
- Liang, Q.; Gongora, A.E.; Ren, Z.; Tiihonen, A.; Liu, Z.; Sun, S.; Deneault, J.R.; Bash, D.; Mekki-Berrada, F.; Khan, S.A.; et al. Benchmarking the performance of Bayesian optimization across multiple experimental materials science domains. NPJ Comput. Mater. 2021, 7, 188. [Google Scholar] [CrossRef]
- Helleckes, L.M.; Hemmerich, J.; Wiechert, W.; von Lieres, E.; Grünberger, A. Machine learning in bioprocess development: From promise to practice. Trends Biotechnol. 2022. [Google Scholar] [CrossRef]
- Baraibar, Á.G.; Von Lieres, E.; Wiechert, W.; Pohl, M.; Rother, D. Effective Production of (S)-α-Hydroxy ketones: An Reaction Engineering Approach. Top. Catal. 2014, 57, 401–411. [Google Scholar] [CrossRef]
- Schmidt, S.; Schallmey, A.; Kourist, R. Multi-Enzymatic Cascades In Vitro. In Enzyme Cascade Design and Modelling; Springer International Publishing: Cham, Switzerland, 2021; pp. 31–48. [Google Scholar]
- Mordhorst, S.; Andexer, J.N. Round, round we go—Strategies for enzymatic cofactor regeneration. Nat. Prod. Rep. 2020, 37, 1316–1333. [Google Scholar] [CrossRef]
- Cho, S.-H.; Tóth, K.; Kim, D.; Vo, P.H.; Lin, C.-H.; Handakumbura, P.P.; Ubach, A.R.; Evans, S.; Paša-Tolić, L.; Stacey, G. Activation of the plant mevalonate pathway by extracellular ATP. Nat. Commun. 2022, 13, 450. [Google Scholar] [CrossRef] [PubMed]
- Rolf, J.; Julsing, M.K.; Rosenthal, K.; Lütz, S. A Gram-Scale Limonene Production Process with Engineered Escherichia coli. Molecules 2020, 25, 1881. [Google Scholar] [CrossRef] [PubMed]
- Dirkmann, M.; Nowack, J.; Schulz, F. An in Vitro Biosynthesis of Sesquiterpenes Starting from Acetic Acid. Chembiochem 2018, 19, 2146–2151. [Google Scholar] [CrossRef] [PubMed]
- Siedentop, R.; Dziennus, M.; Lütz, S.; Rosenthal, K. Debottlenecking of an In Vitro Enzyme Cascade Using a Combined Model- and Experiment-Based Approach. Chem. Ing. Tech. 2023. accepted. [Google Scholar] [CrossRef]
- Shimane, M.; Sugai, Y.; Kainuma, R.; Natsume, M.; Kawaide, H. Mevalonate-Dependent Enzymatic Synthesis of Amorphadiene Driven by an ATP-Regeneration System Using Polyphosphate Kinase. Biosci. Biotechnol. Biochem. 2012, 76, 1558–1560. [Google Scholar] [CrossRef]
- Becker, M.; Nikel, P.; Andexer, J.; Lütz, S.; Rosenthal, K. A Multi-Enzyme Cascade Reaction for the Production of 2′3′-cGAMP. Biomolecules 2021, 11, 590. [Google Scholar] [CrossRef]
- Andexer, J.N.; Richter, M. Emerging Enzymes for ATP Regeneration in Biocatalytic Processes. Chembiochem 2015, 16, 380–386. [Google Scholar] [CrossRef]
- Resnick, S.M.; Zehnder, A.J.B. In Vitro ATP Regeneration from Polyphosphate and AMP by Polyphosphate:AMP Phosphotransferase and Adenylate Kinase from Acinetobacter johnsonii 210A. Appl. Environ. Microbiol. 2000, 66, 2045–2051. [Google Scholar] [CrossRef] [PubMed]
