Abstract
The regioselectivity characteristic of lipases facilitate a wide range of novel molecule unit constructions and fat modifications. Lipases can be categorized as sn-1,3, sn-2, and random regiospecific. Geobacillus zalihae T1 lipase catalyzes the hydrolysis of the sn-1,3 acylglycerol chain. The T1 lipase structural analysis shows that the oxyanion hole F16 and its lid domain undergo structural rearrangement upon activation. Site-directed mutagenesis was performed by substituting the lid domain residues (F180G and F181S) and the oxyanion hole residue (F16W) in order to study their effects on the structural changes and regioselectivity. The novel lipase mutant 3M switches the regioselectivity from sn-1,3 to only sn-3. The mutant 3M shifts the optimum pH to 10, alters selectivity toward p-nitrophenyl ester selectivity to C14-C18, and maintains a similar catalytic efficiency of 518.4 × 10−6 (s−1/mM). The secondary structure of 3M lipase comprises 15.8% and 26.3% of the α-helix and β-sheet, respectively, with a predicted melting temperature (Tm) value of 67.8 °C. The in silico analysis was conducted to reveal the structural changes caused by the F180G/F181S/F16W mutations in blocking the binding of the sn-1 acylglycerol chain and orientating the substrate to bond to the sn-3 acylglycerol, which resulted in switching the T1 lipase regioselectivity.
1. Introduction
T1 lipase is a triacylglycerol acylhydrolase (E.C.3.1.1.3) that belongs to the serine hydrolysis class. It originates from the bacterium Geobacillus zalihae and, due to its unique characteristics, it is considered as an important biocatalyst [1]. Lipases are able to catalyse the transesterification and synthesis of the esters as well as hydrolysis reactions. They can be implemented when performing particular biotransformation reactions and are widely exploited in the detergent, organic synthesis, cosmetics, food, and pharmaceutical industries [2,3].
Lipases can be divided into three groups according to their regioselectivity sn-1, 3, sn-2, or random sn-1, 2, 3 [4,5,6]. Non-specific or random lipases catalyse the hydrolysis of tryglycerols into free fatty acids and glycerols via a random process involving mono- and di-glycerides as intermediates. These lipases include those naturally produced by Candida rugosa and Pseudomonas fluorescens, for instance [7,8]. Lipase produced by Candida antarctica is a well-known and extensively researched non-specific lipase [9].
1,3-specific lipases hydrolyse the acylglycerols at the positions sn-1 and sn-3 of the triglycerides producing free fatty acids, 2-monoacylglycerols, and 1,2- or 2,3-diacylglycerols, avoiding the final production of glycerol. It can be extracted naturally from Aspergillus niger and C. rugosa [6,10]. The product is generated in different compositions to those obtained by random lipases. Most triacylglycerol lipases’ regiospecificity is sn-1,3, because they can only hydrolyse external positions, such as primary ester bonds at the sn-1 and sn-3 positions on the triacylglycerol backbone, and can generate either unhydrolysed diacylglycerol and one free fatty acid or unhydrolysed 2-monoacylglycerol and two free fatty acids [11].
Lipase from Thermomyces lanuginose is the most widely sn-2 lipase [12]. This monoacylglycerol lipase catalyses hydrolysis at the specific sn-2 position of 2-monoacylglycerol and trans-fatty acids into free fatty acid and glycerol. There are other lipases, such as fatty acid-specific lipases, which hydrolyse the long-chain fatty acids with double bonds in the cis-position between C-9 and C-10. To date, lipases with sn-3 regioselectivity have been reported in rabbit gastric lipase and insect midguts, as they prefer unsaturated fatty acids and preferentially hydrolyse TAGs on the α-positions [13,14]. However, there is a need for these rare lipases. Using specific lipases, lipids with a specific molecular structure that is not available through chemical interesterification can be produced, which is useful in the food processing and pharmaceutical industries.
The characteristic of regioselectivity seems to be connected to the selectivity of preferring one binding conformation over another, as dictated by the active site size shape [15]. However, lipases that hydrolyse α-positions on the triacylglycerides (TAGs), either sn-1 or sn-3-regioselectivity, are implemented in the production of enantiomerically pure glycerol derivatives, such as S-12-Oisopropylidene, S-12-O-benzylidene, and R-12-O-dibenzyl glycerol. They are useful building blocks in the synthesis of a host with optically active compounds and biological activity [16]. The structured lipid derived from glycerol is considered an important platform for newly structured triglycerol products. The structured triglycerol can then be exploited, especially when delivering the desired fatty acids for treating specific diseases or maintaining healthy nutrition [17]. The lipase’s regioselectivity merits can be used in the production of alpha chiral acylglycerol and fatty acids. These structured chiral acylglycerols have also exhibited a wide range of antimicrobial activity. It has been reported that the antimicrobial growth activity of fatty acids is involved oleic acid, linoleic acid, and arachidonic acid (C20:4) [18]. Fatty acids (FAs) of different chain lengths and saturation degrees have been widely implemented in anticancer drugs by coupling them with anticancer drugs.
Currently, there are few studies on the relationship between lipase structure and regioselectivity [19]. Mostly, research on lipase regioselectivity is centred on the screening of natural enzymes and their implementation in the alteration of oils and fats [20,21,22]. At present, few studies on the effect of mutation on regioselectivity have been reported [23]. The substitution of active site amino acids in the primary sequence may significantly improve the specificity of the enzymes. This substitution of the amino acid near the active site to either small or bulky amino acids was found to change the shape and size of the enzyme, which redesigned the binding site to be convenient for novel substrate structure and orientation. Hence, amino acid substitution to smaller side chain residues also confers more space and reduces the substrate steric hindrance, subsequently altering the enzyme selectivity, stereoselectivity, enantioselectivity, and regioselectivity [24,25,26]. In addition, the lid domain residues can also confer a certain amount of substrate selectivity by reducing steric hindrance [27].
In the present study, sn-3 regiospecific lipase was engineered from Geobacillus zalihae T1 lipase, cloned, and expressed in Escherichia coli to obtain purified recombinant protein to pave the way for more characterisation. The significance of having purified sn-3 regiospecific lipase can be exploited in the production of pure fatty acids on the sn-3 triglycerol backbone in the industrial and medical industries.
2. Results and Discussions
2.1. Preliminary Analysis and Lipase Mutant Development
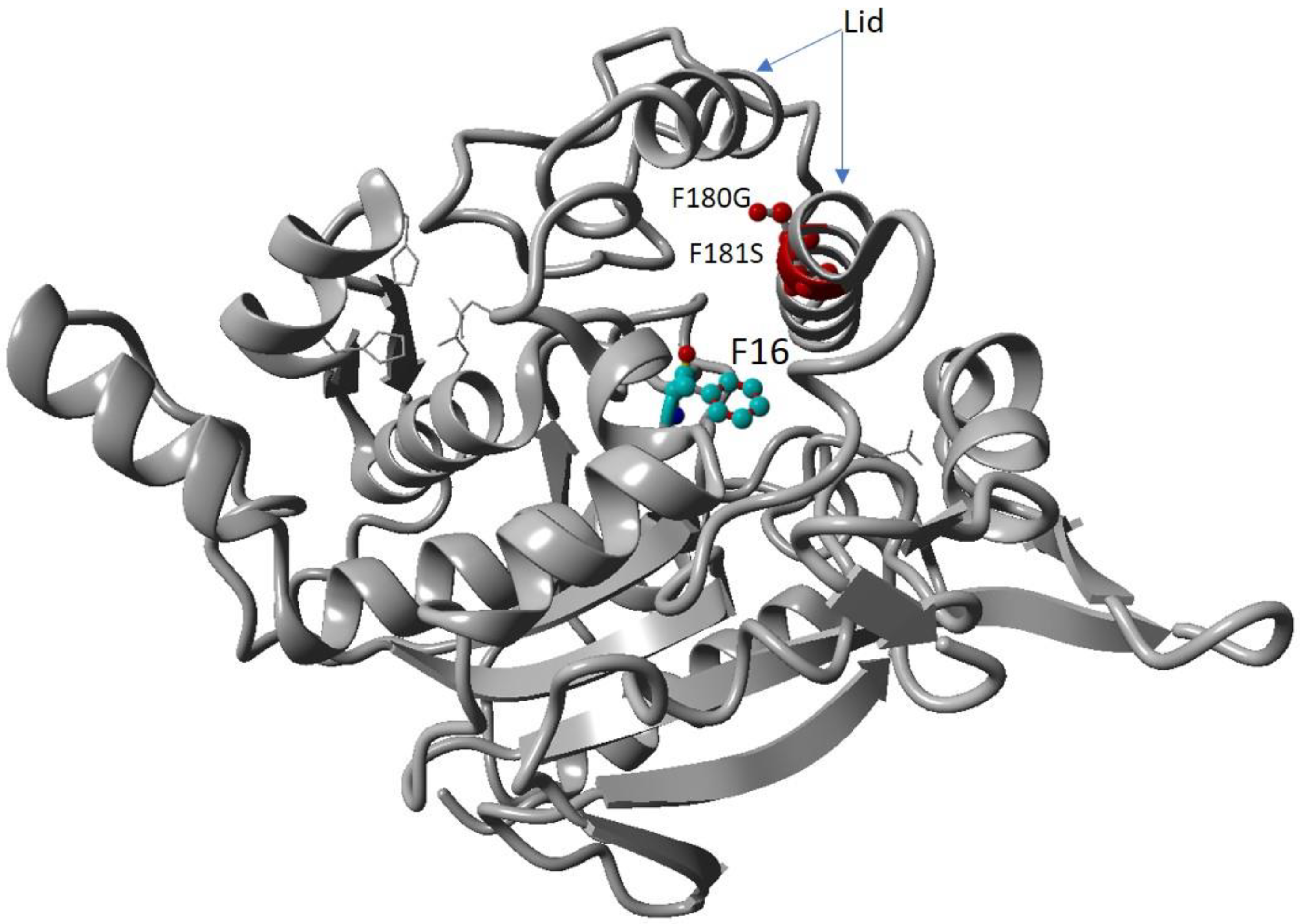
Pseudomonas aeruginosa lipase (PAL) (PDB ID 1EX9) is a non-specific lipase that has three catalytic fitting pockets that accommodate the sn-1, sn-2, and sn-3 fatty acid chains [28]. Based on the sequence alignment and structural analysis of T1 lipase and PAL lipase, it is revealed that acyl chain moieties in the wt-T1 lipase structure coincide with two acyl chains (sn-1 and sn-3) of the triacylglycerol substrate in the PAL. We identified two amino acids at position 180 and 181 at the lid and one oxyanion hole amino acid at position 16. The two amino acids at position 180 and 181 were identified as bulky amino acids in T1 lipase, while in PAL, these amino acids were found to be glycine and serine. Both residues F180 and F181 were located on α-helix 6, which constructs the lid domain structure of T1 lipase. The α-helix 6 is an asymmetric amphiphilic structure facing toward the protein core, with the hydrophobic part in the close conformation and hydrophilic parts facing the surface of the protein in the open conformation. Upon interfacial activation, helix 6 undergoes a sizeable secondary structure rearrangement. However, in the T1 lipase open conformation, the α-helix 6 hydrophobic residues are exposed to solvent. They might have interacted with the lipid which plays a role in the catalytic pocket. The substitution of these two bulky amino acids for simple amino acids might change the regiospecificity of lipase. The T1 lipase crystal structure introduced its closed conformation with the catalytic triad Ser113-Asp317- His359 and oxyanion hole F16, Glu114 (Figure 1), which plays an important role in the substrate hydrolysis process [29]. The oxyanion hole residue F16 (T1 lipase) is located at the enzyme access tunnel and works as a boomerang-like shape on the T1 lipase active site. This oxyanion hole F16 (Figure 1) plays an important role in the substrate hydrolysis process [30]. Hence, the substitution of this amino acid might affect the selectivity of lipase. To identify the most suitable amino acid that might affect regiospecificity, F16 was substituted with 12 other amino acids in combination with F180G/F181S, and the screening of lipase products from these 12 mutations was then conducted. The characteristics of the 12 mutants are presented in Table S1. The preliminary analysis indicates that some lipase variants exhibited sn-1,3-regioselectivity, which is the same as wt-T1, and only six variants (F180G/F181S/F16W, F180G/F181S/F16C, F180G/F181S/F16G, F180G/F181S/F16V, F180G/F181S/F16I, and F180G/F181S/F16S) were detected to have only sn-3 modified regiospecificity, as they displayed different selectivity toward different fatty acid chains (Figure S1). Out of the six, the variant F180G/F181S/F16W, namely the 3M mutant, was selected for further analysis.
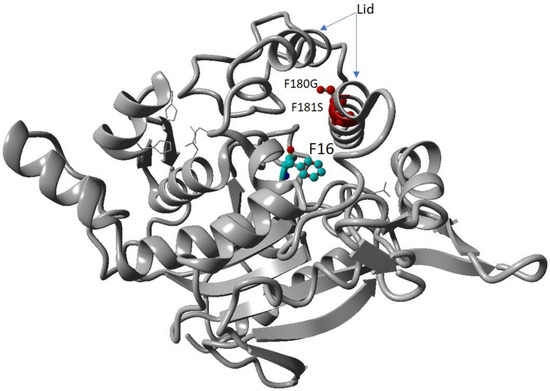
Figure 1.
