Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science
Abstract
:1. Introduction
2. Applications of FMOs in the Biomedical Field
2.1. Applications of FMOs in Antibiotic Research
2.2. Applications of FMOs as Pharmaceutical Targets
2.3. Applications of FMOs in Drug Synthesis
3. Applications of FMOs in the Biomedical Field
3.1. Application of FMOs in the Processing of Pesticides
3.2. Application of FMOs in Plant Life Activities
4. Applications of FMOs in Natural Product Synthesis
4.1. Application of FMOs in the Biosynthesis of Polyether through Epoxidations
4.2. Application of FMOs in the Biosynthesis of Natural Products through Dearomatization
4.3. Application of Baeyer–Villiger Monooxygenases in Natural Product Synthesis
5. Summary and Outlook
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Acronym | Full Name |
| FMOs | Flavin-dependent monooxygenases |
| FAD | Flavin adenine dinucleotide |
| FMN | Flavin mononucleotide |
| CYPs | Cytochrome P450 enzymes |
| KMO | Kynurenine 3-monooxygenase |
| TNBC | Triple-negative breast cancer |
| SQLE | Squalene epoxidase |
| LPS | Lansoprazole sulfide |
| TC2P | 3,5,6-Trichloro-2-pyridinol |
| DBHB | 3,5-Dibromo-4-hydroxybenzoate |
| MEA | 2-Methyl-6-ethyl aniline |
| EDA | 2,6-Diethyl aniline |
| IPA | Indole-3-pyruvic acid |
| IAA | Indole-3-acetic acid |
| SAR | Systemic acquired resistance |
| NPs | Natural products |
| BVMOs | Baeyer–Villiger monooxygenases |
| MRSA | Methicillin-resistant Staphylococcus aureus |
References
- Macheroux, P.; Kappes, B.; Ealick, S.E. Flavogenomics—A genomic and structural view of flavin-dependent proteins. Febs J. 2011, 278, 2625–2634. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, N.; Deng, Y.; Guan, Y.; Xiao, H.; Nitka, T.A.; Yang, H.; Yadav, A.; Vukovic, L.; Mathews, I.I.; et al. Triepoxide formation by a flavin-dependent monooxygenase in monensin biosynthesis. Nat. Commun. 2023, 14, 6273. [Google Scholar] [CrossRef]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.M.; Zhou, Q.; Wu, Y.Z.; Chen, X.; Zhong, F.R. Properties and Mechanisms of Flavin-Dependent Monooxygenases and Their Applications in Natural Product Synthesis. Int. J. Mol. Sci. 2022, 23, 2622. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Miyanaga, A.; Michaudel, Q.; Stull, F.; Louie, G.; Noel, J.P.; Baran, P.S.; Palfey, B.; Moore, B.S. Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 2013, 503, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Eswaramoorthy, S.; Bonanno, J.B.; Burley, S.K.; Swaminathan, S. Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. USA 2006, 103, 9832–9837. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.R. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Krueger, S.K.; Williams, D.E. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005, 106, 357–387. [Google Scholar] [CrossRef]
- Ziegler, D.M. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 2002, 34, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Finefield, J.M.; Frisvad, J.C.; Sherman, D.H.; Williams, R.M. Fungal Origins of the Bicyclo 2.2.2 diazaoctane Ring System of Prenylated Indole Alkaloids. J. Nat. Prod. 2012, 75, 812–833. [Google Scholar] [CrossRef] [PubMed]
- Sehlmeyer, S.; Wang, L.Z.; Langel, D.; Heckel, D.G.; Mohagheghi, H.; Petschenka, G.; Ober, D. Flavin-Dependent Monooxygenases as a Detoxification Mechanism in Insects: New Insights from the Arctiids (Lepidoptera). PLoS ONE 2010, 5, e10435. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Yurimoto, H.; Kato, N.; Sakai, Y. Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5. J. Bacteriol. 2007, 189, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Volkers, G.; Palm, G.J.; Weiss, M.S.; Wright, G.D.; Hinrichs, W. Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase. Febs Lett. 2011, 585, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, e00119-18. [Google Scholar] [CrossRef] [PubMed]
- Huovinen, P. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 2001, 32, 1608–1614. [Google Scholar]