- Frisch, J.; Maršić, T.; Loderer, C. A Novel One-Pot Enzyme Cascade for the Biosynthesis of Cladribine Triphosphate. Biomolecules 2021, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Mordhorst, S.; Singh, J.; Mohr, M.K.F.; Hinkelmann, R.; Keppler, M.; Jessen, H.J.; Andexer, J.N. Several Polyphosphate Kinase 2 Enzymes Catalyse the Production of Adenosine 5′-Polyphosphates. Chembiochem 2019, 20, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, Z.; Ning, X.; Xu, W.; Li, Z. In vitro biosynthesis of ATP from adenosine and polyphosphate. Bioresour. Bioprocess. 2021, 8, 1–10. [Google Scholar] [CrossRef]
- Niederreiter, H. Low-discrepancy and low-dispersion sequences. J. Number Theory 1988, 30, 51–70. [Google Scholar] [CrossRef]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef]
- Siedentop, R.; Rosenthal, K. Industrially Relevant Enzyme Cascades for Drug Synthesis and Their Ecological Assessment. Int. J. Mol. Sci. 2022, 23, 3605. [Google Scholar] [CrossRef]
- Becker, M.; Lütz, S.; Rosenthal, K. Environmental Assessment of Enzyme Production and Purification. Molecules 2021, 26, 573. [Google Scholar] [CrossRef]
- Keppler, M.; Moser, S.; Jessen, H.J.; Held, C.; Andexer, J.N. Make or break: The thermodynamic equilibrium of polyphosphate kinase-catalysed reactions. Beilstein J. Org. Chem. 2022, 18, 1278–1288. [Google Scholar] [CrossRef]
- Tavanti, M.; Hosford, J.; Lloyd, R.C.; Brown, M.J.B. Recent Developments and Challenges for the Industrial Implementation of Polyphosphate Kinases. Chemcatchem 2021, 13, 3565–3580. [Google Scholar] [CrossRef]
- Freier, L.; Hemmerich, J.; Schöler, K.; Wiechert, W.; Oldiges, M.; von Lieres, E. Framework for Kriging-based iterative experimental analysis and design: Optimization of secretory protein production in Corynebacterium glutamicum. Eng. Life Sci. 2016, 16, 538–549. [Google Scholar] [CrossRef]
- Adinarayana, K.; Ellaiah, P.; Srinivasulu, B.; Devi, R.B.; Adinarayana, G. Response surface methodological approach to optimize the nutritional parameters for neomycin production by Streptomyces marinensis under solid-state fermentation. Process Biochem. 2003, 38, 1565–1572. [Google Scholar] [CrossRef]
- Guercio, G.; Perboni, A.; Tinazzi, F.; Rovatti, L.; Provera, S. The Synthesis of GV143253A: A Case Study for the Use of Analytical and Statistical Tools to Elucidate the Reaction Mechanism and to Optimize the Process. Org. Process Res. Dev. 2010, 14, 840–848. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, SciPy, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0 Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]