Modelled mutant lipase with highlighted mutated residues. Red coloured residues are the mutations of F180G and F181S. The oxyanion hole residue F16 is the coloured element.
2.2. Expression and Purification of wt-T1 and 3M Lipases
The site-directed mutagenesis was implemented using a previously constructed pGEX-4 HT1 as a template [30]. The constructed 3M lipase was expressed in E. coli (BL21) fused to the His-tag. The wt-T1 and 3M lipases were purified using Ni-Sepharose affinity chromatography with a high final yield (88.6%) compared to wt-T1 lipase (54.8%), whereas the specific activity of the 3M lipase showed a lower range of specific activity (76.4 U/mg) compared to wt-T1 lipase (305.2 U/mg) (Table S2). Both purified proteins showed single bands of the expected size of 43 kDa, compared to the standard protein markers (Figure S2). The modified regiospecific mutant 3M has lower specific activity, emphasising the role of lid and oxyanion hole residues in lipase activity. The reason for this is that it is related to interfacial activity and the formation of hydrogen bonds to stabilize high-energy oxyanion intermediates [2,31]. The substitution of aromatic residues in the binding site can affect lipase activity because of their hydrophobicity and bulky size, which affects the shape and size of the binding site.
2.3. Biochemical Characterisation of wt-T1 and 3M Lipases
The TLC analysis of the purified wt-T1 and 3M lipases demonstrates the altered 3M regioselectivity from an sn-1,3 to only an sn-3-regioselectivity preference, compared to the wt-T1 [32] (Figure S3). This changes the T1 lipase regioselectivity from an sn-1,3-regioselectivity to only an sn-3-regioselectivity due to the substitution of the F16 residue, which works as a boomerang-like shape on the active site. This residue allows the access of the substrate to the catalytic (Ser), which subsequently changes its conformation to reorient its Oγ group toward the substrate. In a study conducted by Carrasco-Lopez et al. [33], which used BTL2 lipase (sequence identity 96% with T1 lipase), it was reported that the movement of Phe-17 in BTL2 is opposite to F16 in wt-T1, as it involves a change in its side-chain torsion angle from the closed to the open state. Thus, altering this residue to a bulky residue, such as Trp, would block the sn-1 binding pocket, thus allowing only the sn-3 acylglycerol chain to enter its binding pocket. Altering Penicillium expansum lipase regioselectivity was achieved by substituting residues at the C-terminal loop domain, which shows obvious sn-1,3 regioselectivity compared to the wild type [23].
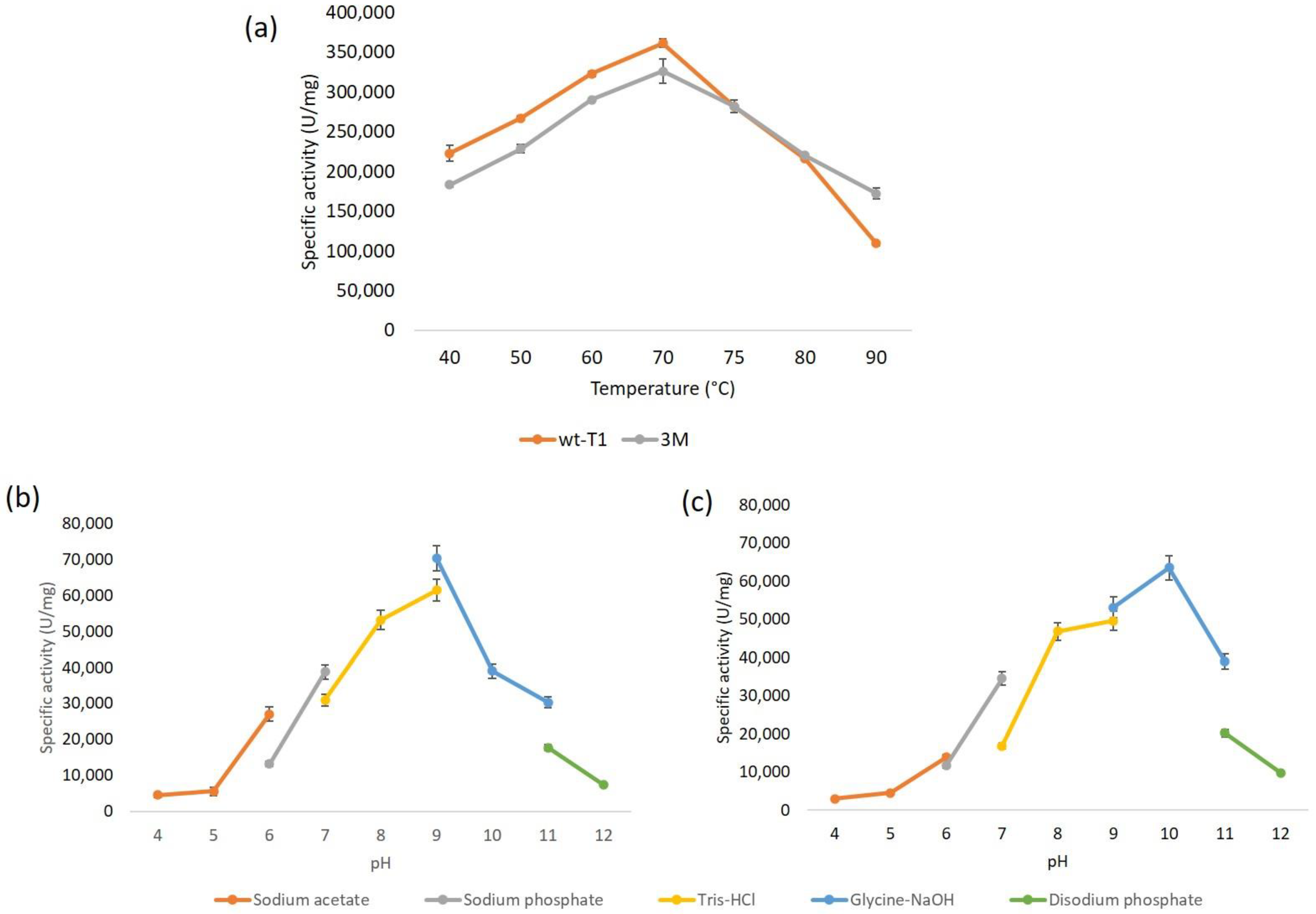
The wt-T1 and 3M lipase’s optimal temperature and pH were determined spectrophotometrically using the p-nitrophenyl-dodecanoate (pNP-C12) as a substrate (Figure 2). The characterization results are shown in Table S3. The maximal activity for both the wt-T1 and 3M lipases was at 70 °C, and the optimum pH for wt-T1 was at 9.0, whereas for the 3M, the pH was shifted to 10.0. A study conducted on T1 lipase reported that altering the second oxyanion hole, Q114, to Leu significantly improved T1 lipase folding and hydrophobic interaction, which in turn improved lipase stability, whereas exchanging Q114 to Phe and Trp disrupted enzyme stability [34]. Many studies reported that particular point mutations in the active sites were able to change the pH dependence of enzymatic activity [35]. Furthermore, the pKa value is generally determined by the catalytic group ionization and is influenced by the microenvironments of the enzyme and substrate interactions [36]. Hence, the shifting of pKa might occur due to the acidic and basic amino acids of the active site interaction comprising the alkaline substrate (fatty acids). This presumably creates a relatively favourable electrostatic binding environment [37]. A previous study reported that altering F17 to Ser in the Bacillus thermocatenulatus lipase BTL2, which has a 96% identity with T1 lipase, shifted the optimum pH from 9 to 10 [38].
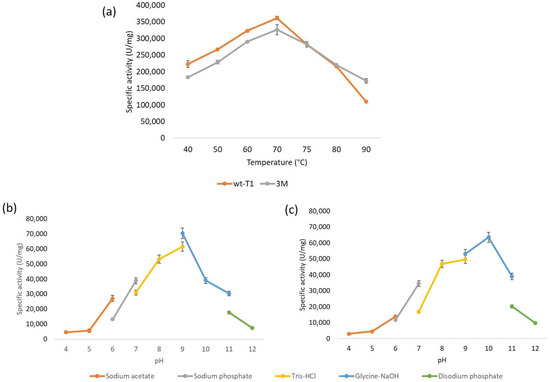
Figure 2.
Biochemical characterization of lipases. (a) The temperature profile of wt-T1 and 3M lipases. (b,c) pH profile on the activity of wt-T1 and 3M lipases, respectively.
The determination of secondary structural elements and the thermal denaturation of wt-T1 and 3M lipases, both proteins were subjected to circular dichroism (CD) spectroscopy analysis, which is a powerful tool for determining the secondary structure and folding properties of proteins. CD spectra allowed the rapid determination of the secondary structure elements of lipase in an aqueous environment. As shown in Table S3, the melting temperature (Tm) of wt-T1 was 73.5 °C and 67.8 °C for the 3M lipase. A 5.7% decline in the denaturation point of the 3M lipase mutant illustrated the weakening of interactions among buried residues. As mentioned by [39], losing the intramolecular hydrogen bonds required for buried residues exposes the packing of close-packed spheres and groups in the interior of a protein to protein unfolding or denaturation. The substitution of the lid and oxyanion hole residues buried in the core of the protein might affect the intramolecular interactions and partly cause the decrease in the thermal denaturation of the mutant by 5.7%. The alteration of the protein surface residues and charge distribution in certain cases could be responsible for the retention of the enhanced activity upon thermal treatment [40] and, vice versa, could decrease protein thermal stability because of protein misfolding and the disruption of the hydrogen network [41,42].
The CD wavelength scan in the far ultraviolet region (190–240 nm) at pH 7.4 revealed a similar curve for both lipases, with two negative bands, as expected for an ɑ/β-protein (Figure S4). At 193–194 nm, the CD spectrum revealed a sharp positive band and a strong negative band at 195 to 235 nm. The results showed that wt-T1 and 3M lipases have a substantial similarity in the structure composition of both the α-helix and random coil. The CD-spectral scans of wt-T1 and 3M did not completely overlap at the negative band, indicating that certain secondary structural changes occurred upon mutation. Structurally, 3M was found to have a lower α-helical ratio of 16% with a concomitant increase in the β-sheet ratio to 26%. As for wt-T1 lipase, it was detected to have a 20% α-helical and 13% of β-sheet. The slight changes in tertiary and secondary structures were reflected in optimal catalytic parameters, such as pH. The optimal pH for activity increased from pH 9 to pH 10, indicating that the protein folded into a structure that required alkaline pH to achieve an absolute functional form after mutation. Jiang et al. [43] demonstrated that the mutation cause changes in the secondary structure of the lipase. The study of Lipr27RCL-K64N and Lipr27RCL-K68T by the CD spectra also indicates there were some differences in the thermal stability of the two mutants.
Both lipases were subjected to Michaelis–Menten kinetics to determine substrate affinity (KM), maximum velocity (Vmax), turnover number (kcat) and kcat/KM. According to the kinetic analyses, 3M lipase showed a catalytic efficiency (kcat/Km) of 6.98 × 107 s−1/mM with enhanced substrate affinity (Km) to 1.47 ± 0.29 mM and a decline in the maximum velocity (Vmax) to 119.33 ± 18.44 µmol/min/mL compared to wt-T1, which displayed catalytic efficiency, the Km and Vmax of 1.23 × 108 s−1/mM, 1.14 ± 0.29 mM and 163.24 ± 8.16 µmol/min/mL, respectively (Figure S5 and Table 1). Thus, the substitution of bulky amino acids with small residues in the lid domain F180S/F181G and oxyanion hole F16W with larger amino acids changed the binding site conformation and improved the substrate affinity and caused a decline in maximum velocity. The catalytic efficiency of Candida antarctica lipase B (CALB) increased the hydrolysing 4-nitrophenyl benzoate by 40-fold more than the wild type by substituting the binding site residues W104V/A281Y/A282Y/V149G, as these substituted residues reshaped the binding site and facilitated the entrance of this substrate [44].

Table 1.
Michaelis–Menten kinetics measuring the KM and kcat of the wt-T1 and 3M variant lipases.