- Yang, W.R.; Moore, I.F.; Koteva, K.P.; Bareich, D.C.; Hughes, D.W.; Wright, G.D. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 2004, 279, 52346–52352. [Google Scholar] [CrossRef]
- Kim, D.W.; Thawng, C.N.; Lee, K.; Wellington, E.M.H.; Cha, C.J. A novel sulfonamide resistance mechanism by two-component flavin-dependent monooxygenase system in sulfonamide-degrading actinobacteria. Environ. Int. 2019, 127, 206–215. [Google Scholar] [CrossRef]
- Hashimoto, M.; Taguchi, T.; Ishikawa, K.; Mori, R.; Hotta, A.; Watari, S.; Katakawa, K.; Kumamoto, T.; Okamoto, S.; Ichinose, K. Unveiling Two Consecutive Hydroxylations: Mechanisms of Aromatic Hydroxylations Catalyzed by Flavin-Dependent Monooxygenases for the Biosynthesis of Actinorhodin and Related Antibiotics. ChemBioChem 2020, 21, 623–627. [Google Scholar] [CrossRef]
- Mole, D.J.; Webster, S.P.; Uings, I.; Zheng, X.Z.; Binnie, M.; Wilson, K.; Hutchinson, J.P.; Mirguet, O.; Walker, A.; Beaufils, B.; et al. Kynurenine-3-monooxygenase inhibition prevents multiple organ failure in rodent models of acute pancreatitis. Nat. Med. 2016, 22, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.L.; Lei, H.L.; Liao, A.T.C.; Chu, R.M.; Lin, C.S. KMO as a novel diagnostic and prognostic biomarker in canine mammary gland tumors. Cancer Res. 2012, 72, 4511A. [Google Scholar] [CrossRef]
- Huang, T.T.; Tseng, L.M.; Chen, J.L.; Chu, P.Y.; Lee, C.H.; Huang, C.T.; Wang, W.L.; Lau, K.Y.; Tseng, M.F.; Chang, Y.Y.; et al. Kynurenine 3-monooxygenase upregulates pluripotent genes through β-catenin and promotes triple-negative breast cancer progression. Ebiomedicine 2020, 54, 102717. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.R.; Guillemin, G.J.; Lovejoy, D.B. Development of a Rapid Fluorescence-Based High-Throughput Screening Assay to Identify Novel Kynurenine 3-Monooxygenase Inhibitor Scaffolds. Slas Discov. 2018, 23, 554–560. [Google Scholar] [CrossRef]
- Phillips, R.S.; Iradukunda, E.C.; Hughes, T.; Bowen, J.P. Modulation of Enzyme Activity in the Kynurenine Pathway by Kynurenine Monooxygenase Inhibition. Front. Mol. Biosci. 2019, 6, 3. [Google Scholar] [CrossRef]
- Padyana, A.K.; Gross, S.; Jin, L.; Cianchetta, G.; Narayanaswamy, R.; Wang, F.; Wang, R.; Fang, C.; Lv, X.B.; Biiler, S.A.; et al. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat. Commun. 2019, 10, 97. [Google Scholar] [CrossRef]
- Brown, A.J.; Chua, N.K.; Yan, N. The shape of human squalene epoxidase expands the arsenal against cancer. Nat. Commun. 2019, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.G.; da Rocha, G.O.; de Andrade, J.B. Occurrence of the potent mutagens 2-nitrobenzanthrone and 3-nitrobenzanthrone in fine airborne particles. Sci. Rep. 2019, 9, 1. [Google Scholar] [CrossRef]
- Furuya, T.; Kino, K. Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 2014, 98, 1145–1154. [Google Scholar] [CrossRef]
- Nakagawa, A.; Nakamura, S.; Matsumura, E.; Yashima, Y.; Takao, M.; Aburatani, S.; Yaoi, K.; Katayama, T.; Minami, H. Selection of the optimal tyrosine hydroxylation enzyme for (S)-reticuline production in Escherichia coli. Appl. Microbiol. Biotechnol. 2021, 105, 5433–5447. [Google Scholar] [CrossRef]
- Deng, Y.F.; Faivre, B.; Back, O.; Lombard, M.; Pecqueur, L.; Fontecave, M. Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli. ChemBioChem 2020, 21, 163–170. [Google Scholar] [CrossRef]
- Herrmann, S.; Dippe, M.; Pecher, P.; Funke, E.; Pietzsch, M.; Wessjohann, L.A. Engineered Bacterial Flavin-Dependent Monooxygenases for the Regiospecific Hydroxylation of Polycyclic Phenols. ChemBioChem 2022, 23, e202100480. [Google Scholar] [CrossRef] [PubMed]
- Fraley, A.E.; Tran, H.T.; Kelly, S.P.; Newmister, S.A.; Tripathi, A.; Kato, H.; Tsukamoto, S.; Du, L.; Li, S.Y.; Williams, R.M.; et al. Flavin-Dependent Monooxygenases NotI and NotI′ Mediate Spiro-Oxindole Formation in Biosynthesis of the Notoamides. ChemBioChem 2020, 21, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.G.; Chen, D.; Parales, R.E.; Jiang, J.D. Oxygenases as Powerful Weapons in the Microbial Degradation of Pesticides. Annu. Rev. Microbiol. 2022, 76, 325–348. [Google Scholar] [CrossRef]