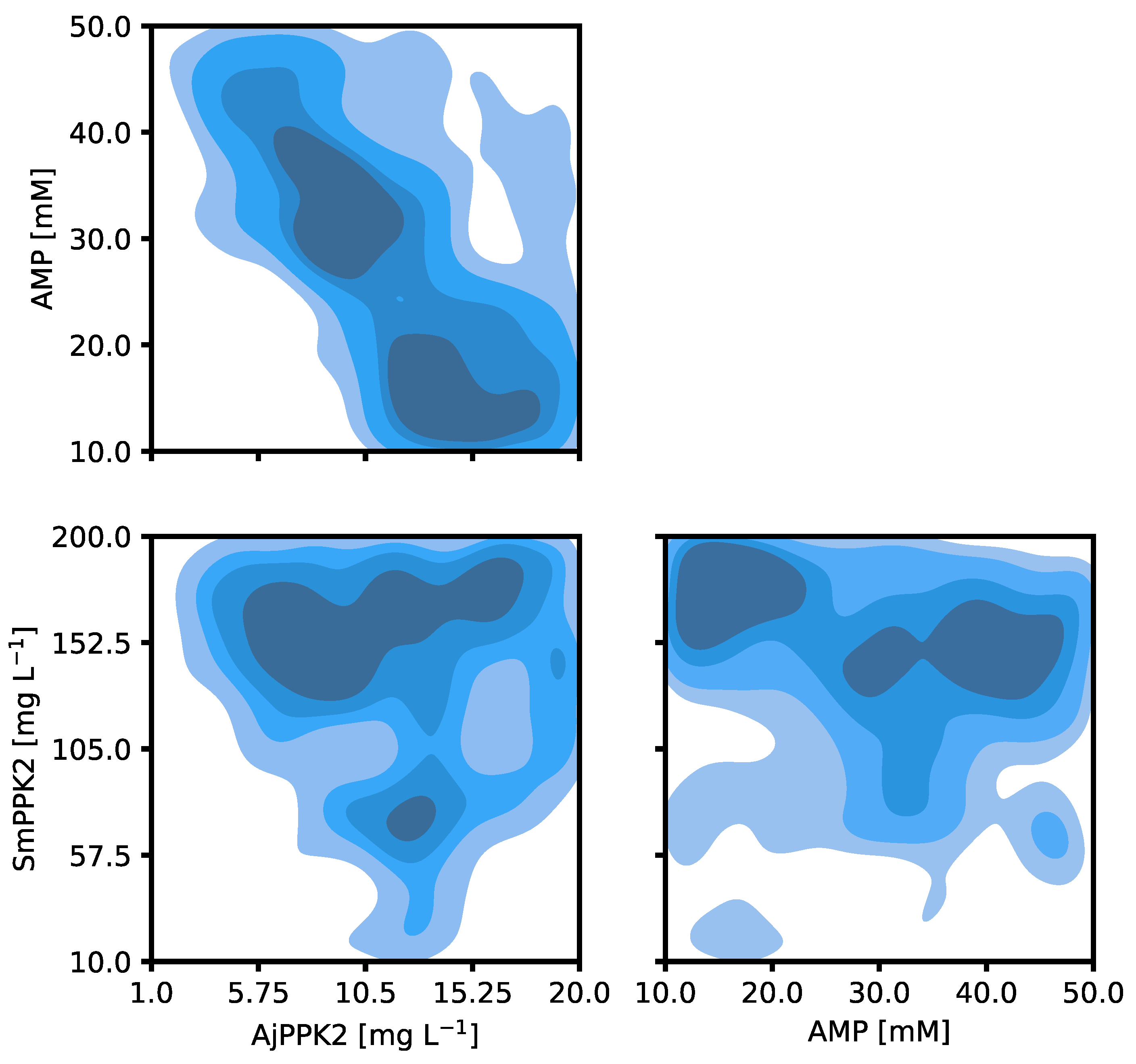
| Parameter | Tested Range |
|---|---|
| AMP concentration | 10–50 mM |
| AjPPK2 concentration | 1–20 mg L−1 |
| SmPPK2 concentration | 10–200 mg L−1 |
| Experiment | Conditions | Results | |||
|---|---|---|---|---|---|
| AMP [mM] | AjPPK2 [mg L−1] | SmPPK2 [mg L−1] | Average Specific Activity [U mg−1] | MVAP Concentration after 24 h [mM] | |
| Reference | - | - | - | 8.8 ± 1.4 | 44.0 ± 5.9 |
| Sobol 1 | 27.5 | 10.5 | 105.0 | 9.2 ± 0.5 | 33.9 ± 14.9 |
| Sobol 2 | 38.8 | 5.8 | 152.5 | 10.2 ± 0.3 | 35.7 ± 8.9 |
| Sobol 3 | 16.3 | 15.3 | 57.5 | 8.5 ± 0.6 | 26.9 ± 1.9 |
| Sobol 4 | 44.4 | 17.6 | 33.8 | 4.9 ± 2.9 | 23.3 ± 13.7 |
| Sobol 5 | 10.6 | 12.9 | 176.3 | 9.9 ± 1.8 | 26.9 ± 1.0 |
| Sobol 6 | 47.2 | 2.2 | 116.9 | 7.2 ± 0.6 | 44.2 ± 10.6 |
| Experiment | Conditions | Results | |||
|---|---|---|---|---|---|
| AMP [mM] | AjPPK2 [mg L−1] | SmPPK2 [mg L−1] | Average Specific Activity [U mg−1] | MVAP Concentration after 24 h [mM] | |
| Reference | - | - | - | 8.8 ± 1.4 | 44.0 ± 5.9 |
| Iteration 1.1 | 13.2 | 8.1 | 118.1 | 10.2 ± 1.5 | 34.8 ± 5.0 |
| Iteration 1.2 | 49.7 | 18.8 | 11.4 | 3.9 ± 0.3 | 22.6 ± 1.4 |
| Iteration 1.3 | 50.0 | 1.0 | 199.9 | 6.8 ± 0.4 | 18.5 ± 0.4 |
| Iteration 2.1 | 24.2 | 5.0 | 31.8 | 4.1 ± 0.8 | 31.7 ± 4.5 |
| Iteration 2.2 | 18.2 | 4.8 | 100.6 | 3.9 ± 0.3 | 20.9 ± 0.7 |
| Iteration 2.3 | 37.0 | 3.1 | 107.1 | 4.5 ± 2.0 | 18.4 ± 0.4 |
| Iteration 3.1 | 25.8 | 13.1 | 198.0 | 8.1 ± 1.0 | 50.8 ± 5.6 |
| Iteration 3.2 | 15.8 | 9.0 | 138.0 | 8.0 ± 0.8 | 51.9 ± 3.6 |
| Iteration 3.3 | 29.4 | 16.2 | 120.6 | 5.8 ± 0.5 | 52.0 ± 2.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siedentop, R.; Siska, M.; Möller, N.; Lanzrath, H.; von Lieres, E.; Lütz, S.; Rosenthal, K. Bayesian Optimization for an ATP-Regenerating In Vitro Enzyme Cascade. Catalysts 2023, 13, 468. https://doi.org/10.3390/catal13030468
Siedentop R, Siska M, Möller N, Lanzrath H, von Lieres E, Lütz S, Rosenthal K. Bayesian Optimization for an ATP-Regenerating In Vitro Enzyme Cascade. Catalysts. 2023; 13(3):468. https://doi.org/10.3390/catal13030468
Chicago/Turabian StyleSiedentop, Regine, Maximilian Siska, Niklas Möller, Hannah Lanzrath, Eric von Lieres, Stephan Lütz, and Katrin Rosenthal. 2023. "Bayesian Optimization for an ATP-Regenerating In Vitro Enzyme Cascade" Catalysts 13, no. 3: 468. https://doi.org/10.3390/catal13030468
APA StyleSiedentop, R., Siska, M., Möller, N., Lanzrath, H., von Lieres, E., Lütz, S., & Rosenthal, K. (2023). Bayesian Optimization for an ATP-Regenerating In Vitro Enzyme Cascade. Catalysts, 13(3), 468. https://doi.org/10.3390/catal13030468