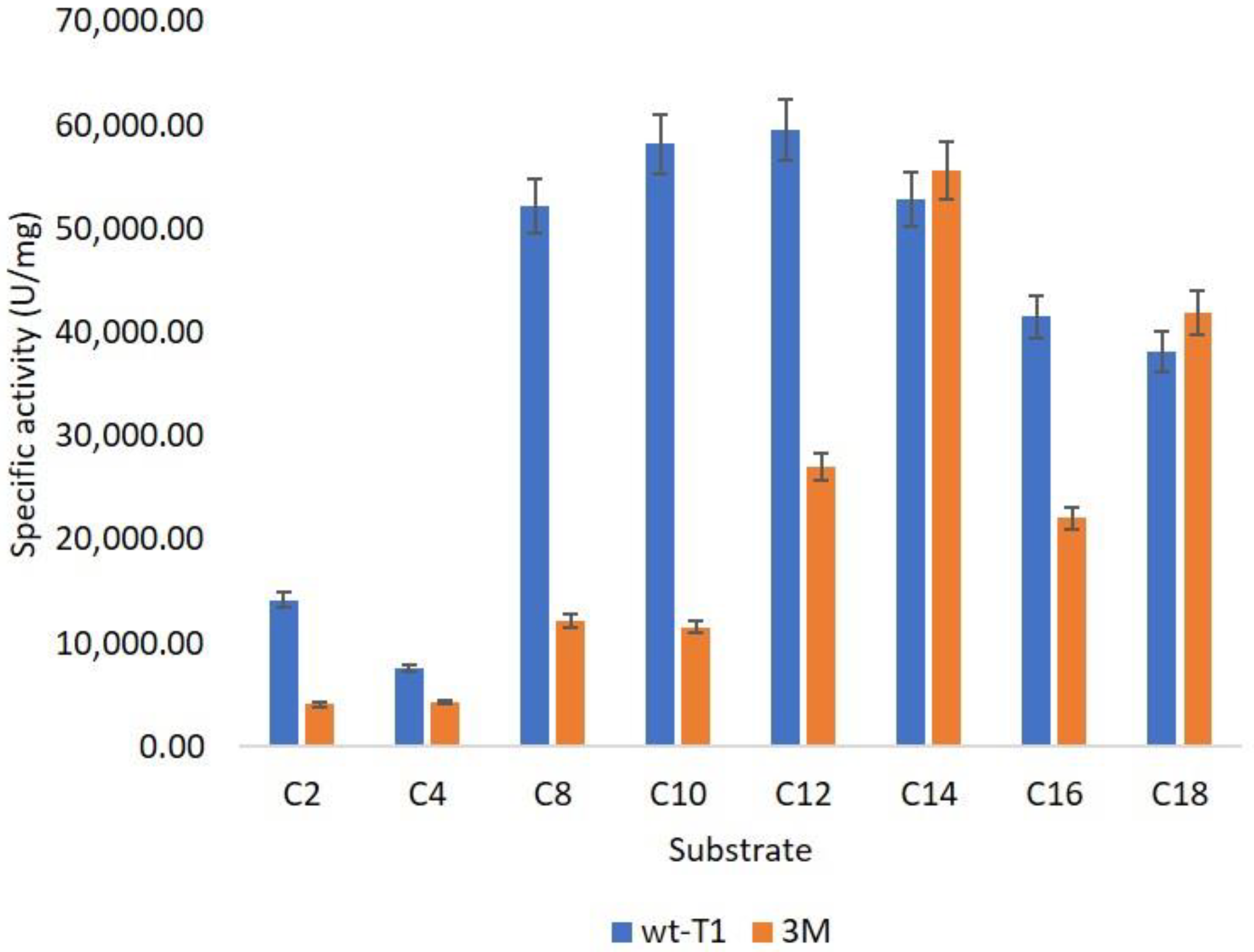
The substrate selectivity (pNP-ester) of 3M is also influenced by enhancing the long-chain preference from (C14–C18) compared to wt-T1 (C12–C14), as displayed in Figure 3. Thus, reshaping the binding site altered enzyme size and shape to be convenient for the lengthy carbon chain. Targeting the binding site access channel altered Candida antarctica lipase A (CAL-A) specificity for different chain-length fatty acids [45,46,47].
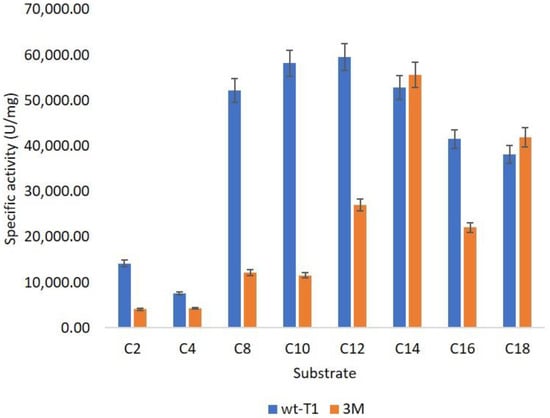
Figure 3.
Substrate specificity of wt-T1 and 3M lipases toward different pNP-ester carbon lengths.
2.4. Hydrolysis of Palm Stearin by wt-T1 and 3M Lipases
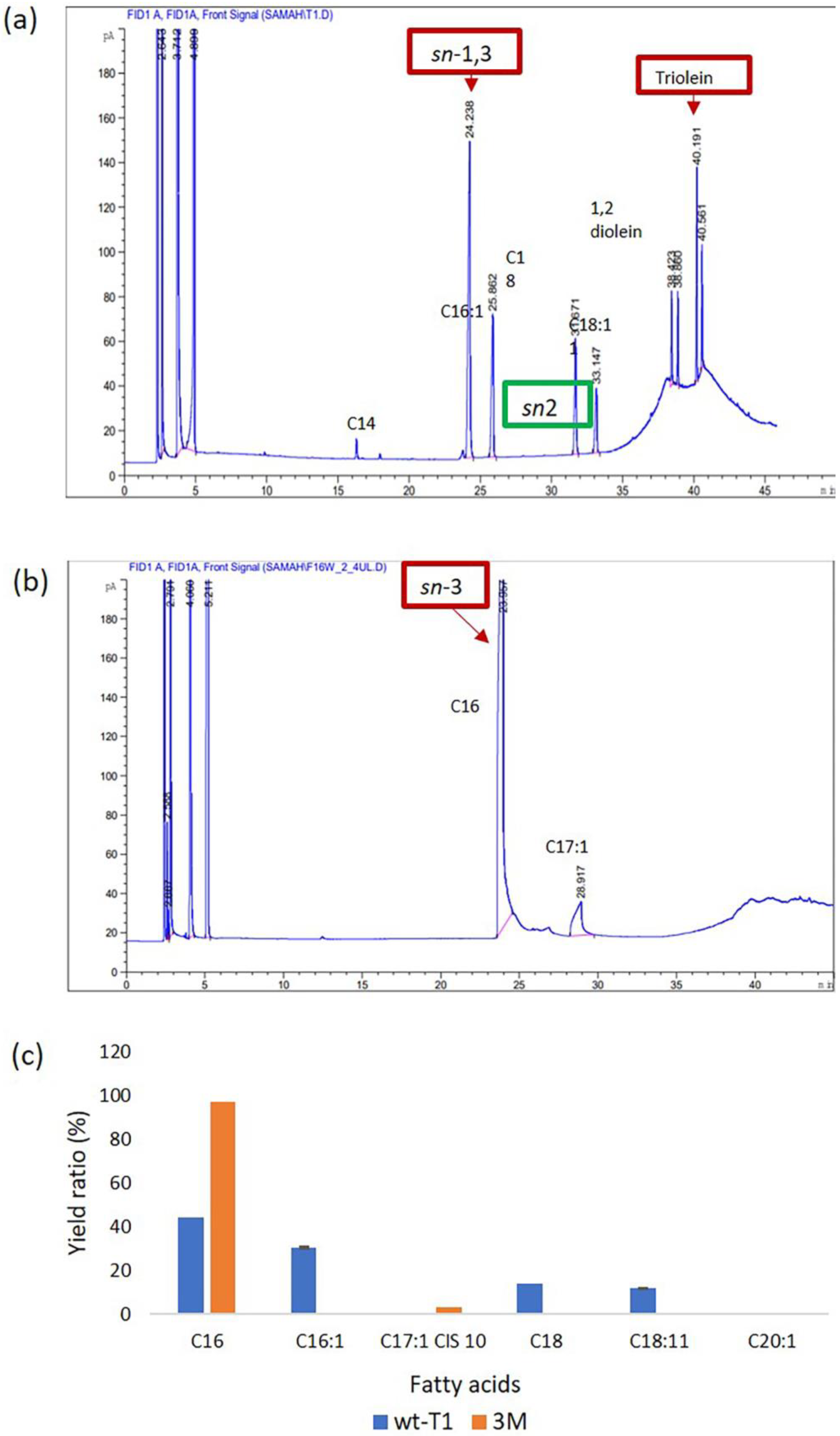
The wt-T1 and 3M lipase affinity toward C12–C14 of pNP-ester and their abilities to hydrolyse long-chain triacylglycerols (TAGs) was investigated using palm stearin substrate as it is composed of different fatty acids, ranging from C12–C20. The wt-T1 lipase exhibited the ability to hydrolyse C16 (43.9%), C16:1 (30.4%), C18 (13.6%) and C18:11 (11.8%). Whereas the 3M lipase displayed high selectivity toward C16 (97%) and C17:1 cis 10 (2.9%). Reactions were carried out under the optimized temperature and pH of each lipase. The fatty acid (FA) content released from palm stearin and its rate are shown in Figure 4. Engineering T1 lipase resulted in altering both regioselectivities toward sn-3 and its selectivity as palmitic acid C16 allocated on the sn-1 and sn-3 of palm stearin backbone. Using palm stearin as a substrate demonstrated that the 3M lipase has higher selectivity towards C16 and C17:1 cis 10 compared to wt-T1, which has a selectivity towards C16, C16:1, C18 and C18:11. A previous study reported that targeting Candida antarctica (CAL-A) lipase acyl-binding tunnel improved the esterification of elaidic acid (C18:11 trans fatty acid) reaction rate by 15-fold [45]. Altering T1 regiospecificity also depends on the constructed shape and space of the substituted residue, which in turn alters the steric hindrance of the active site. Thus, 3M lipase can be exploited in the hydrolysing sn-3 region, with either saturated or unsaturated fatty acids. This enzyme could be utilized in anticancer medication by coupling it with anticancer drugs to improve tissue selectivity and enhance drug stability and bioavailability. These combinations also reduce the undesirable interactions of the used drugs, as these fatty acids are not cytotoxic for normal cells [48].
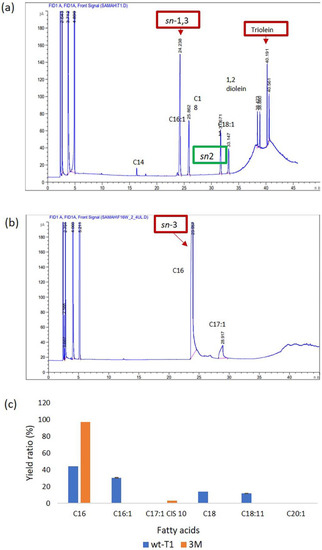
Figure 4.
Gas chromatography flame ionisation detector (GC FID) analysis for (a) wt-T1 lipase and (b) for 3M lipase. The acylglycerol position is highlighted by the red box. (c) Different fatty acid yields in percentages produced by wt-T1 and 3M lipases. The experiment was conducted using palm stearin as a substrate and single-factor ANOVA analysis with n = 3.
2.5. Computational Analysis
2.5.1. Homology Modelling and Docking Analysis of the Open Conformation of Lipases
The open structure of lipases (3M and wt-T1) was developed using the available crystal structure of BTL2 lipase (PDB ID: 2W22) as a template. Both modelled structures show a high similarity and the superimposed of RMSD values of 0.4555 Å over 5936 matched atoms (Figure S6). The validation data performed by online software indicates that both structures are of good quality (Table 2). According to Laskowski et al. [49], a good quality model is obtained if the model has more than 90% of residues in the most favoured region in the Ramachandran plot. As shown in Figure S7, both structures recorded more than 90% of all residues in the most favoured region.

Table 2.
Summary of validation data of lipases.