- Chu, C.W.; Liu, B.; Li, N.; Yao, S.G.; Cheng, D.; Zhao, J.D.; Qiu, J.G.; Yan, X.; He, Q.; He, J. A Novel Aerobic Degradation Pathway for Thiobencarb Is Initiated by the TmoAB Two-Component Flavin Mononucleotide-Dependent Monooxygenase System in Acidovorax sp. Strain T1. Appl. Environ. Microbiol. 2017, 83, e01490-17. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.J.; Zhou, X.Y. Experiment and Simulation on Adsorption of 3,5,6-Trichloro-2-Pyridinol in Typical Farmland of Purple Soil, Southwestern China. Soil Sediment Contam. 2017, 26, 345–363. [Google Scholar] [CrossRef]
- Fang, L.C.; Shi, T.Z.; Chen, Y.F.; Wu, X.W.; Zhang, C.; Tang, X.Y.; Li, Q.X.; Hua, R.M. Kinetics and Catabolic Pathways of the Insecticide Chlorpyrifos, Annotation of the Degradation Genes, and Characterization of Enzymes TcpA and Fre in Cupriavidus nantongensis X1T. J. Agric. Food Chem. 2019, 67, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Mu, Y.; Jian, S.S.; Zang, X.X.; Chen, Q.; Jia, W.B.; Ke, Z.; Gao, Y.Z.; Jiang, J.D. Comparative Transcriptome Analysis Reveals the Mechanism Underlying 3,5-Dibromo-4-Hydroxybenzoate Catabolism via a New Oxidative Decarboxylation Pathway. Appl. Environ. Microbiol. 2018, 84, e02467-17. [Google Scholar] [CrossRef]
- Yan, X.; Jin, W.; Wu, G.; Jiang, W.K.; Yang, Z.G.; Ji, J.B.; Qiu, J.G.; He, J.; Jiang, J.D.; Hong, Q. Hydrolase CehA and Monooxygenase CfdC Are Responsible for Carbofuran Degradation in Sphingomonas sp. Strain CDS-1. Appl. Environ. Microbiol. 2018, 84, e00805-18. [Google Scholar] [CrossRef]
- Cheng, M.G.; Meng, Q.; Yang, Y.J.; Chu, C.W.; Chen, Q.; Li, Y.; Cheng, D.; Hong, Q.; Yan, X.; He, J. The Two-Component Monooxygenase MeaXY Initiates the Downstream Pathway of Chloroacetanilide Herbicide Catabolism in Sphingomonads. Appl. Environ. Microbiol. 2017, 83, e03241-16. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, T.; Li, J.; Wang, Y.R.; Wang, G.L.; Li, F.; Liu, Y.; Xiong, M.H.; Ma, Y.Q. Two dcm Gene Clusters Essential for the Degradation of Diclofop-methyl in a Microbial Consortium of Rhodococcus sp. JT-3 and Brevundimonas sp. JT-9. J. Agric. Food Chem. 2018, 66, 12217–12226. [Google Scholar] [CrossRef] [PubMed]
- Takeo, M.; Murakami, M.; Niihara, S.; Yamamoto, K.; Nishimura, M.; Kato, D.; Negoro, S. Mechanism of 4-Nitrophenol Oxidation in Rhodococcus sp. Strain PN1: Characterization of the Two-Component 4-Nitrophenol Hydroxylase and Regulation of Its Expression. J. Bacteriol. 2008, 190, 7367–7374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.D.; Ke, Z.J.; Ye, H.T.; Hong, M.T.; Xu, Y.F.; Zhang, M.L.; Jiang, W.K.; Hong, Q. Hydrolase CehA and a Novel Two-Component 1-Naphthol Hydroxylase CehC1C2 are Responsible for the Two Initial Steps of Carbaryl Degradation in Rhizobium sp. X9. J. Agric. Food Chem. 2020, 68, 14739–14747. [Google Scholar] [CrossRef] [PubMed]
- Riebel, A.; de Gonzalo, G.; Fraaije, M.W. Expanding the biocatalytic toolbox of flavoprotein monooxygenases from Rhodococcus jostii RHA1. J. Mol. Catal. B Enzym. 2013, 88, 20–25. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Deng, Z.; Zhu, D. Baeyer-Villiger monooxygenases in the biosynthesis of microbial secondary metabolites. Sheng Wu Gong Cheng Xue Bao = Chin. J. Biotechnol. 2019, 35, 351–362. [Google Scholar] [CrossRef]
- Romero, E.; Castellanos, J.R.G.; Gadda, G.; Fraaije, M.W.; Mattevi, A. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. Chem. Rev. 2018, 118, 1742–1769. [Google Scholar] [CrossRef]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef]
- Thodberg, S.; Neilson, E.J.H. The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins. Catalysts 2020, 10, 329. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Kamerbeek, N.M.; van Berkel, W.J.H.; Janssen, D.B. Identification of a Baeyer-Villiger monooxygenase sequence motif. Febs Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef]
- Malito, E.; Alfieri, A.; Fraaije, M.W.; Mattevi, A. Crystal structure of a Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. USA 2004, 101, 13157–13162. [Google Scholar] [CrossRef]