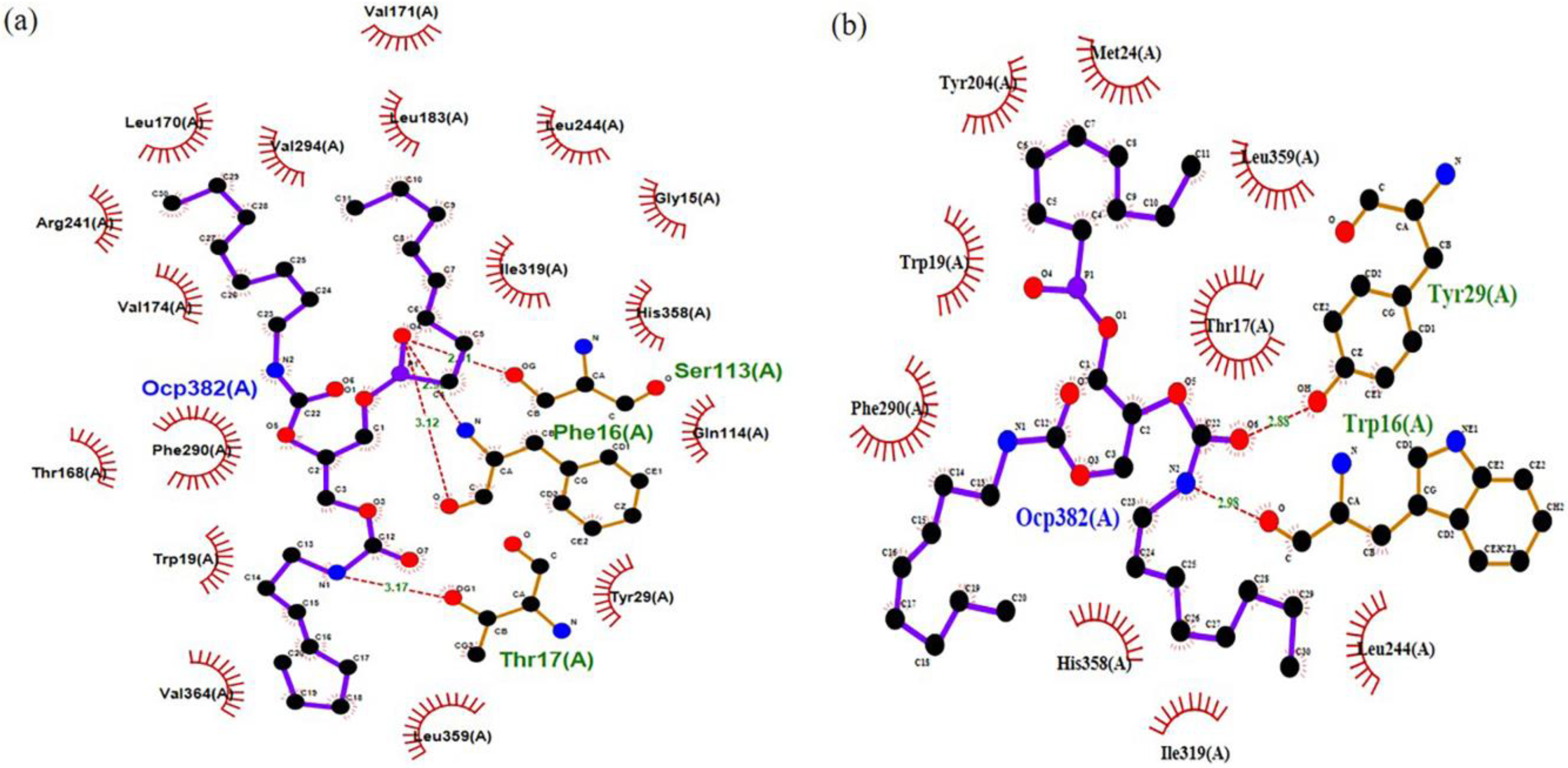
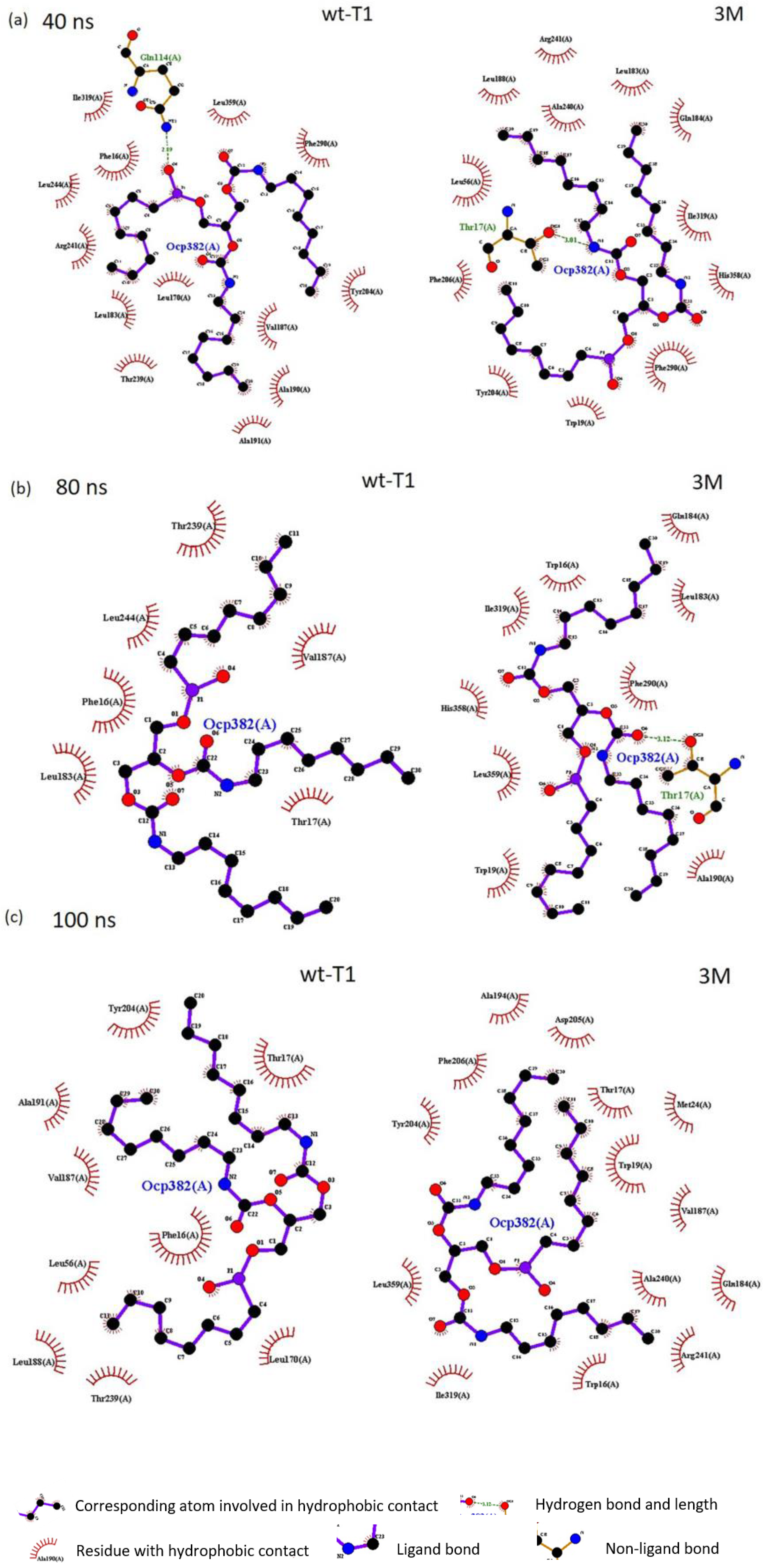
The docking analysis of wt-T1_OCP and 3M_OCP complexes determined the binding energy of 7.236 kcal/mol and 5.644 kcal/mol, respectively (Table S3). The complex 3M_OCP recorded low binding energy as compared to the wt-T1_OCP complex. Further analysis was carried out to investigate the enzyme–ligand molecular interaction changes, including the substituted oxyanion hole Phe16 to Trp and catalytic residues Ser113, His358 and Asp317. According to Figure 5, four hydrogen bonds were formed between the catalytic residue Ser113 OG group of wt-T1 and O4 atom of the substrate acylglycerol chain, along with a couple of hydrogen bonds formed by the O and N atoms of Phe16, which represented the oxyanion hole with the substrate O4 atoms. The fourth hydrogen bond was formed by the OG group of the Thr17 residue next to the oxyanion hole with the substrate N1 atom, which belongs to the second acylglycerol chain. The distances of the four generated hydrogen bonds ranged from 2.31 to 3.17 Å. The modified 3M sn-3 regiospecific structure was observed to have a distinct binding mode accompanied by the rearrangement of the active site. The imidazole moiety of Trp16 swings upwards from the lipase binding site entrance and blocks the acyl glycerol cavity sn1, leading to the active site structural rearrangement. A stable hydrogen bond was formed between the imidazole ring of both Trp16 and Tyr29 and the substrate carboxyl group. The acyl glycerol chain of the substrate was attacked by the two benzene rings of Trp16 and Tyr29. Overall, the acyl glycerol chain located in the sn-3 trench appeared to have more extensive interactions with the surrounding residues in 3M lipase compared to the wt-T1 lipase.
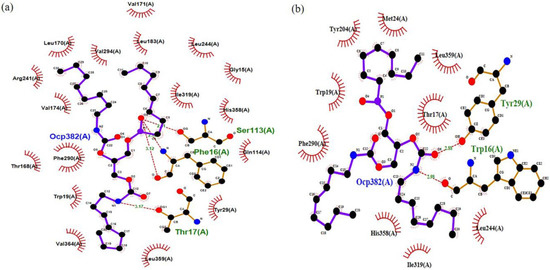
Figure 5.
The 2D structure complex of wt-T1 and 3M variants with a substrate analogue OCP (RC-(RP, SP)-1,2-dioctylcarbamoyl-glycero-3-O-octylphosphonate). (a) wt-T1 lipase-OCP. (b) 3M lipase-OCP (sn-3 modified regiospecific lipase). Hydrogen–bond interactions are shown as red dotted lines and ligands as purple and black.
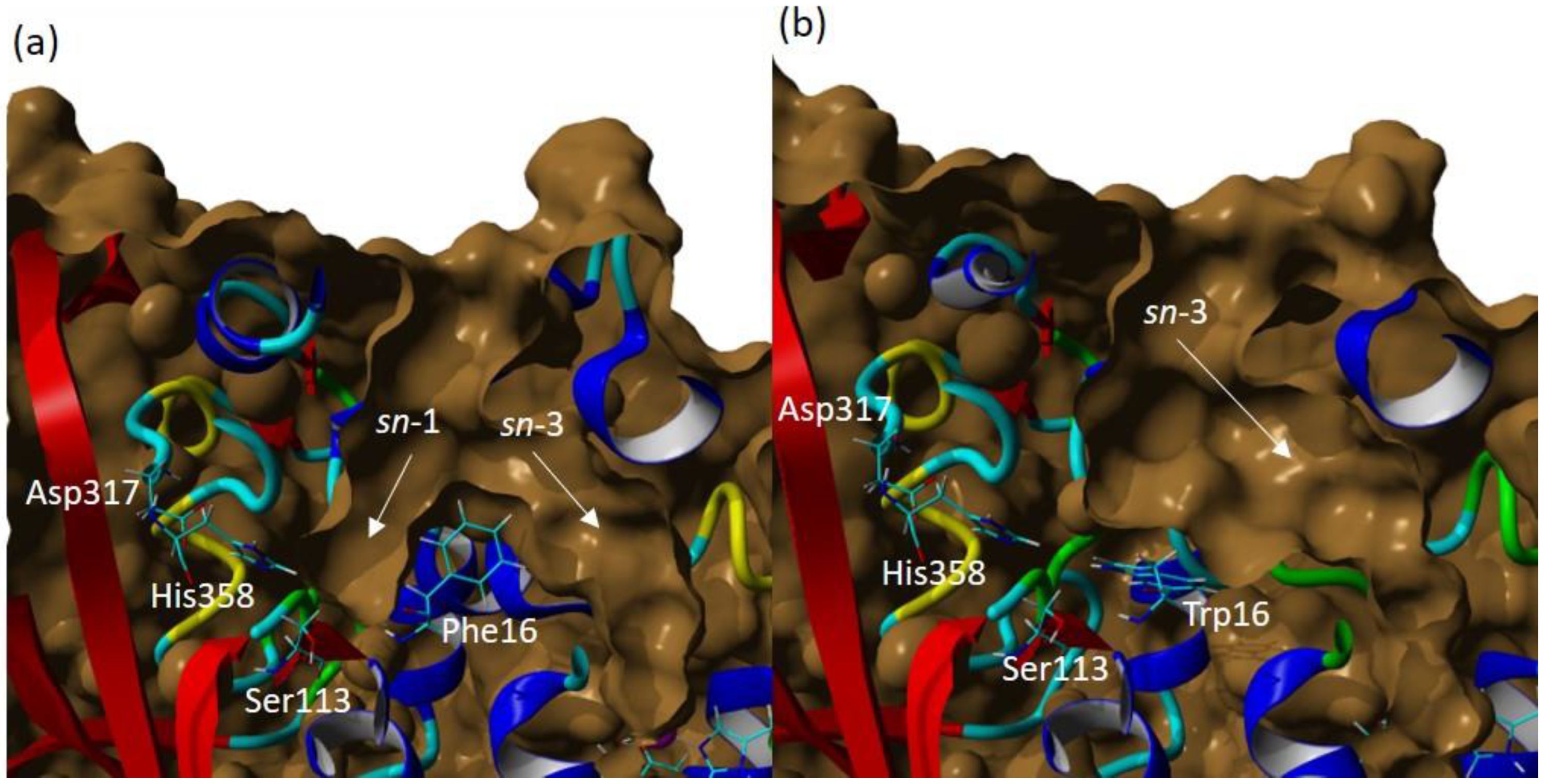
The volume of the wt-T1 lipase binding site was detected to be around 3082.41 Å and a depth of 22 Å. In contrast, the volume of the 3M lipase was detected to be 2587.75Å, and the depth of 30 Å. Figure 6 shows the difference in the shape of both structures and emphasised the blocking of one of the acylglycerol trenches, assumed to be the sn-1 binding pocket by the altered F16 to W 16 residue. It is worth mentioning that the measured binding site was set up using the specific identical residues for both structures and these residues are Phe16, Thr17, Ser113, Asp317 and His358. However, both structures were plotted and measured by using open conformation.
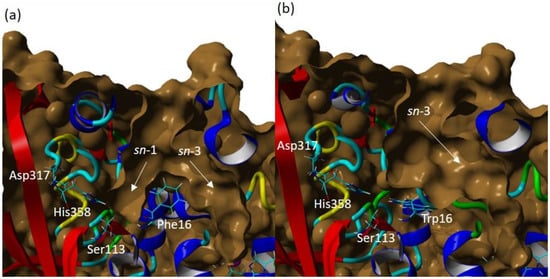
Figure 6.
The surface of wt-T1 and 3M variant lipase binding sites. (a) wt-T1 lipase. (b) 3M mutant lipase. The catalytic residues are shown in a cyan colour and labelled in a white colour and the light grey surface represents the open conformation of the binding site, binding pocket sn-1 and sn-3 highlighted by the yellow arrow.
2.5.2. Molecular Dynamics (MD) Simulation Analysis
In order to predict the effect of substituted residues on the 3M lipase stability structure, a molecular dynamics simulation (MD) study was conducted and several parameters, such as the root mean square deviation (RMSD), the root mean square fluctuation (RMSF), the radius of gyration and solvent accessible surface area (SASA) (Figure S8) for a simulation of 100 ns at 70 °C, were analysed.
In Figure S8a, the RMSD value of the 3M structure starting configuration was slightly increased during the initial equilibration phase but quickly converged after 11 ns over the 90 ns of MD simulations and reduced again at the end of the 100 ns. The 3M lipase system displayed slightly higher RMSD values of 0.55 Å and 4.1 Å, followed by wt-T1, which displayed the RMSD of 0.58 Å and 3 Å, respectively.
Supplementary Figure S8b represents the SASA value of wt-T1 and mutant lipases over a 100 ns simulation period. The wt-T1 displayed a slightly higher value of 15543.6 to 16597.8 Å2 compared to the mutant lipase structure of 15359.8 to 15543.3 Å2. However, the value of mutant lipase fluctuated between 10 to 20 ns and 50 to 98 ns, corresponding to the same period structure deviation based on the RMSD analysis.
Supplementary Figure S8c represents the compactness displayed in the radius of gyration (Rg) of the wt-T1 lipase, starting with 20.67 Å and ending with 20.98 Å over the 100 ns. The 3M structure starts with 20.62 Å and ends with 20.74 Å. In terms of the SASA and gyration values, it could be proposed that the structure modification has affected the structure compactness and secondary structure folding of the mutant lipase but in a narrow range, which might refer to the stabilised protein–ligand complex.
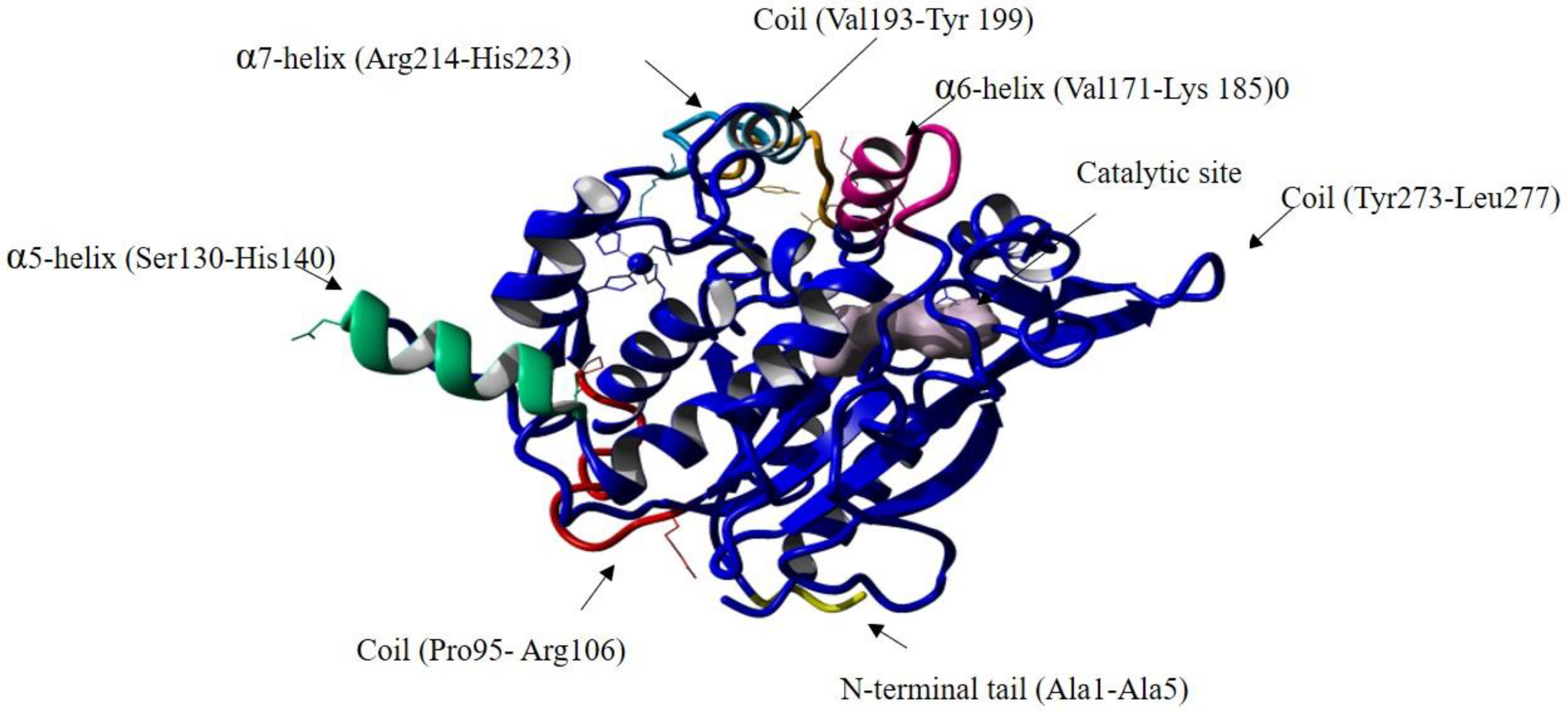
Figure S8d reveals that the RMSF overall value during the simulation was maintained between 0.5Å and 2 Å, except for many individual residues that were revealed to have high peaks, as listed in Supplementary Table S4. These residues are depicted in Figure 7. The residues that displayed flexibility were Pro95 and Arg106, which construct a coil that connects the α-helix 3 and α-helix 4, and both were observed to have a high deviation. These residues are (Pro95 and Arg106) close to the catalytic Ser113, which could increase the active site flexibility and facilitate substrate accommodation. A study conducted on the cold-adapted Pseudomonas mandelii Esterase EstK reported that a mutation close to the active site’s trait loop increased the flexibility of the active site as it is involved in the hydrogen bond formation [50]. The second structure deviation was on the α-helix 6 (Val171-Lys 185), α-helix 7 (Arg214-His223) and the turn domain connected them, which might be due to their function, as they comprise the lid domain in the wt-T1 lipase, and their role is involved in the interfacial activation process.
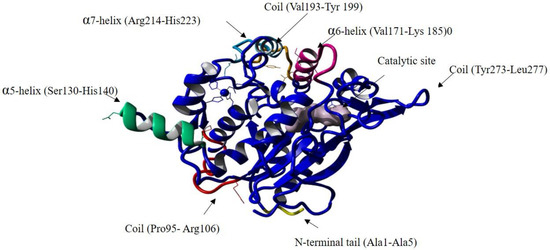
Figure 7.
The structure of the wt-T1 lipase with the positions of amino acids residues with high RMSF values highlighted. The residues are indicated by the arrows.
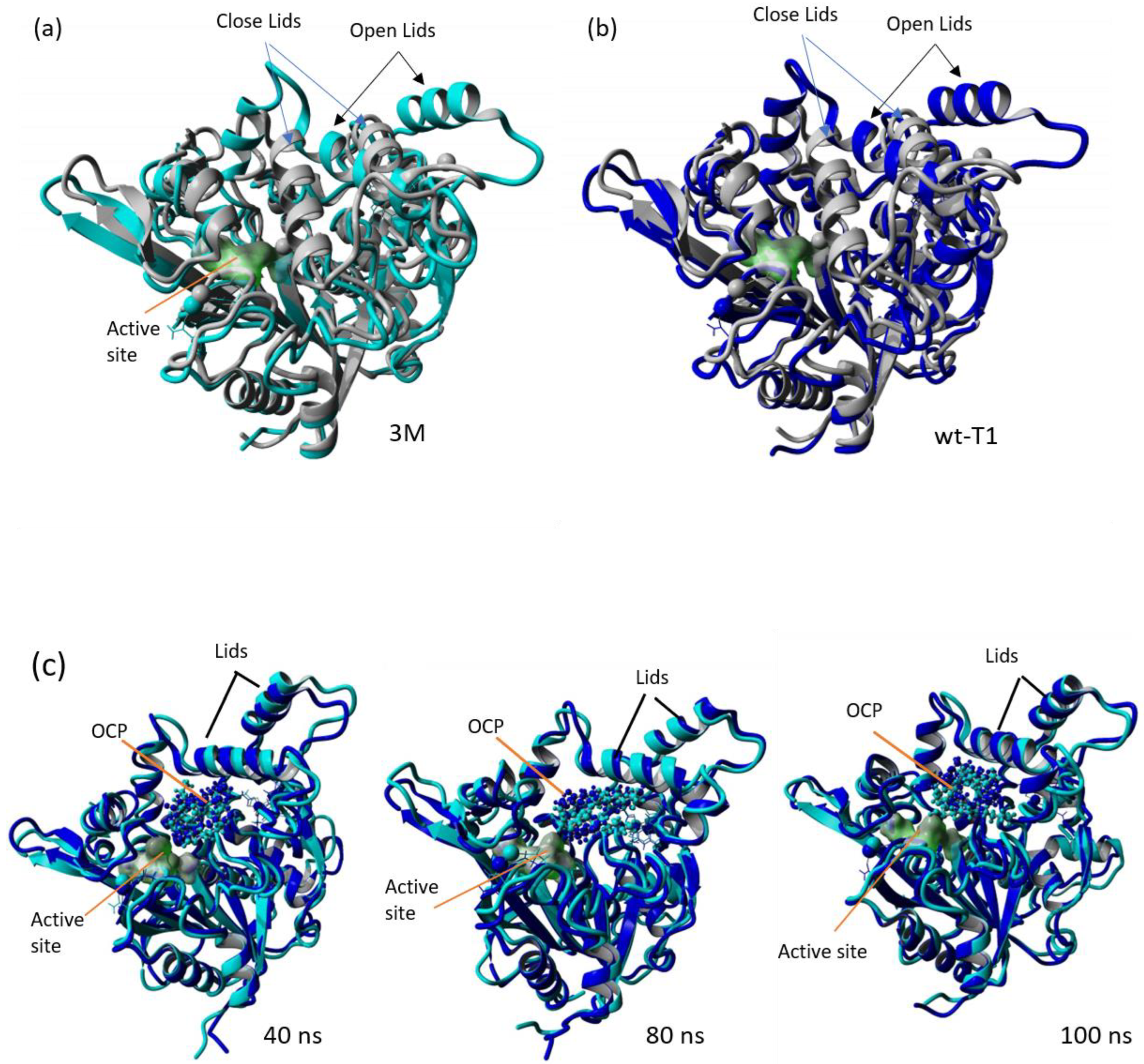
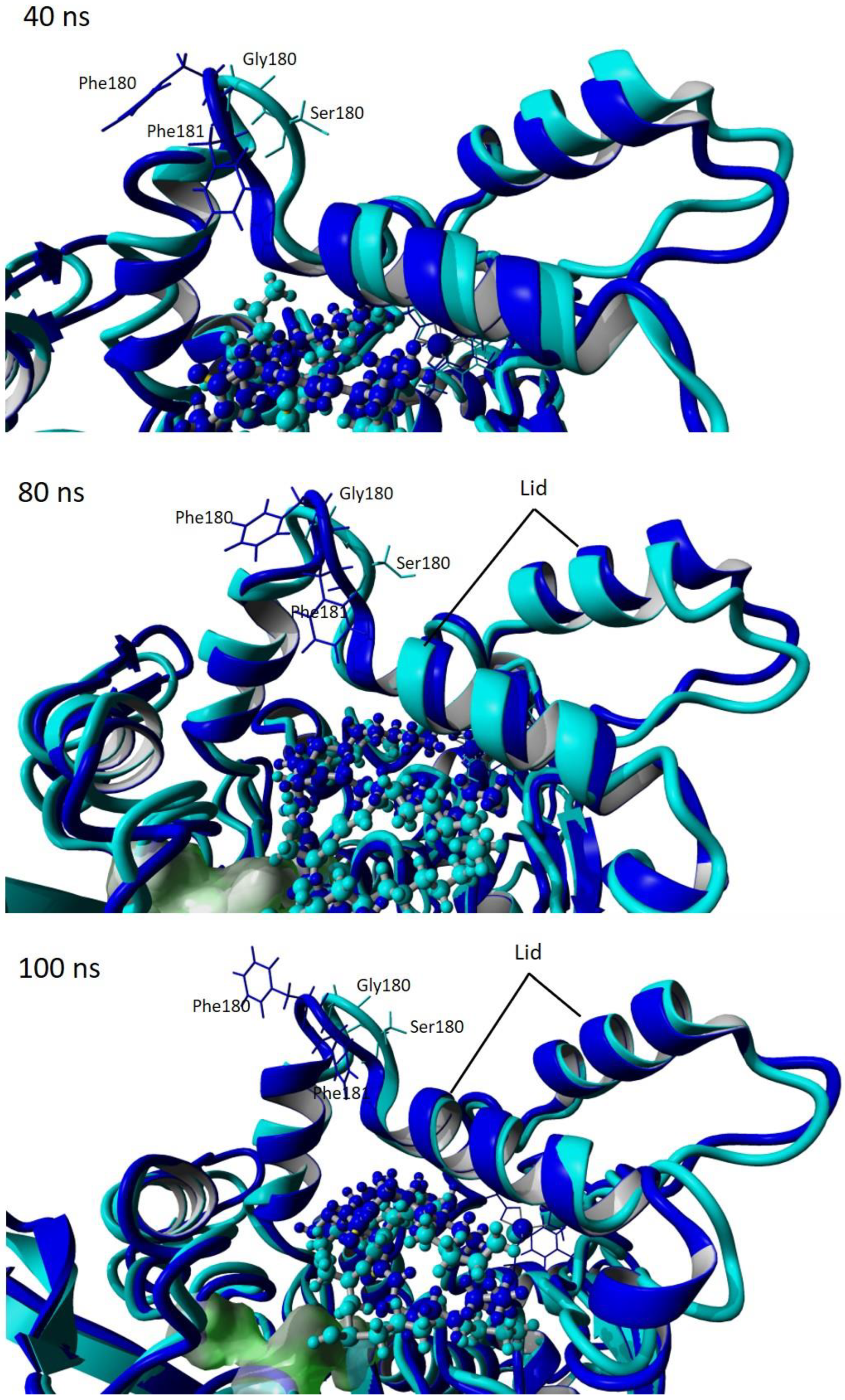
In the study conducted by Carrasco-López et al. [33], it was reported that the structural rearrangement of α-helix 6 and α-helix 7 in BTL2 opened conformation, which facilitated the substrate entrance. The zinc-binding domain played a role in stabilising the oxyanion hole; thus, altering the oxyanion hole residue F16 increased the flexibility of this domain. The role of the Zn2+ ion-binding domain in the subfamily I.5 lipases has been investigated and approved for its role in the opening of the lid and maintaining active conformation at high temperatures [33,51]. The snapshots of the trajectories of MD simulations for the wt-T1_OCP and 3M_OCP variant lipases are given in Figure 8, Figure 9 and Figure 10, demonstrating the motion of the lids and the OCP substrate throughout the simulation. The comparison of the complex structures of wt-T1_OCP and 3M_OCP showed several differences noticeable through the superposition of the modelled structures; however, the most notable difference is the structural transformation of the lid domain and around the Zn+2 atom coordination domain of the 3M lipase. Further analysis showed the structure deviation of the α-helix 5 and the partial construction of the lid domain α-helix 6 and α-helix 7. In addition, a partial structure transformation that rotated from helix–coil–helix–coil through the simulation was noticed on the α-helix3, α-helix 8 and α-helix 2, which comprise the Zn+2 atom coordination domain. The domain that constructs residues around Zn+2, which is involved in hydrogen bond formation, was noticed to have a secondary structure transformation, and this might be due to the oxyanion hole Phe16 alteration to Trp. The Phe16 residue movement is proposed to contribute to the formation of the oxyanion-binding pocket, and the altering of this residue might distort the hydrogen bonds between the Zn+2 atom coordination residues Asp61, Tyr77, His87 and Asp238. These residues are located on the α-helix 2 and α-helix 3. In the bacterial lipases of subfamily I.5, the Arg63 is placed just after the Zn+2 atom coordination Asp62, which is assumed to play a role in stabilising the oxyanion binding pocket, as is reinforced by the Zn+2- binding domain [33]. Higher flexibility was noticed on both the α-helix 6 and α-helix 7, which forms the lid domain of the T1 lipase, and this flexibility increased because of the replacement of the bulky hydrophobic residues Phe180 and Phe181 located on α-helix 6 with small residues Gly and Ser, respectively. The substitution of Phe for Gly and Ser enhances the conformational flexibility of the lipase structure according to the conformational flexibility scale, whereas introducing larger residues leads to lower flexibility because of the lower collision frequencies [52]. The α-helix 7 molecular hinge is located at residue Asp239, which is involved in the Zn+2 atom coordination residues, and this may contribute to the α-helix 7 flexibility as it is involved in the formation of the oxyanion-binding pocket. The α-helix 5 showed a higher deviation compared to other enzyme parts due to its location, which is between the α-helix 3, which forms the oxyanion hole, and α-helix 6, which forms the lid domain, and both were subjected to the modification as it involved F16W, F180G and F181S. The active site superposition of both wt-T1 and 3M modelled lipases reveals a significant change to the position of the catalytic site His residue and the oxyanion hole Phe16 residue, while a minor change was noticed in the catalytic residues Ser and Asp (Figure S9).