- Beneventi, E.; Niero, M.; Motterle, R.; Fraaije, M.; Bergantino, E. Discovery of Baeyer-Villiger monooxygenases from photosynthetic eukaryotes. J. Mol. Catal. B Enzym. 2013, 98, 145–154. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Ayub, M.J.; Furnham, N.; Thornton, J.M.; Laskowski, R.A. Chopping and Changing: The Evolution of the Flavin-dependent Monooxygenases. J. Mol. Biol. 2016, 428, 3131–3146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jung, J.H.; Han, D.Y.; Seo, P.J.; Park, W.J.; Park, C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Shen, X.L.; Mashiguchi, K.; Zheng, Z.Y.; Dai, X.H.; Cheng, Y.F.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y.D. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.L.; Shang, C.Q.; Ma, S.; Liu, L.; Cheng, J.L. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Wu, R.R.; Chen, J.B.; Lin, Y.; Jia, Q.Y.; Guo, Y.J.; Liu, J.Y.; Yan, Q.; Xue, C.C.; Chen, X.; Yuan, X.X. Genome-Wide Identification, Expression Analysis, and Potential Roles under Abiotic Stress of the YUCCA Gene Family in Mungbean (Vigna radiata L.). Int. J. Mol. Sci. 2023, 24, 1603. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.H.; Mashiguchi, K.; Chen, Q.G.; Kasahara, H.; Kamiya, Y.; Ojha, S.; DuBois, J.; Ballou, D.; Zhao, Y.D. The Biochemical Mechanism of Auxin Biosynthesis by an Arabidopsis YUCCA Flavin-containing Monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Kim, W.Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, M.R.; Jung, I.J.; Kang, S.B.; Park, H.J.; Kim, M.G.; Yun, D.J.; Kim, W.Y. The Thiol Reductase Activity of YUCCA6 Mediates Delayed Leaf Senescence by Regulating Genes Involved in Auxin Redistribution. Front. Plant Sci. 2016, 7, 626. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Saito, K. S-Alk(en)ylcysteine sulfoxides in the genus Allium: Proposed biosynthesis, chemical conversion, and bioactivities. J. Exp. Bot. 2019, 70, 4123–4137. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Onuma, M.; Sugino, Y.; Nakabayashi, R.; Imai, S.; Tsuneyoshi, T.; Sumi, S.; Saito, K. Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic. Plant J. 2015, 83, 941–951. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006, 141, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469.e16. [Google Scholar] [CrossRef] [PubMed]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef] [PubMed]
- Tintore, M.; Vidal-Jordana, A.; Sastre-Garriga, J. Treatment of multiple sclerosis—Success from bench to bedside. Nat. Rev. Neurol. 2019, 15, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Waltenberger, B.; Mocan, A.; Smejkal, K.; Heiss, E.H.; Atanasov, A.G. Natural Products to Counteract the Epidemic of Cardiovascular and Metabolic Disorders. Molecules 2016, 21, 807. [Google Scholar] [CrossRef]
- Hu, Z.F.; Gu, A.D.; Liang, L.; Li, Y.; Gong, T.; Chen, J.J.; Chen, T.J.; Yang, J.L.; Zhu, P. Construction and optimization of microbial cell factories for sustainable production of bioactive dammarenediol-II glucosides. Green Chem. 2019, 21, 3286–3299. [Google Scholar] [CrossRef]
- Zhong, F.; Xing, J.; Li, X.; Liu, X.; Fu, Z.; Xiong, Z.; Lu, D.; Wu, X.; Zhao, J.; Tan, X.; et al. Artificial intelligence in drug design. Sci. China Life Sci. 2018, 61, 1191–1204. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Vogt, M.; Stenzel, A.; Gätgens, J.; Bott, M.; Marienhagen, J. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 2016, 38, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.L.; Araújo, R.G.; Prather, K.L.J.; Kluskens, L.D.; Rodrigues, L.R. Production of curcuminoids from tyrosine by a metabolically engineered Escherichia coli using caffeic acid as an intermediate. Biotechnol. J. 2015, 10, 599–609. [Google Scholar] [CrossRef]
- Liu, L.Q.; Liu, H.; Zhang, W.; Yao, M.D.; Li, B.Z.; Liu, D.; Yuan, Y.J. Engineering the Biosynthesis of Caffeic Acid in Saccharomyces cerevisiae with Heterologous Enzyme Combinations. Engineering 2019, 5, 287–295. [Google Scholar] [CrossRef]