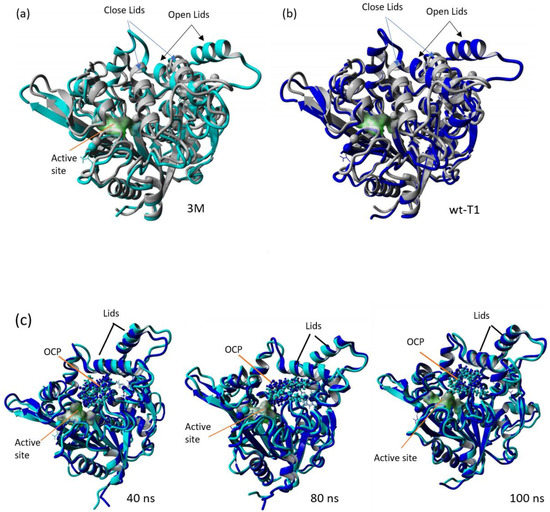
Figure 8.
Superposition of wt_T1 closed structure (grey), wt-T1_OCP (blue) and 3M_OCP (cyan). (a) Superimposition of the closed structure of T1 lipase with wt-T1_OCP extracted from MD simulation trajectories at 100 ns. (b) Superimposition of the closed structure of T1 lipase with 3M_OCP extracted from MD simulation trajectories at 100 ns. (c) Superimposition of complexed wt-T1_OCP and 3M_OCP at 40, 80 and 100 ns.
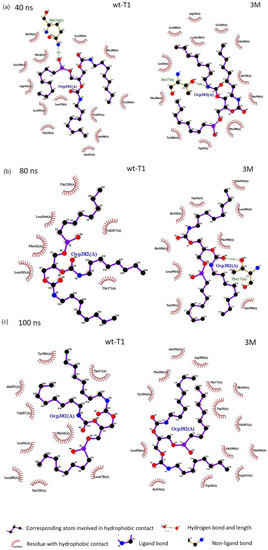
Figure 9.
Snapshot of the structure complex of wt-T1 and 3M variant with a substrate analogue OCP (RC-(RP, SP)-1,2-dioctylcarbamoyl-glycero-3-O-octylphosphonate (purple) extracted from MD simulations trajectories showing ligand interaction with amino acid surrounding the active site: (a) 40 ns. (b) 80 ns. (c) 100 ns. Hydrogen–bond interactions were shown as green dotted lines. Hydrophobic interactions are represented by the red spike. The figures were generated by LigPlot+ (version 2.2).
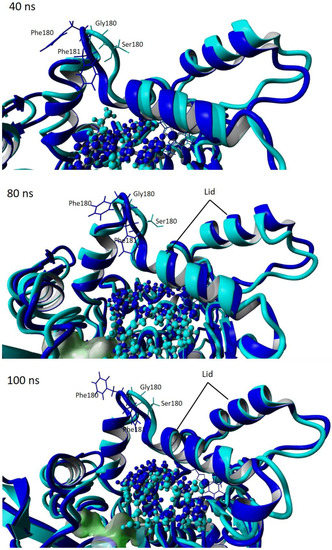
Figure 10.
Snapshot of simulation trajectories at 40, 80 and 100 ns of wt-T1 and 3M variant lipases showing the motion of the lid. Cyan represents the 3M variant. Blue represents the wt-T1.
3. Materials and Methods
3.1. Mutant Construction, Expression and Purification
Based on the structural analyses of Pseudomonas aeruginosa lipase (PAL) (PDB ID 1EX9) and Geobacillus zalihae T1 lipase, three mutation sites have been identified. Two of the mutations were located on the lid structure (amino acids 180 and 181), while the other is the oxyanion hole amino acid, which is important during the docking process of the substrate on the active site. Amino acids 180 and 181 were identified as bulky amino acids in T1 lipase, while in PAL, these amino acids were identified as G180 and S180. Based on PAL structure, F180 and F181 in T1 lipase were mutated simultaneously to glycine and serine, respectively. The combination of the mutant was used as a template to screen the most suitable amino acid to replace F16 with 12 other amino acids (Phe, Asp, Leu, Ile, Val, Gly, Cys, Asn, Ser, Tyr, His and Trp). The lipase variants were generated using the QuickChange Lightning Mutagenesis kit (Agilent, Santa Clara, CA, USA), following the manufacturer’s protocol and using the corresponding mutations primer and the template of a modified pGEX-4T1 with His-tag/T1 plasmid. The primer was designed using the primer design website, 10 October 2020 and the oligonucleotide with desired mutations was synthesised by Bio Basic Inc. (Markham, ON, Canada). PCR products of the T1 gene containing the desired mutated site were subjected to the Dpn I enzyme to digest the parental methylated DNA. To confirm the success of mutations, the plasmids were sequenced using the forward primer, 5′ CGG TGC ACC AAT GCT TCT GGC 3′, and reverse primer, 5′ GGG AGC TGC ATG TGT CAG AGG 3′. The plasmid contained the correct sequence of the mutated lipase and was transformed into expression vector E. coli BL21(DE3). The expressed mutated lipase was later identified as F180G/F181S/F16X (X = Phe, Asp, Leu, Ile, Val, Gly, Cys, Asn, Ser, Tyr, His and Trp), and unmutated T1 lipase was identified as wt-T1. The cultures were grown on LB–tributyrin agar media (BD, Franklin Lakes, NJ, USA) supplemented with ampicillin 100 µg/mL (Gold Biotechnology, Olivette, MO, USA). The expression and purification of wt-T1 and variant lipases were carried out according to previously described methods [53]. In brief, T1 and variant lipases were expressed as fusion proteins with a His tag for a one-step protein purification strategy using His-tag Sepharose Ni2+ affinity beads (GE Healthcare, Chicago, IL, USA) chromatography. The purified proteins in the supernatant were collected by centrifugation and were analysed by SDS-PAGE. Protein concentrations were determined by the Bradford assay before and after the experiment [54]. The preliminary analysis of all F180G/F181S/F16X variants was conducted using gas chromatography to verify their altered regioselectivity. The preliminary analysis indicates that the combination of F180G/F181S/F16W, namely as 3M, has a positive effect on the alteration of lipase regiospecificity.
3.2. Regioselectivity Analysis of 3M Lipase Using Thin Layer Chromatography (TLC)
The 3M lipase was examined for its regioselectivity modification using TLC analysis by hydrolysing triolein. The wt-T1 enzyme, lipases from Candida rugosa (Merck, Darmstadt, Germany) and Candida antarctica (Merck, Germany), were used as controls. The experiment started with the dissolving of 1 mg of glyceryl trioleate with 99% purity (Sigma-Aldrich, St. Louis, MO, USA) in an aqueous solution of 20 g/L polyvinyl alcohol (Sigma-Aldrich, USA). For the total volume of the 1 mL reaction, 0.05 mL of homogenised triolein was added to 0.04 mL of 50 mM glycine–NaOH buffer, pH 9. The reaction was triggered by the addition of 0.01 mL of the purified enzyme with 20 µL of 20 mM CaCL2. The reaction mixture was incubated at 70 °C for 30 min under 150 rpm shaking. The reaction was halted by the addition of 0.01 mL of 6 N HCl, and 0.01 mL of n-Hexane was added to the reaction to extract the released product. The TLC mobile phase mixture was composed of n-Hexane/Diethyl ether/Acetic acid in the ratio of 45:54:1 (Merck, Germany). Silica gel 60 TLC (Sigma-Aldrich, USA) was marked horizontally at 1 cm intervals, and aliquots of the products were pinned on the marked TLC paper. The plate containing the free fatty acids was dipped into the mobile phase and left to dry. The spots of the glycerides and fatty acids were visualised by exposure to iodine vapour (Merck, Germany).
3.3. Biochemical Properties of wt-T1 and 3M Lipases
The wt-T1 and 3M lipase hydrolytic activity was detected via a spectrophotometric method with p-nitrophenyl laurate pNP-C12 (Sigma-Aldrich, USA) as a substrate using the method by Vorderwülbecke et al. [55]. The mixture of 0.01 mL of 5 mM pNP-C12, glycine-NaOH buffer pH 9 and 1 mg/mL gum arabic (R&M Chemicals, London, UK) was pre-warmed to the desired temperature before starting the reaction. The reaction was initiated by adding 2 µL of 1 mg/mL of enzyme solutions and was incubated for 10 min at 70 °C. It was terminated by adding 100 μL of 1% SDS (Merck, Germany), and the absorbance was measured at 410 nm according to Vorderwülbecke et al. [55]. One unit of enzyme activity (U) was defined as the amount of enzymes releasing 1 µmol of p-nitrophenol per min under the given experimental conditions.
The wt-T1 and 3M lipases optimum temperatures were detected by measuring the enzyme activity at temperatures ranging from 40 to 90 °C, at pH 9.0. Meanwhile, the optimum pH for wt-T1 and 3M lipases were detected using a buffer system of different pH ranging from 4.0 to 12.0. Buffers used in this study were as follows: sodium acetate pH 4–6, sodium phosphate pH 6–8, Tris-HCl pH 8–9, glycine-NaOH pH 9–11, and disodium phosphate, pH 11–12. The reaction mixture contained 1% (w/v) gum arabic, 10 µL of 5 mM pNP-dodecanoate (C12:0) and 2 µL of 1 mg/mL of the enzyme to explore the optimum pH for each lipase. The reaction mixture was incubated at 70 °C for 10 min with a shaking rate of 200 rpm and was terminated by adding 100 μL of 1% SDS. The substrate specificity for wt-T1 and 3M lipases were determined using pNP-esters with different chain lengths, including pNP-acetate (C2), pNP-butyrate (C4), pNP-caprylate (C8), pNP-caprate (C10), pNP-laurate (C12), pNP-myristate (C14), pNP-palmitate (C16) and pNP-stearate (C18) [55].
3.4. Determination of Kinetic Constants
The kinetic constant of wt-T1 and 3M lipases was determined using a modified method by Tang et al. [16]. The pNP-laurate (C12) was used as a substrate. The amount of 0.05 µg of lipase was added to the reaction and assayed at 70 °C. The pNP-laurate initial hydrolysis rate was detected over the substrate concentration, ranging from 0.01 to 7.00 mM. The experiment was carried out on a 96-MTP plate, and the absorption was recorded at 410 nm. The rate of reaction was calculated by using the pNP-ester standard curve experimentally determined in the given buffer. The obtained data were used to determine the Michaelis–Menten by utilizing Microsoft Excel to investigate the Vmax and Km values of each sample. The Vmax value was converted to kcat by dividing the enzyme concentration by the kinetic constant of wt-T1.
3.5. Circular Dichroism Spectral Analysis
The circular dichroism (CD) was used to study the folding–unfolding state of a protein to explore the enzyme conformational dynamics [56]. The wt-T1 and 3M lipase thermal unfolding was monitored by a spectropolarimeter (J-810, JASCO, Tokyo, Japan). The concentration of stock enzyme was determined by the Bradford assay [54] and enzyme concentration was adjusted to 0.1 mg/mL in 10 mM sodium phosphate buffer, pH 7.4. The warm-up period was set from 20 to 95 °C with a heating rate of 1 °C/min. In this experiment, a 1 cm optical path length was used and was scanned at a 222 nm wavelength. The determination of the denaturation point of lipases was adopted based on the protocol of Greenfield [56]. The melting temperatures (Tm) of each sample were measured using Spectra Manager™ Suite Software (J-810, JASCO, Japan).
The secondary structural elements of lipases were measured based on the far-UV spectrum (190–240 nm) using the 1 mm path cell length at a temperature of 20 °C. The protein sample was diluted to a final concentration of 0.1 mg/mL in 10 mM sodium phosphate buffer pH 7.4. Each spectrum was the average of three successive scans, and a reference sample containing the corresponding buffer was subtracted from the CD signal. The preparation of protein was employed based on the protocol of Greenfield [57]. The analysis of structural elements was measured using Spectra Manager™ Suite Software (JASCO, Japan). Yang [58] was used as reference to calculate the secondary structural analysis of the CD spectral, which showed ɑ-helix (190–220), beta-sheet (194–204), turn (199–240) and random coil (190–193).
3.6. Analysis of Fatty Acids Composition by Gas Chromatography
The esters and free fatty acids of wt-T1 and 3M lipases were quantified according to Xu et al. [32] using gas chromatography (GC) FID. The substrate solution was prepared by dissolving 7 g of palm stearin in 30% H2O (concerning the oil mass), followed by heating in a water bath at 70 °C. The mixture of 1 mL substrate solution with a 0.5 mL enzyme, 0.5 mL of 50 Mm glycine–NaOH buffer pH 9 and 0.02 mL of 50 mM CaCl2 was incubated at 70 °C for 1 h and halted by adding 1 mL of ethanol:sulphuric acid (100:0.8, v/v) mixture. The top layer containing hexane and fatty acids (200 μL) was dispensed into a GC vial equipped with a 0.3 mL glass insert. Fatty acids were derivatised by sialylation and by adding 25 μL pyridine and 25 μL N-methyl-N-tri (methyl silyl)- trifluoroacetamide (MSTFA) to the vial, then heating at 50 °C for 20 min before analysis using an Agilent Technologies 7890 GC system equipped with a flame ionisation detector. The parameters used for the experiment for the capillary column were a BPX70- 60 m, 0.25 mm and SGE Analytical Science, Ringwood Victoria, Australia), and they were driven from GC application note/analysis of fatty acid compounds on an A PBX70/Column Part No.: 054606/SGE analytical science device. The helium carrier flow was 2.2 mL/min, and the split ratio was 58:1. Temperatures were as follows: injector 250 °C, detector 300 °C, oven and 80–130 °C at 10 °C/min.
The hydrolysed fatty acid yield was calculated as a percentage by using the following equation:
where represents the area under the peak, corresponding to the component, while i and represent the sum of the areas under all the peaks. The correction factor was calculated according to the equation as mi represents the mass of palmitic acid in the standard, and represents the total mass of fatty acids in the standard. The correction factor is expressed as relative to palmitic acid kC16, so that it is calculated as , this calculation was taken from [59].
3.7. Homology Modelling and Docking Analysis of the Open Structure of Lipases
The wt-T1 and 3M lipases were remodelled using an open structure of BTL2 lipase (PDB ID: 2W22) as a template, which have a sequence identity with wt-T1 lipase of more than 96%. The prediction of the wt-T1 and 3M open structures was conducted using the automated homology model by Yet Another Scientific Artificial Reality Application (YASARA) [60]. The energy minimization was performed on the modelled structures. The modelled structures were validated using available online software, ERRAT, Verify-3D and PROCHECK [49,61,62,63]. The predicted mutated open conformation of lipases was used in a molecular dynamics simulation and docking study.
The volume of the binding site was measured by MOLE 2.0 software [64]. The residues of the binding site were manually identified and plotted using YASARA software.
The molecular docking of the open structures of wt-T1 and 3M lipases was performed using the YASARA (Yet Another Scientific Artificial Reality Application) program, using the VINA and AMBER03 force field approach. The triglyceride analogue RC-(RP, SP)-1,2-dioctylcarbamoyl-glycero-3-o-octylphosphonate (OCP) was retrieved from the protein data bank (PDB) database 5th December 2020 [28]. The specific docking analysis was performed by generating the cubic simulation cell around the catalytic site of the lipase. The simulation box with a dimension of 5 Å × 5 Å × 5 Å was designed to cover the catalytic triad (Ser113, Asp317 and His358) of lipase. The grid box size was set in the centre to fit the active site cleft and allow nonconstructive binding. The docking analysis was performed for 25 runs. The possible binding interactions between ligand OCP and amino acid residues present in the catalytic pocket of the lipase were analysed. The B-factor and atomic properties were used to estimate the binding energy and dissociation constant (kd). More positive binding energies indicate stronger binding, and negative energies mean no binding. All figures were prepared using YASARA software and LigPlot+ (version 2.2) [65].
3.8. Molecular Dynamics Simulation of Predicted sn-3 Modified Regioselectivity Mutant (3M) and wt-T1 Lipase Structures
Molecular dynamic (MD) simulation was implemented in the protein–ligand complexes of lipases extracted from molecular docking analysis. The complexes of wt-T1_OCP and 3M_OCP structure simulations were obtained using the YASARA software package and the AMBER 14 force field at a temperature of 70 °C (343.15 K). The cubic simulation box with dimensions 50 Å × 50 Å ×50 Å was filled with water and neutralized by sodium ions. Energy minimization was performed, and the system was equilibrated with the steepest descent parameters for 1 ns, followed by the simulation runs of 100 ns each in triplicate.
Several parameters, such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), a radius of gyration and solvent accessible surface area (SASA), were analysed using tools within the YASARA simulation package. The starting structure was used as a reference for evaluating the RMSD calculation. By using the final trajectories from the simulations, the hydrogen bond networks in wt-T1_OCP and 3M_OCP were explored. The YASARA program was used for visualising the structure, analysis and figures preparation.
3.9. Statistical Analysis
The wt-T1 and 3M lipases activity, pH, substrate specificity, kinetic study and gas chromatography tests were carried out in triplicate (n = 3). The ANOVA single factor was used for the analysis by utilizing Microsoft Excel to perform the statistical analysis. The results were considered statistically significant at a p-value < 0.05.
4. Conclusions
In this study, we constructed a modified regiospecific sn-3 lipase (3M lipase), and this provided insight into the oxyanion hole and lid domain structure/function relationship. The result suggested that the bulky residues of the oxyanion hole and lid domain affected the T1 lipase regiospecificity, selectivity, activity and thermostability. We detected that F180/F181/F16 was an important residue for the reshaping enzyme binding site. The 3M lipase displayed different regiospecificity and preference toward long-chain selectivity. This finding lays the framework for further T1 lipase engineering for implementation in the drug and food industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal13020416/s1, Figure S1: Gas chromatography flame ionization detector (GC FID) analysis for sn-3 modified regiospecific variants. (a) wt-T1 (b) F16C (c) F16G (d) F16V (e) F16I (f) F16S respectively. The acylglycerol position highlighted with red box; Figure S2: Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified wt-T1 and 3M variant lipases; Figure S3: Thin-layer chromatography (TLC) sheet of purified wt-T1 and 3M variant lipases: Cr = Candida rugosa lipase, Ca = Candida antarctica lipase, T1= wt-T1 lipase, 3M = 3M variant lipase, respectively; Figure S4: Circular dichroism (CD) spectra analysis of wt-T1 and 3M lipases at 190 to 240 nm. All scans were performed at temperature of 20 °C. Analysis of CD spectral reference based on Yang [58], showing α-helix (190–220), beta sheet (194–204), turn (199–240) and random coil (190–193); Figure S5: The change in the initial rate of the reaction at different concentrations of pNP-12 for wt-T1 (purple line) and 3M (green line) are shown. The figure is generated from online tool http://www.ic50.tk/kmvmax.html, accessed on 8 December 2022; Figure S6: Superimposition of open conformation wt-T1 (blue) and 3M variant (cyan) structures. The green surface residues represent the catalytic triad; Figure S7: Ramachandran plot of wt-T1 (a) and 3M (b) lipases. The images generated using PDBsum (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html, accessed on 8 December 2022); Figure S8: Molecular dynamics simulation analyses of lipases-complexed with the substrate (OCP). (a) Root Mean Square Deviation (RMSD). (b) Accessible surface area (SASA). (c) Radius of gyration. (d) Root means square fluctuation (RMSF); Figure S9: Superimposition of wt-T1 and 3M variant structure complexed with the substrate (OCP). (a) Global structure of wt-T1 and sn-3-regiospecific variant structure (b) Close up of the catalytic site residues (Asp317, His358, and Ser113). (c) The ribbon of binding site residues (d) Close up of the lid structure of lipases. The wt-T1 coloured in blue and 3M coloured in cyan. Table S1: Characteristic of lipase variants; Table S2: Purification table of wt-T1 and its 3M variant lipases; Table S3: Biochemical characterization of wt-T1 and its variant 3M; Table S4: Residues with higher RMSF value peak and their corresponding location on wt-T1 lipase structure.
Author Contributions
S.H.A. and M.M. data; A.T.C.L. assisted in software usage; S.H.A. designed Figures. S.H.A. and R.N.Z.R.A.R. wrote the manuscript; S.N.H.I., M.S.M.A., F.M.S. and N.D.M.N. read and contributed to the manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Education, Malaysia. (FRGS/1/2019/STGO5/UPM/01/1/01-01-19-2178FR).