- Campbell, A.; Bauchart, P.; Gold, N.D.; Zhu, Y.; De Luca, V.; Martin, V.J.J. Engineering of a Nepetalactol-Producing Platform Strain of Saccharomyces cerevisiae for the Production of Plant Seco-Iridoids. ACS Synth. Biol. 2016, 5, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Interrante, M.F.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2015, 349, 1095–1100. [Google Scholar] [CrossRef]
- Reis, R.A.G.; Li, H.; Johnson, M.; Sobrado, P. New frontiers in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 2021, 699, 108765. [Google Scholar] [CrossRef]
- Cane, D.E. Unified Stereochemical Model of Polyether Antibiotic Structure and Biogenesis. J. Am. Chem. Soc. 1983, 105, 3594–3600. [Google Scholar] [CrossRef]
- Leadlay, P.F.; Staunton, J.; Oliynyk, M.; Bisang, C.; Cortés, J.; Frost, E.; Hughes-Thomas, Z.A.; Jones, M.A.; Kendrew, S.G.; Lester, J.B.; et al. Engineering of complex polyketide biosynthesis: Insights from sequencing of the monensin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2001, 27, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, R.D.; Garrett, J.E.; Gast, D.R.; Kirick, M.A.; Larson, D.A.; Meiske, J.C. Influence of monensin on the performance of cattle. J. Anim. Sci. 1984, 58, 1484–1498. [Google Scholar] [CrossRef]
- Bhatt, A.; Stark, C.B.W.; Harvey, B.M.; Gallimore, A.R.; Demydchuk, Y.A.; Spencer, J.B.; Staunton, J.; Leadlay, P.F. Accumulation of an E,E,E-triene by the monensin-producing polyketide synthase when oxidative cyclization is blocked. Angew. Chem.-Int. Ed. 2005, 44, 7075–7078. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Minami, A.; Shimaya, M.; Kodama, T.; Morimoto, Y.; Oguri, H.; Oikawa, H. Analysis of Enantiofacial Selective Epoxidation Catalyzed by Flavin-containing Monooxygenase Lsd18 Involved in Ionophore Polyether Lasalocid Biosynthesis. Chem. Lett. 2014, 43, 1779–1781. [Google Scholar] [CrossRef]
- Pinkerton, D.M.; Banwell, M.G.; Willis, A.C. Chemoenzymatic Access to Versatile Epoxyquinol Synthons. Org. Lett. 2009, 11, 4290–4293. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Romero, E.O.; Pyser, J.B.; Yazarians, J.A.; Narayan, A.R.H. Chemoenzymatic Total Synthesis of Natural Products. Acc. Chem. Res. 2021, 54, 1374–1384. [Google Scholar] [CrossRef]
- Davison, J.; al Fahad, A.; Cai, M.H.; Song, Z.S.; Yehia, S.Y.; Lazarus, C.M.; Bailey, A.M.; Simpson, T.J.; Cox, R.J. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 7642–7647. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Takada, S.; Mori, M.; Ishiyama, A.; Namatame, M.; Nishihara-Tsukashima, A.; Nonaka, K.; Masuma, R.; Otoguro, K.; Shiomi, K.; et al. In vitro and in vivo antimalarial activity of puberulic acid and its new analogs, viticolins A-C, produced by Penicillium sp. FKI-4410. J. Antibiot. 2011, 64, 183–188. [Google Scholar] [CrossRef]
- Zabala, A.O.; Xu, W.; Chooi, Y.H.; Tang, Y. Characterization of a Silent Azaphilone Gene Cluster from Aspergillus niger ATCC 1015 Reveals a Hydroxylation-Mediated Pyran-Ring Formation. Chem. Biol. 2012, 19, 1049–1059. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Szewczyk, E.; Davidson, A.D.; Keller, N.; Oakley, B.R.; Wang, C.C.C. A Gene Cluster Containing Two Fungal Polyketide Synthases Encodes the Biosynthetic Pathway for a Polyketide, Asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 2009, 131, 2965–2970. [Google Scholar] [CrossRef]
- al Fahad, A.; Abood, A.; Fisch, K.M.; Osipow, A.; Davison, J.; Avramovic, M.; Butts, C.P.; Piel, J.; Simpson, T.J.; Cox, R.J. Oxidative dearomatisation: The key step of sorbicillinoid biosynthesis. Chem. Sci. 2014, 5, 523–527. [Google Scholar] [CrossRef]
- Shu, X.; Wei, G.Z.; Qiao, Y.B.; Zhang, K.X.; Zhang, J.; Ai, G.M.; Tang, M.C.; Zhang, Y.H.; Gao, S.S. TerC Is a Multifunctional and Promiscuous Flavoprotein Monooxygenase That Catalyzes Bimodal Oxidative Transformations. Org. Lett. 2021, 23, 8947–8951. [Google Scholar] [CrossRef]
- Leisch, H.; Morley, K.; Lau, P.C.K. Baeyer-Villiger Monooxygenases: More Than Just Green Chemistry. Chem. Rev. 2011, 111, 4165–4222. [Google Scholar] [CrossRef] [PubMed]
- Tolmie, C.; Smit, M.S.; Opperman, D.J. Native roles of Baeyer-Villiger monooxygenases in the microbial metabolism of natural compounds. Nat. Prod. Rep. 2019, 36, 326–353. [Google Scholar] [CrossRef]
- van Berkel, W.J.H.; Kamerbeek, N.M.; Fraaije, M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006, 124, 670–689. [Google Scholar] [CrossRef] [PubMed]
- Yachnin, B.J.; Sprules, T.; McEvoy, M.B.; Lau, P.C.K.; Berghuis, A.M. The Substrate-Bound Crystal Structure of a Baeyer-Villiger Monooxygenase Exhibits a Criegee-like Conformation. J. Am. Chem. Soc. 2012, 134, 7788–7795. [Google Scholar] [CrossRef] [PubMed]
- Isupov, M.N.; Schröder, E.; Gibson, R.P.; Beecher, J.; Donadio, G.; Saneei, V.; Dcunha, S.A.; McGhie, E.J.; Sayer, C.; Davenport, C.F.; et al. The oxygenating constituent of 3,6-diketocamphane monooxygenase from the CAM plasmid of Pseudomonas putida: The first crystal structure of a type II Baeyer-Villiger monooxygenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2344–2353. [Google Scholar] [CrossRef]
- Song, Y.X.; Li, Q.L.; Qin, F.X.; Sun, C.L.; Liang, H.; Wei, X.Y.; Wong, N.K.; Ye, L.; Zhang, Y.; Shao, M.W.; et al. Neoabyssomicins A-C, polycyclic macrolactones from the deep-sea derived Streptomyces koyangensis SCSIO 5802. Tetrahedron 2017, 73, 5366–5372. [Google Scholar] [CrossRef]
- Wang, Q.; Song, F.H.; Xiao, X.; Huang, P.; Li, L.; Monte, A.; Abdel-Mageed, W.M.; Wang, J.; Guo, H.; He, W.N.; et al. Abyssomicins from the South China Sea Deep-Sea Sediment Verrucosispora sp.: Natural Thioether Michael Addition Adducts as Antitubercular Prodrugs. Angew. Chem. Int. Ed. 2013, 52, 1231–1234. [Google Scholar] [CrossRef]
- Tu, J.J.; Li, S.T.; Chen, J.; Song, Y.X.; Fu, S.B.; Ju, J.H.; Li, Q.L. Characterization and heterologous expression of the neoabyssomicin/abyssomicin biosynthetic gene cluster from Streptomyces koyangensis SCSIO 5802. Microb. Cell Factories 2018, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Q.; Tu, J.J.; Song, Y.X.; Zhang, C.Y.; Wang, L.Y.; Li, Q.L.; Ju, J.H. A Luciferase-Like Monooxygenase and Flavin Reductase Pair AbmE2/AbmZ Catalyzes Baeyer-Villiger Oxidation in Neoabyssomicin Biosynthesis. ACS Catal. 2020, 10, 2591–2595. [Google Scholar] [CrossRef]
- Fass, R.; Frazier, R. The role of dexlansoprazole modified-release in the management of gastroesophageal reflux disease. Ther. Adv. Gastroenterol. 2017, 10, 243–251. [Google Scholar] [CrossRef]
- Mejia, A.; Kraft, W.K. Acid peptic diseases: Pharmacological approach to treatment. Expert Rev. Clin. Pharmacol. 2009, 2, 295–314. [Google Scholar] [CrossRef]
- Robinson, M. Proton pump inhibitors: Update on their role in acid-related gastrointestinal diseases. Int. J. Clin. Pract. 2005, 59, 709–715. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Xu, N.; Wu, Y.Q.; Zheng, Y.C.; Zhao, Q.; Lin, G.Q.; Yu, H.L.; Xu, J.H. Discovery of Two Native Baeyer-Villiger Monooxygenases for Asymmetric Synthesis of Bulky Chiral Sulfoxides. Appl. Environ. Microbiol. 2018, 84, e00638-18. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Shou, C.; Geng, Q.; Zhao, C.; Xu, J.H.; Yu, H.L. A Baeyer-Villiger monooxygenase from Cupriavidus basilensis catalyzes asymmetric synthesis of (R)-lansoprazole and other pharmaco-sulfoxides. Appl. Microbiol. Biotechnol. 2021, 105, 3169–3180. [Google Scholar] [CrossRef] [PubMed]
- Malca, S.H.; Scheps, D.; Kühnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115–5117. [Google Scholar] [CrossRef] [PubMed]
- Scheps, D.; Malca, S.H.; Richter, S.M.; Marisch, K.; Nestl, B.M.; Hauer, B. Synthesis of ω-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb. Biotechnol. 2013, 6, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.H.; Ness, J.E.; Xie, W.C.; Zhang, X.Y.; Minshull, J.; Gross, R.A. Biosynthesis of Monomers for Plastics from Renewable Oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef]
- Rehdorf, J.; Kirschner, A.; Bornscheuer, U.T. Cloning, expression and characterization of a Baeyer-Villiger monooxygenase from Pseudomonas putida KT2440. Biotechnol. Lett. 2007, 29, 1393–1398. [Google Scholar] [CrossRef]