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the members of Enzyme and Microbial Technology Research Centre, UPM, and all the principal investigators.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leow, T.C.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. High level expression of thermostable lipase from Geobacillus sp. strain T1. Biosci. Biotechnol. Biochem. 2004, 68, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An overview. Lipases Phospholipases 2018, 1835, 3–38. [Google Scholar]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Lanser, A.C.; Manthey, L.K.; Hou, C.T. Regioselectivity of new bacterial lipases determined by hydrolysis of triolein. Curr. Microbiol. 2002, 44, 336–340. [Google Scholar] [CrossRef]
- Alnoch, R.C.; Martini, V.P.; Glogauer, A.; Costa, A.C.D.S.; Piovan, L.; Muller-Santos, M.; de Souza, E.M.; de Oliveira Pedrosa, F.; Mitchell, D.A.; Krieger, N. Immobilization and characterisation of a new regioselective and enantioselective lipase obtained from a metagenomic library. PLoS ONE 2015, 10, e0114945. [Google Scholar] [CrossRef]
- Laguerre, M.; Nlandu Mputu, M.; Brïys, B.; Lopez, M.; Villeneuve, P.; Dubreucq, E. Regioselectivity and fatty acid specificity of crude lipase extracts from Pseudozyma tsukubaensis, Geotrichum candidum, and Candida rugosa. Eur. J. Lipid Sci. Technol. 2017, 119, 1600302. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Han, S.O.; Park, C.; Kim, S.W. Co-immobilization of Candida rugosa and Rhyzopus oryzae lipases and biodiesel production. Korean J. Chem. Eng. 2013, 30, 1335–1338. [Google Scholar] [CrossRef]
- Miotti, R.H., Jr.; Cortez, D.V.; De Castro, H.F. Transesterification of palm kernel oil with ethanol catalyzed by a combination of immobilized lipases with different specificities in continuous two-stage packed-bed reactor. Fuel 2022, 310, 122343. [Google Scholar] [CrossRef]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One biocatalyst–many applications: The use of Candida antarctica B-lipase in organic synthesis. Biocatal. Biotransform. 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Xing, S.; Zhu, R.; Li, C.; He, L.; Zeng, X.; Zhang, Q. Gene cloning, expression, purification and characterization of a sn-1, 3 extracellular lipase from Aspergillus niger GZUF36. J. Food Sci. Technol. 2020, 57, 2669–2680. [Google Scholar] [CrossRef]
- Toldrá-Reig, F.; Mora, L.; Toldrá, F. Developments in the use of lipase transesterification for biodiesel production from animal fat waste. Appl. Sci. 2020, 10, 5085. [Google Scholar] [CrossRef]
- Wei, W.; Sun, C.; Wang, X.; Jin, Q.; Xu, X.; Akoh, C.C.; Wang, X. Lipase-catalyzed synthesis of sn-2 palmitate: A review. Engineering 2020, 6, 406–414. [Google Scholar] [CrossRef]
- Villeneuve, P.; Pina, M.; Montet, D.; Graille, J. Determination of lipase specificities through the use of chiral triglycerides and their racemics. Chem. Phys. Lipids 1995, 76, 109–113. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C. Biochemistry of digestion. Compr. Mol. Insect Sci. 2005, 4, 171–224. [Google Scholar]
- Yang, Y.; Wong, S.E.; Lightstone, F.C. Understanding a substrate’s product regioselectivity in a family of enzymes: A case study of acetaminophen binding in cytochrome P450s. PLoS ONE 2014, 9, e87058. [Google Scholar] [CrossRef]
- Mikkilineni, A.B.; Kumar, P.; Abushanab, E. The chemistry of l-ascorbic and d-isoascorbic acids. 2. R and S glyceraldehydes from a common intermediate. J. Org. Chem. 1988, 53, 6005–6009. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, L.; Wang, J.; Hu, Y.; Chen, S. Enzymatic synthesis of functional structured lipids from glycerol and naturally phenolic antioxidants. In Glycerine Production and Transformation—An Innovative Platform for Sustainable Biorefinery and Energy; IntechOpen: London, UK, 2019. [Google Scholar]
- Yoon, B.K.; Jackman, J.A.; Valle-González, E.R.; Cho, N.J. Antibacterial free fatty acids and monoglycerides: Biological activities, experimental testing, and therapeutic applications. Int. J. Mol. Sci. 2018, 19, 1114. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Hou, S.; Lan, D.; Wang, X.; Liu, J.; Khan, F.I.; Wang, Y. Crystal structure of a lipase from Streptomyces sp. strain W007–implications for thermostability and regiospecificity. FEBS J. 2017, 284, 3506–3519. [Google Scholar] [CrossRef] [PubMed]
- Hasnaoui, I.; Dab, A.; Mechri, S.; Abouloifa, H.; Saalaoui, E.; Jaouadi, B.; Noiriel, A.; Asehraou, A.; Abousalham, A. Purification, Biochemical and Kinetic Characterization of a Novel Alkaline sn-1, 3-Regioselective Triacylglycerol Lipase from Penicillium crustosum Thom Strain P22 Isolated from Moroccan Olive Mill Wastewater. Int. J. Mol. Sci. 2022, 23, 11920. [Google Scholar] [CrossRef]
- Cao, X.; Liao, L.; Feng, F. Purification and characterization of an extracellular lipase from Trichosporon sp. and its application in enrichment of omega-3 polyunsaturated fatty acids. LWT 2020, 1, 108692. [Google Scholar] [CrossRef]
- Gao, K.; Chu, W.; Sun, J.; Mao, X. Identification of an alkaline lipase capable of better enrichment of EPA than DHA due to fatty acids selectivity and regioselectivity. Food Chem. 2020, 15, 127225. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Bai, Q.; Yang, L.; Tang, L.; Su, M. Substitution and mutation of the C-terminal loop domain of Penicillium expansum lipase significantly changed its regioselectivity and activity. Biochem. Eng. J. 2022, 1, 108313. [Google Scholar] [CrossRef]
- Crotti, M.; Parmeggiani, F.; Ferrandi, E.E.; Gatti, F.G.; Sacchetti, A.; Riva, S.; Brenna, E.; Monti, D. Stereoselectivity switch in the reduction of α-alkyl-β-arylenones by structure-guided designed variants of the ene reductase OYE1. Front. Bioeng. Biotechnol. 2019, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Balke, K.; Schmidt, S.; Genz, M.; Bornscheuer, U.T. Switching the regioselectivity of a cyclohexanone monooxygenase toward (+)-trans-dihydrocarvone by rational protein design. ACS Chem. Biol. 2016, 11, 38–43. [Google Scholar] [CrossRef]
- Moharana, T.R.; Rao, N.M. Substrate structure and computation guided engineering of a lipase for omega-3 fatty acid selectivity. PLoS ONE 2020, 15, e0231177. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Lan, D.; Yang, B.; Khan, F.I.; Wang, Y. Site-directed mutagenesis studies of hydrophobic residues in the lid region of T1 lipase. Eur. J. Lipid Sci. Technol. 2017, 119, 1600107. [Google Scholar] [CrossRef]
- Nardini, M.; Lang, D.A.; Liebeton, K.; Jaeger, K.E.; Dijkstra, B.W. Crystal structure of Pseudomonas aeruginosa lipase in the open conformation: The prototype for family I. 1 of bacterial lipases. J. Biol. Chem. 2000, 275, 31219–31225. [Google Scholar] [CrossRef]
- Matsumura, H.; Yamamoto, T.; Leow, T.C.; Mori, T.; Salleh, A.B.; Basri, M.; Inoue, T.; Kai, Y.; Rahman, R.N.Z.R.A. Novel cation-π interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins Struct. Funct. Bioinform. 2008, 70, 592–598. [Google Scholar] [CrossRef]
- Hussain, C.H.A.C.; Rahman, R.N.Z.R.A.; Chor, A.L.T.; Salleh, A.B.; Ali, M.S.M. Enhancement of a protocol purifying T1 lipase through molecular approach. PeerJ 2018, 6, e5833. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, S.; Wang, W.; Wang, Y.; Yang, B. Enzymatic hydrolysis of palm stearin to produce diacylglycerol with a highly thermostable lipase. Eur. J. Lipid Sci. Technol. 2013, 115, 564–570. [Google Scholar] [CrossRef]
- Carrasco-López, C.; Godoy, C.; de Las Rivas, B.; Fernández-Lorente, G.; Palomo, J.M.; Guisán, J.M.; Fernández-Lafuente, R.; Martínez-Ripoll, M.; Hermoso, J.A. Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 2009, 284, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Wahab, R.A.; Basri, M.; Rahman, M.B.A.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Leow, T.C. Combination of oxyanion Gln114 mutation and medium engineering to influence the enantioselectivity of thermophilic lipase from Geobacillus zalihae. Int. J. Mol. Sci. 2012, 13, 11666–11680. [Google Scholar] [CrossRef] [PubMed]
- Tynan-Connolly, B.M.; Nielsen, J.E. Redesigning protein pKa values. Protein Sci. 2007, 16, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Mullaney, E.J.; Porres, J.M.; Roneker, K.R.; Crowe, S.; Rice, S.; Ko, T.; Ullah, A.H.; Daly, C.B.; Welch, R.; et al. Shifting the pH profile of Aspergillus niger PhyA phytase to match the stomach pH enhances its effectiveness as an animal feed additive. Appl. Environ. Microbiol. 2006, 72, 4397–4403. [Google Scholar] [CrossRef] [PubMed]
- Kanicky, J.R.; Shah, D.O. Effect of degree, type, and position of unsaturation on the pKa of long-chain fatty acids. J. Colloid Interface Sci. 2002, 256, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Khaleghinejad, S.H.; Motalleb, G.; Karkhane, A.A.; Aminzadeh, S.; Yakhchali, B. Study the effect of F17S mutation on the chimeric Bacillus thermocatenulatus lipase. J. Genet. Eng. Biotechnol. 2016, 14, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Trevino, S.; Prabhakaran, E.; Martin Scholtz, J. Protein structure, stability and solubility in water and other solvents. Philosophical Transactions of the Royal Society of London. Ser. B Biol. Sci. 2004, 359, 1225–1235. [Google Scholar] [CrossRef]
- Augustyniak, W.; Brzezinska, A.A.; Pijning, T.; Wienk, H.; Boelens, R.; Dijkstra, B.W.; Reetz, M.T. Biophysical characterisation of mutants of Bacillus subtilis lipase evolved for thermostability: Factors contributing to increased activity retention. Protein Sci. 2012, 21, 487–497. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, Y. The relation between lipase thermostability and dynamics of hydrogen bond and hydrogen bond network based on long time molecular dynamics simulation. Protein Pept. Lett. 2017, 24, 643–648. [Google Scholar] [CrossRef]
- Rathi, P.C.; Jaeger, K.E.; Gohlke, H. Structural rigidity and protein thermostability in variants of lipase A from Bacillus subtilis. PLoS ONE 2015, 10, e0130289. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, C.; Tang, M.; Xu, B.; Wang, L.; Qian, W.; He, J.; Zhao, Z.; Wu, Q.; Mu, Y.; et al. Improving the thermostability of Rhizopus chinensis lipase through site-directed mutagenesis based on B-factor analysis. Front. Microbiol. 2020, 3, 346. [Google Scholar] [CrossRef] [PubMed]
- Cen, Y.; Singh, W.; Arkin, M.; Moody, T.S.; Huang, M.; Zhou, J.; Wu, Q.; Reetz, M.T. Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution. Nat. Commun. 2019, 10, 3198. [Google Scholar] [CrossRef] [PubMed]
- Brundiek, H.; Padhi, S.K.; Kourist, R.; Evitt, A.; Bornscheuer, U.T. Altering the scissile fatty acid binding site of Candida antarctica lipase A by protein engineering for the selective hydrolysis of medium chain fatty acids. Eur. J. Lipid Sci. Technol. 2012, 114, 1148–1153. [Google Scholar] [CrossRef]
- Zorn, K.; Oroz-Guinea, I.; Brundiek, H.; Dörr, M.; Bornscheuer, U.T. Alteration of chain length selectivity of Candida antarctica lipase a by semi-rational design for the enrichment of erucic and gondoic fatty acids. Adv. Synth. Catal. 2018, 360, 4115–4131. [Google Scholar] [CrossRef]
- Quaglia, D.; Alejaldre, L.; Ouadhi, S.; Rousseau, O.; Pelletier, J.N. Holistic engineering of Cal-A lipase chain-length selectivity identifies triglyceride binding hot-spot. PLoS ONE 2019, 14, e0210100. [Google Scholar] [CrossRef]
- Chrzanowska, A.; Olejarz, W.; Kubiak-Tomaszewska, G.; Ciechanowicz, A.K.; Struga, M. The Effect of Fatty Acids on Ciprofloxacin Cytotoxic Activity in Prostate Cancer Cell Lines—Does Lipid Component Enhance Anticancer Ciprofloxacin Potential? Cancers 2022, 14, 409. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. A Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Truongvan, N.; Jang, S.H.; Lee, C. Flexibility and stability trade-off in active site of cold-adapted Pseudomonas mandelii esterase EstK. Biochemistry 2016, 55, 3542–3549. [Google Scholar] [CrossRef]
- Bukhari, N.; Leow, A.T.C.; Abd Rahman, R.N.Z.R.; Mohd Shariff, F. Single Residue Substitution at N-Terminal Affects Temperature Stability and Activity of L2 Lipase. Molecules 2020, 25, 3433. [Google Scholar] [CrossRef]
- Huang, F.; Nau, W.M. A conformational flexibility scale for amino acids in peptides. Angew. Chem. Int. Ed. 2003, 42, 2269–2272. [Google Scholar] [CrossRef]
- Ishak, S.N.H.; Masomian, M.; Kamarudin, N.H.A.; Ali, M.S.M.; Leow, T.C.; Rahman, R.N.Z.R.A. Changes of thermostability, organic solvent, and pH stability in Geobacillus zalihae HT1 and its mutant by calcium ion. Int. J. Mol. Sci. 2019, 20, 2561. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vorderwülbecke, T.; Kieslich, K.; Erdmann, H. Comparison of lipases by different assays. Enzym. Microb. Technol. 1992, 14, 631–639. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2006, 1, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.T.; Wu, C.S.; Martinez, H.M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986, 130, 208–269. [Google Scholar]
- Kuntom, A.; Aini Idris, N.; Amri Ibrahim, N.; Thin Sue, M.; Wai Lin, S.; Yew Ai, T. A Compendium of Test on Palm Oil Products, Palm Kernel Products, Fatty Acids, Food Related Products and Others; Malaysian Palm Oil Board, Ministry of Plantation Industries and Commodities Malaysia: Kuala Lumpur, Malaysia, 2005.
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterising force field. Proteins Struct. Funct. Bioinform. 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 12, 164–170. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Sehnal, D.; Svobodová Vařeková, R.; Berka, K.; Pravda, L.; Navrátilová, V.; Banáš, P.; Ionescu, C.M.; Otyepka, M.; Koča, J. MOLE 2.0: Advanced approach for analysis of biomacromolecular channels. J. Cheminform. 2013, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).