- Kirschner, A.; Altenbuchner, J.; Bornscheuer, U.T. Cloning, expression, and characterization of a Baeyer-Villiger monooxygenase from Pseudomonas fluorescens DSM 50106 in E-coli. Appl. Microbiol. Biotechnol. 2007, 73, 1065–1072. [Google Scholar] [CrossRef]
- Yu, J.M.; Liu, Y.Y.; Zheng, Y.C.; Li, H.; Zhang, X.Y.; Zheng, G.W.; Li, C.X.; Bai, Y.P.; Xu, J.H. Direct Access to Medium-Chain α,ω-Dicarboxylic Acids by Using a Baeyer-Villiger Monooxygenase of Abnormal Regioselectivity. ChemBioChem 2018, 19, 2049–2054. [Google Scholar] [CrossRef]
- Zhang, G.X.; You, Z.N.; Yu, J.M.; Liu, Y.Y.; Pan, J.; Xu, J.H.; Li, C.X. Discovery and Engineering of a Novel Baeyer-Villiger Monooxygenase with High Normal Regioselectivity. ChemBioChem 2021, 22, 1190–1195. [Google Scholar] [CrossRef]
- Qi-yue, Y.; Ting, Z.; Ya-nan, H.; Sheng-jie, H.; Xuan, D.; Li, H.; Chun-guang, X. From natural dye to herbal medicine: A systematic review of chemical constituents, pharmacological effects and clinical applications of indigo naturalis. Chin. Med. 2020, 15, 127. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Wu, J.; Heuts, D.; van Hellemond, E.W.; Spelberg, J.H.L.; Janssen, D.B. Discovery of a thermostable Baeyer-Villiger monooxygenase by genome mining. Appl. Microbiol. Biotechnol. 2005, 66, 393–400. [Google Scholar] [CrossRef]
- Parra, L.P.; Acevedo, J.P.; Reetz, M.T. Directed evolution of phenylacetone monooxygenase as an active catalyst for the baeyer-villiger conversion of cyclohexanone to caprolactone. Biotechnol. Bioeng. 2015, 112, 1354–1364. [Google Scholar] [CrossRef]
- Núñez-Navarro, N.; Muñoz, J.S.; Castillo, F.; Ramírez-Sarmiento, C.A.; Poblete-Castro, I.; Zacconi, F.C.; Parra, L.P. Discovery of New Phenylacetone Monooxygenase Variants for the Development of Substituted Indigoids through Biocatalysis. Int. J. Mol. Sci. 2022, 23, 12544. [Google Scholar] [CrossRef]
- Hansen, C.C.; Sorensen, M.; Veiga, T.A.M.; Zibrandtsen, J.F.S.; Heskes, A.M.; Olsen, C.E.; Boughton, B.A.; Moller, B.L.; Neilsono, E.H.J. Reconfigured Cyanogenic Glucoside Biosynthesis in Eucalyptus cladocalyx Involves a Cytochrome P450 CYP706C55. Plant Physiol. 2018, 178, 1081–1095. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamamoto, K.; Asano, Y. Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in L-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume Sieb. et Zucc. Plant Mol. Biol. 2014, 86, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Toplak, M.; Matthews, A.; Teufel, R. The devil is in the details: The chemical basis and mechanistic versatility of flavoprotein monooxygenases. Arch. Biochem. Biophys. 2021, 698, 108732. [Google Scholar] [CrossRef]
- Toplak, M.; Saleem-Batcha, R.; Piel, J.; Teufel, R. Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis. Angew. Chem.-Int. Ed. 2021, 60, 26960–26970. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, F.; Taglieber, A.; Schulz, F.; Reetz, M.T. A light-driven stereoselective biocatalytic oxidation. Angew. Chem.-Int. Ed. 2007, 46, 2903–2906. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, F.; Hofstetter, K.; Schmid, A. Non-enzymatic regeneration of nicotinamide and flavin cofactors for monooxygenase catalysis. Trends Biotechnol. 2006, 24, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Cryo-clectron microscopy: Commentary on the 2017 Nobel Prize in Chemistry. Sci. Technol. Rev. 2017, 35, 16–21. [Google Scholar]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Bunzel, H.A.; Anderson, J.L.R.; Mulholland, A.J. Designing better enzymes: Insights from directed evolution. Curr. Opin. Struct. Biol. 2021, 67, 212–218. [Google Scholar] [CrossRef] [PubMed]
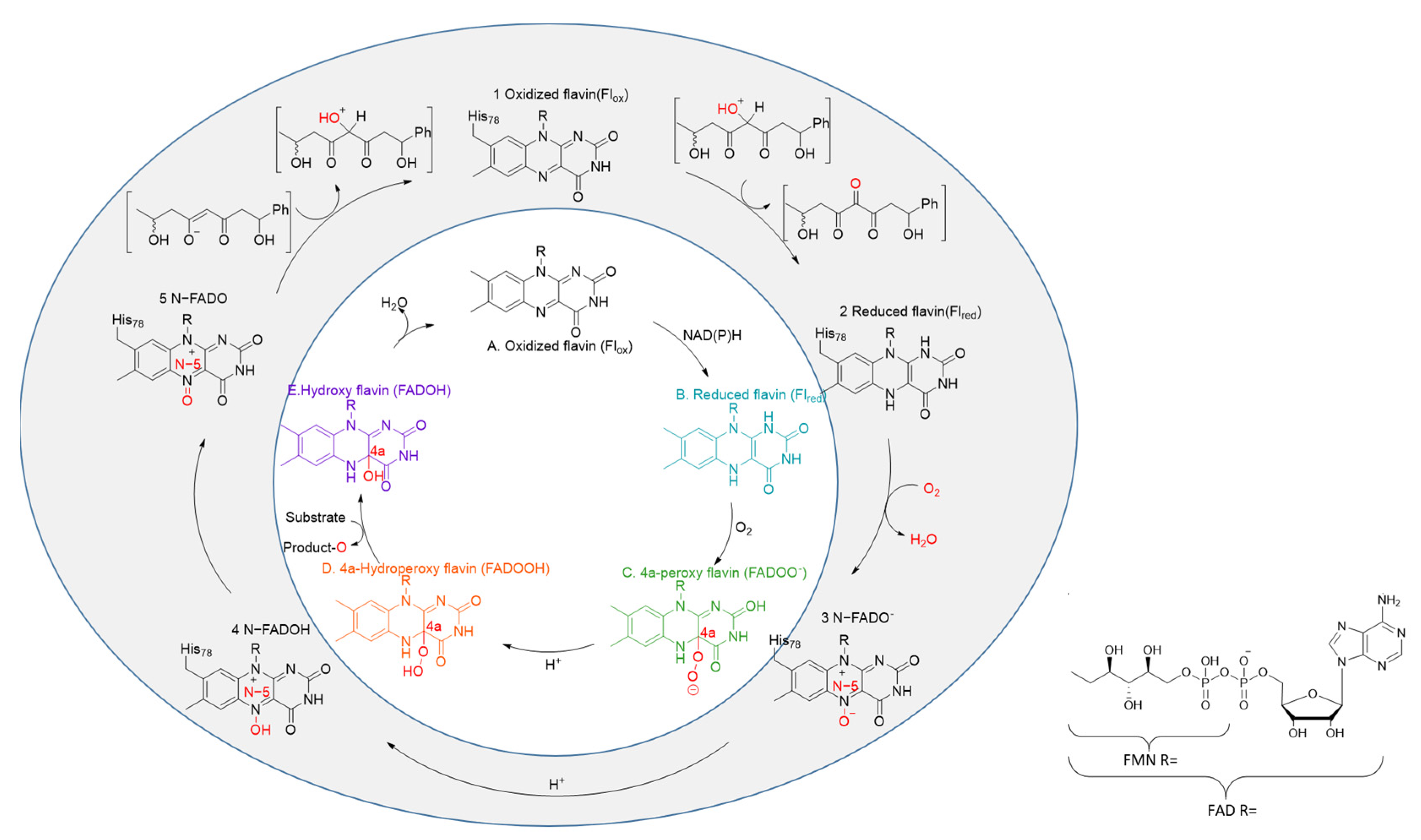

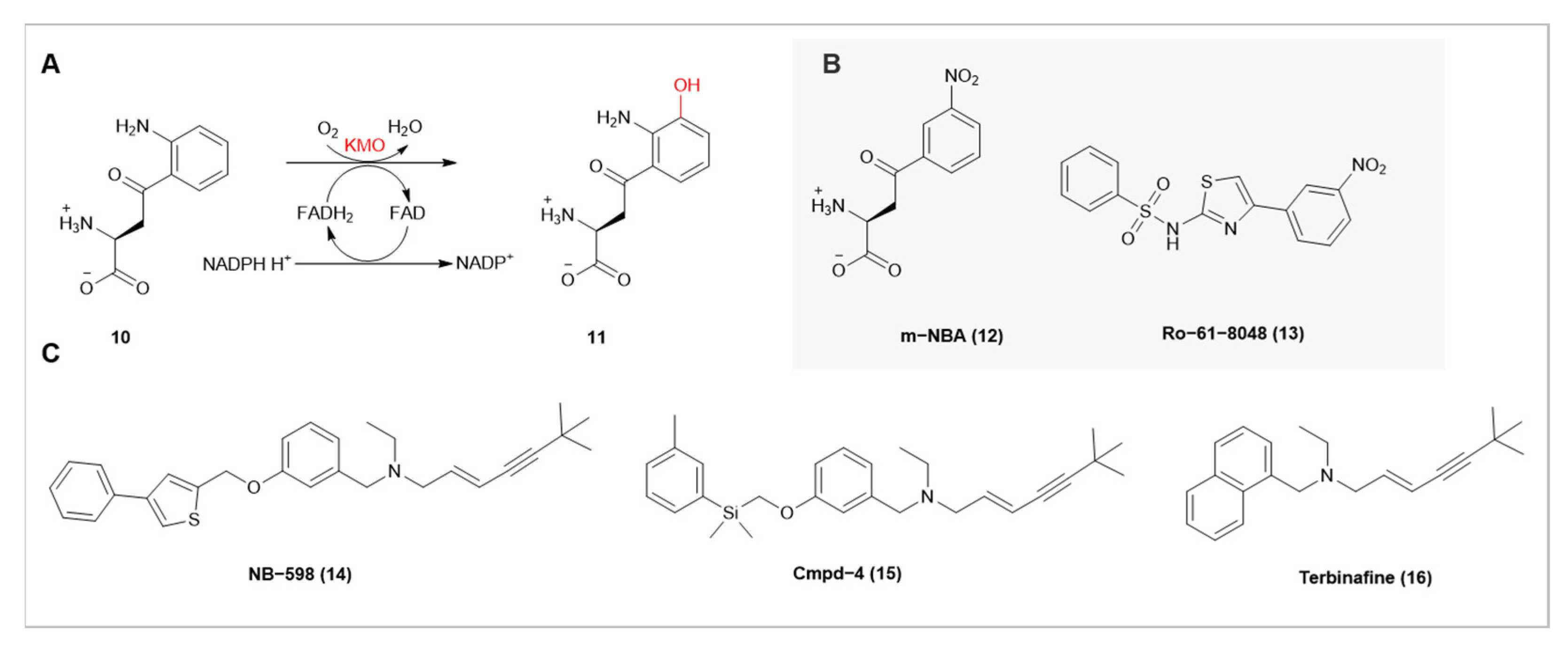
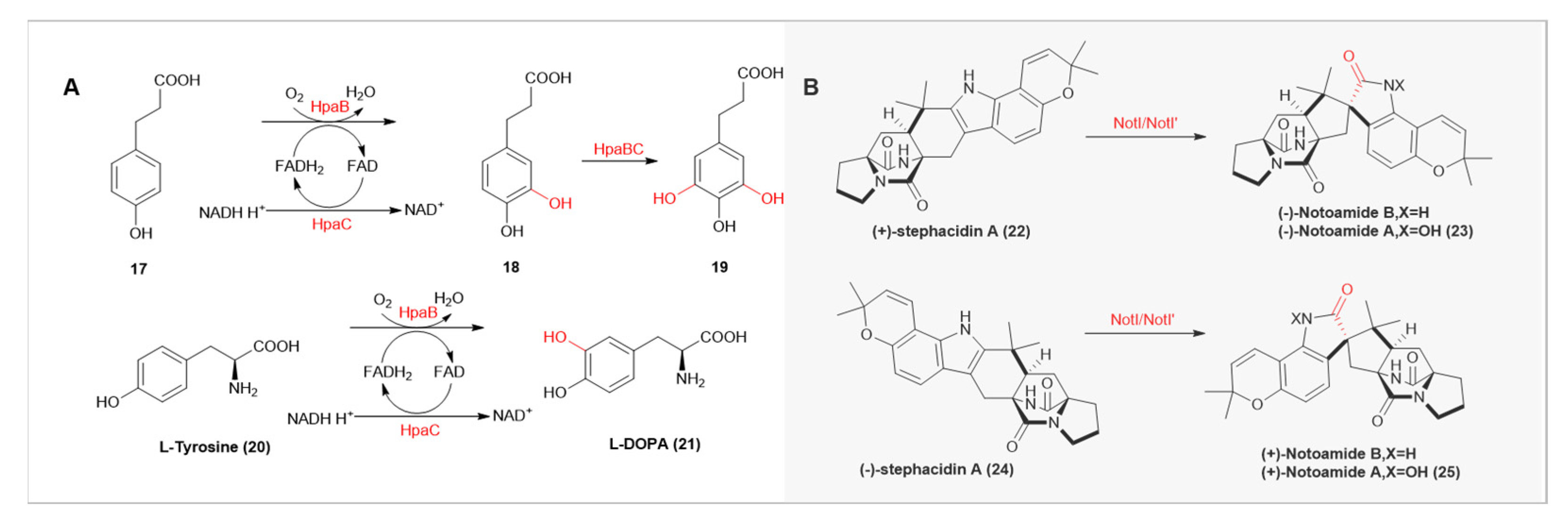


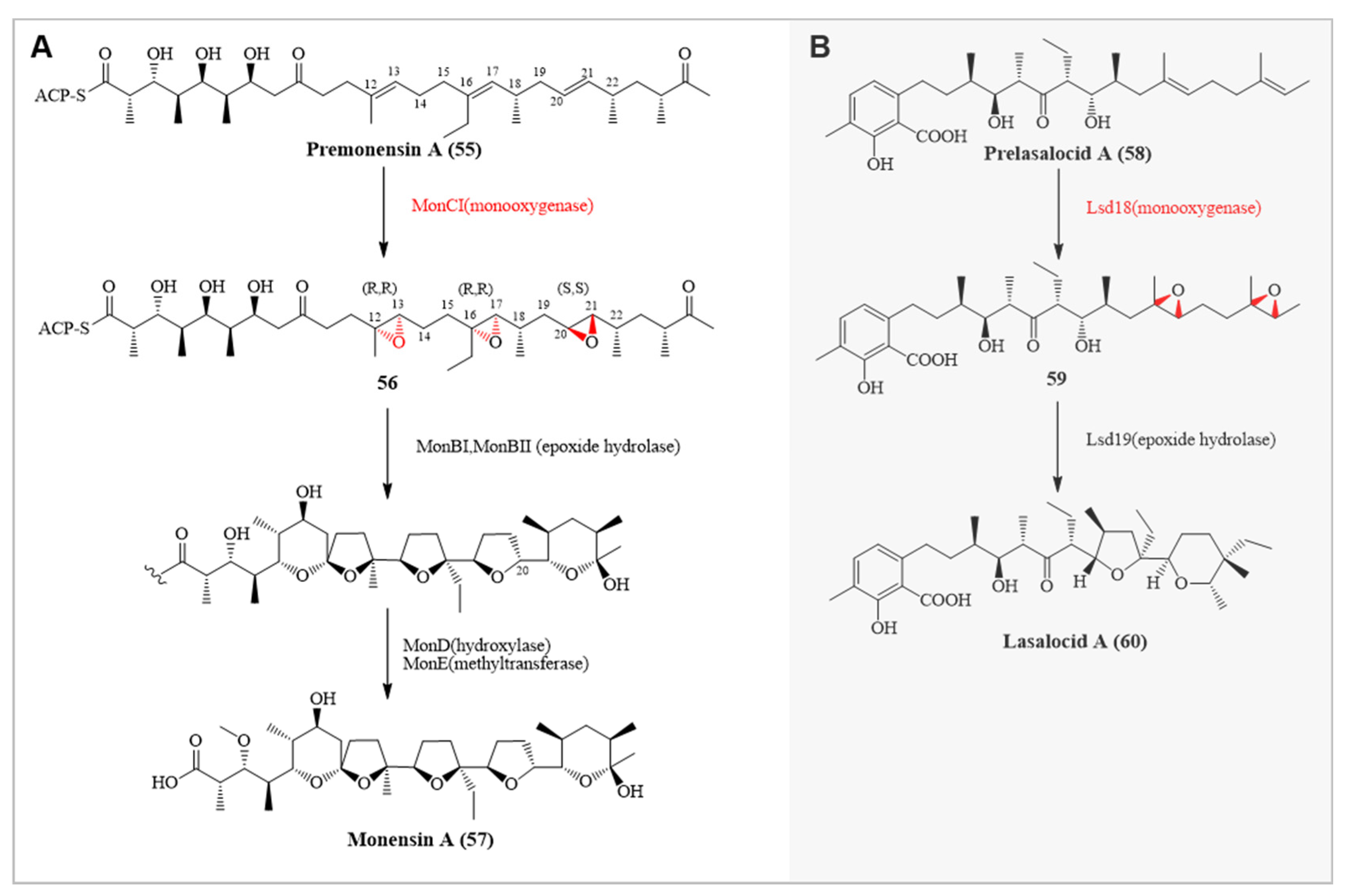

| Enzyme Name | Source Organism | Catalytic Function |
|---|---|---|
| MabTetX | Mycobacterium abscessus | Catalyzes the modification of tetracycline and doxycycline |
| SadA | Microbacterium sp. CJ77 | Responsible for the initial ipso-hydroxylation reaction of sulfonamides |
| KMO | Human | Catalyzes the hydroxylation of L-kynurenine, producing 3-hydroxykynurenine |
| SQLE | Human | Potential drug targets |
| HpaBC | Pseudomonas aeruginosa | Catalyzes the hydroxylation of cinnamic acid derivatives |
| TomAB | Acidovorax sp. | Involved in the C-S bond cleavage of the thiocarbamate herbicide thiobencard |
| TcpA | X1T strain | Progressively dechlorinates TC2P in the presence of a redox partner protein (Fre) |
| OdcA | Pigmentiphaga sp. | Exhibits decarboxylation activity, converting DBHB to 2,6-dibromohydroquinone |
| CfdCCDS-1 | Sphingomonas sp. | Capable of hydroxylating furan phenol, a hydrolysis product of furan |
| MeaXY | Sphingobium baderi DE-13 | Degrades 2-methyl-6-ethyl aniline and hydroxylates MEA and its metabolic product EDA |
| DcmB1B2 | Brevundimonas sp. JT-9 | Ortho-hydroxylates 4-chlorophenol to form 4-chlorocatechol |
| NphA1A2 | Rhodococcus sp | Converts 4-nitrophenol to 4-nitrocatechol |
| CehC1C2 | Rhizobium sp. X9 | Catalyzes the conversion of 1-naphthol to 1,2-dihydroxynaphthalene |
| Pp-BVMO | Physcomitrella patens (Pp) | Inserts oxygen atoms into C-C bonds, catalyzing the conversion of phenylacetone to benzyl acetate |
| YUCCA | Arabidopsis | Involved in the synthesis of the hormone auxin (IAA) |
| AsFMO1 | Garlic | Exhibits high stereoselective S-oxygenation activity towards (+)-alliin |
| MonCI | Streptomyces cinnamonensis | Involved in the stereoselective epoxidation of three double bonds in premonensin A |
| Lsd18 | Streptomyces lasalocidi | Catalyzes the conversion of prelasalocid A to bisepoxyprelasalocid A |
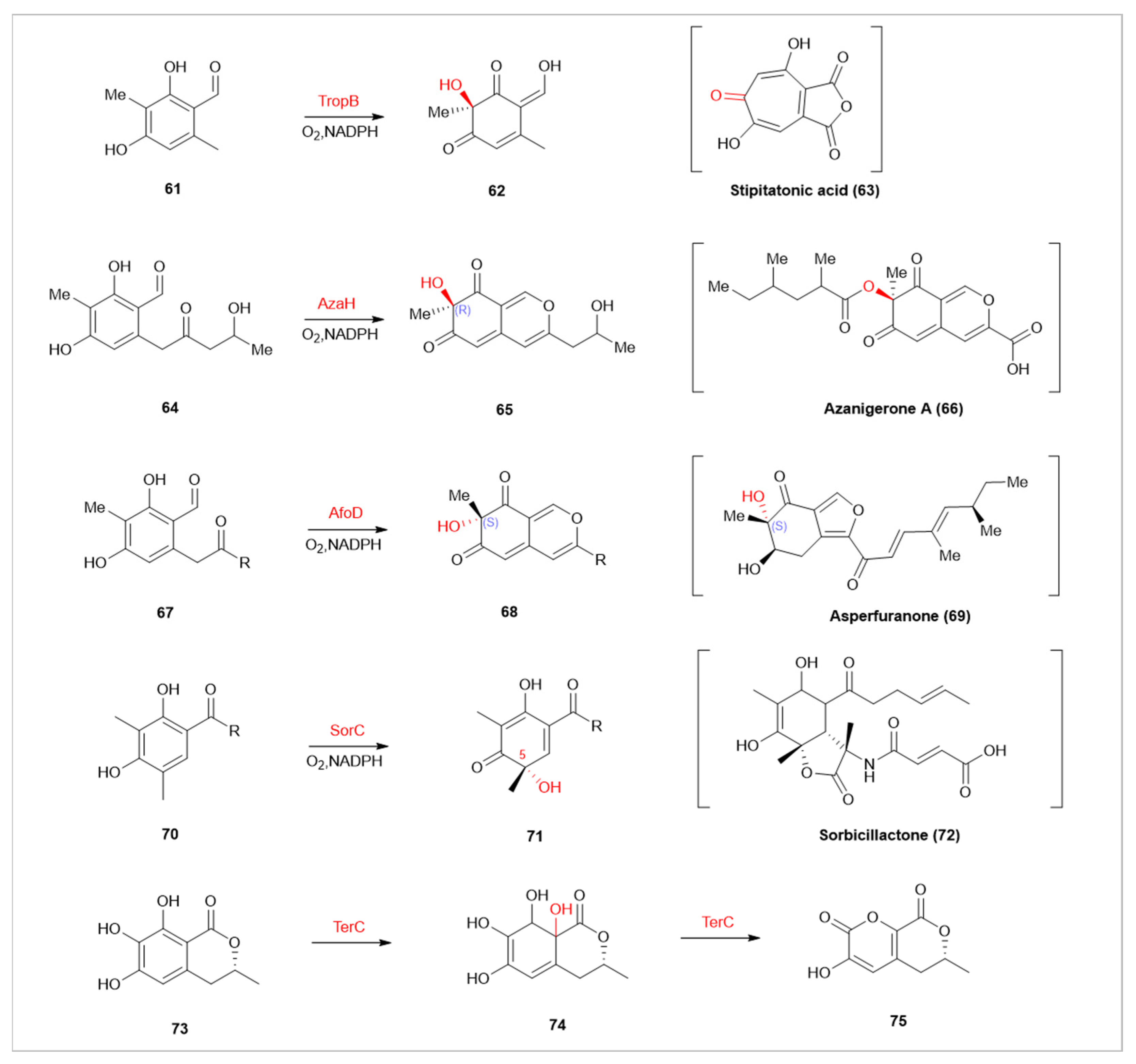
| TropB | Ttopolone | Selectively hydroxylates 3-methyl-octanal at the C-3 position for dearomatization in stipitatonic acid synthesis |
| AzaH | silent Aspergillus niger | Participates in the synthesis of azanigerone A, inducing an ‘R’ configuration at the newly formed stereocenter |
| AfoD | Aspergillus nidulans | Involved in asperfuranone synthesis, producing a complementary ‘S’ configuration |
| SorbC | Penicillium chrysogenum E01-10/3 | Involved in sorbicillactone A synthesis, differs in site selectivity from AzaH, AfoD, and TropB |
| TerC | terCDEF | Catalyzes the dearomatization of 6-hydroxymellein to form 1,4-benzoquinone |
| AbmE2/AbmZ | Verrucosispora and Streptomyces species | Involved in the catalytic conversion of abyssomicin 2 to neoabyssomicin B |
| BoBVMO | Bradyrhizobium oligotrophicum | Catalyzes the asymmetric sulfoxidation of bulky prazole thioethers |
| CbBVMO | Cupriavidus basilensis | Exhibits a high specific activity toward lansoprazole sulfide |
| PpBVMO | Pseudomonas putida KT2440 | Catalyzes the insertion of oxygen atoms in asymmetric linear ketones |
| PfBVMO | Pseudomonas fluorescens DSM 50106 | Catalyze the formation of α,ω-dicarboxylic acids |
| PaBVMO | Pseudomonas aeruginosa | Demonstrates high regioselectivity toward long-chain ketones |
| GsBVMO | G. sihwensis | Exhibits high selectivity in catalyzing medium-to-long-chain ketone acids |
| PAMO | Thermobifida fusca | Catalyzes the conversion of phenylacetone to lactone |
| PAMOHPCD/PAMOHPED | Thermobifida fusca | Optimized mutant of PAMO, effective in the hydroxylation of indole and in the formation of indigo |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Y.; Chen, X. Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science. Catalysts 2023, 13, 1495. https://doi.org/10.3390/catal13121495
Guan Y, Chen X. Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science. Catalysts. 2023; 13(12):1495. https://doi.org/10.3390/catal13121495
Chicago/Turabian StyleGuan, Yuze, and Xi Chen. 2023. "Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science" Catalysts 13, no. 12: 1495. https://doi.org/10.3390/catal13121495
APA StyleGuan, Y., & Chen, X. (2023). Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science. Catalysts, 13(12), 1495. https://doi.org/10.3390/catal13121495







